Abstract
Host genetic variations may influence a changing profile of biochemical markers and outcome in patients with trauma/injury. The objective of this study was to assess clinical associations of single nucleotide polymorphisms (SNPs) in the genes of cytokines in critically ill patients. A total of 430 patients were genotyped for SNPs in the genes of pro- (IL1B, IL6, IL8) and anti-inflammatory (IL4, IL10, IL13) cytokines. The main end-points were sepsis, mortality and adult respiratory distress syndrome (ARDS). We evaluated the dynamic levels of bilirubin, blood urea nitrogen, creatine kinase, creatinine and lactate dehydrogenase in five points of measurements (between 1 and 14 days after admission) and correlated them with SNPs. High-producing alleles of proinflammatory cytokines protected patients against sepsis (IL1B −511A and IL8 —251A) and mortality (IL1B −511A). High-producing alleles of anti-inflammatory cytokines IL4 —589T and IL13 431A (144Gln) were less frequent in ARDS patients. The carriers of IL6 —174C/C genotypes were prone to the increased levels of biochemical markers and acute kidney and liver insufficiency. Genotype-dependent differences in the levels of biochemical indicators gradually increased to a maximal value on the 14th day after admission. These findings suggest that genetic variability in pro- and anti-inflammatory cytokines may contribute to different clinical phenotypes in patients at high risk of critical illness.
Keywords: adult respiratory distress sybdrome, cytokines, genetic association studies, sepsis, single nucleotide polymorphism
Introduction
An unbalanced immune reaction is considered to be responsible for a substantial amount of poor outcome in severely injured patients. The protective role of proinflammatory cytokines in early innate defence against extracellular bacteria is well established, but it is recognized that excessive production of these cytokines may also be responsible for the development of severe complications [systemic inflammatory response syndrome, sepsis, adult respiratory distress syndrome (ARDS), multiple organ dysfunction syndrome (MODS)]1. However, an anti-inflammatory therapy in sepsis clinical trials was unsuccessful, and the assumption that poor outcome in patients with or at high risk of critical conditions derives from an uncontrolled proinflammatory response is questionable2. Having survived the initial, hyperinflammatory phase of sepsis, patients may suffer from immunosuppression due to the T helper type 2 (Th2) cell response in the later stages of sepsis3.
Polymorphisms in cytokine genes may influence the corresponding proteins' quantity, activity and stability, and this may have an impact upon the development of critical conditions. Numerous assays were performed to seek the genetic markers in the genes of cytokines in association with sepsis and other severe acute complications (reviewed in4–6). Given the complexity and heterogeneity of the problem, it is not surprising that the results are contradictory. With regard to the most studied genetic variations, despite the total number of publications, data concerning the same single nucleotide polymorphisms (SNPs) in the same populations and clinical conditions are scarce. Moreover, some relevant genes and SNPs are unconsidered in the field.
Nowadays, numerous assays consider multiple biomarkers of sepsis and organ failure. For example, a review of sepsis biomarkers has mentioned 178 different biomarkers7. Parameters of routine biochemical tests may also be useful as biomarkers of sepsis and organ insufficiency, as they are measured sequentially to capture a changing profile which reflects the severity of illness and target organ damage. Bilirubin, blood urea nitrogen (BUN) and creatinine are included in multiple scoring systems for post-injury multiple organ failure8. Moreover, bilirubin is elevated in patients with severe infections, and its measurement may be performed to discriminate between patients with and without bacteraemia9. Hyperbilirubinaemia is both a risk factor and complication of sepsis, which is associated with a reduction of the bile flow in hepatocytes9. In people with severe sepsis, hyperbilirubinaemia has been correlated with worse outcomes and predicted the development of ARDS10–13. Serum creatinine levels are elevated not only in patients with renal failure, but also in patients with ARDS14. A high lactate dehydrogenase (LDH) level has been linked to infection as a marker for severe prognosis, in-hospital major complications and high mortality, probably related to tissue destruction from immune hyperactivity12,15,16. LDH activity level was shown to predict pathological conditions in the lungs, such as cell damage or inflammation17. Elevation of creatine kinase (CK) was associated with more complications in patients with pH1N1 influenza A infection admitted to the intensive care unit (ICU) for severe acute respiratory insufficiency (SARI)18. In animal experiments, concentrations of creatinine, BUN, CK and LDH were already increased markedly at the early stage of multiple organ dysfunction syndrome caused by trauma and infection19. It has also been shown that creatinine, BUN and CK levels were correlated negatively with the survival outcome in sepsis20,21.
The dynamics of the variations of biochemical parameters may depend particularly upon host genetic resistance to the infection. To the best of our knowledge, little or no research has been performed to date to understand the relationships between the genetic variability in the acute-phase response genes and the dynamic pattern of routine biochemical indicators. In light of these challenging issues, we performed an association study of the outcome in patients with severe trauma/injury and well-studied functional polymorphic variations in the genes of pro- (IL1B, IL6, IL8) and anti-inflammatory (IL4, IL10, IL13) cytokines. Single-nucleotide polymorphisms (SNPs) (IL1B −511 G>A, rs16944; IL6 –174 G>C, rs1800795; IL8 −251 T>A, rs4073; IL4 −589 C>T, rs2243250; IL10 −1082 G>A, rs1800896; IL13 431 A>G, 144Gln/Arg, rs20541) in the above genes have been shown to influence the level of the gene product (missense SNP in the IL13 gene) or the activity of gene promoter (SNPs in other genes). In addition, these SNPs have been demonstrated to be associated with susceptibility to sepsis and/or organ failure (in particular, SNPs in IL1B, IL6, IL8, IL10) and a number of infectious and inflammatory diseases22–26. The main end-points studied in association with SNPs were sepsis, mortality and ARDS. The dynamic levels of biochemical markers (bilirubin, BUN, CK, creatinine and LDH), shown to be related to these end-points, were evaluated in five points of measurements (between 1 and 14 days after ICU admission) and correlated with gene polymorphisms. Based on the findings in the set of biochemical indicators, we performed additional genetic association studies of liver and renal insufficiency.
Materials and methods
Study subjects
From January 2008 to February 2013 we selected a group of accident victims with severe physical trauma and patients with acute diseases requiring extensive surgery. A total of 430 patients at risk of critical illness [81% males; among these, 193 patients (55%) were workers of the Rescue Service; mean age 42.12 ± 18.56 years], hospitalized at the clinical bases of V. A. Negovsky Research Institute of General Reanimatology, Moscow, Russia were included in the study. The severity of each patient was evaluated on the basis of the Acute Physiology and Chronic Health Evaluation (APACHE) II score within the first 24 h after ICU admission27. The Sequential Organ Failure Assessment (SOFA) score was calculated consecutively as an indicator of organ dysfunction28. Exclusion criteria for the group under study consisted of age less than 18 years, lack of informed consent, defined immunodeficiency, corticosteroid administration less than 6 weeks previously, final stage of chronic disease, decompensated heart failure [New York Heart Association (NYHA) Class IV], decompensated diabetes, severe neurological deficit (Glasgow Coma Scale ≤ 8), addiction, alcoholism, AIDS and pregnancy. Patients with chronic respiratory, kidney and liver diseases were also excluded. Treatment decisions for all study participants were standardized according to the conciliatory guidelines of the Russian Respiratory Society and Interregional Association of Clinical Microbiology and Antimicrobial Chemotherapy in all patients (http://webmed.irkutsk.ru/doc/pdf/cap.pdf). Sepsis and multiple organ dysfunction syndrome (particularly, renal and liver insufficiency) were diagnosed according the definitions of the ACCP-SCCM consensus conference on sepsis and organ failure29 and SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions30. The diagnostics of the ARDS is similar to the ARDS Berlin definition31.
All biochemical analyses were performed using Cobas 6000, ABX Pentra 400 and Mythic 22 analysers (Roche, Boulogne-Billancourt, France) according to the manufacturer's instructions. Measurements were carried out within the first day of ICU admission and on the subsequent third, fifth, seventh and 14th days.
The study protocol was approved by the Ethics Committee of V. A. Negovsky Research Institute of General Reanimatology RAMS (with Institutional Review Board approval number 2/6/2012), and adhered to the tenets of the Declaration of Helsinki.
Genotyping
DNA was isolated from 200 μl of blood using the gDNA purification kit Diatom DNA Prep 200 (Isogene Laboratory, Moscow, Russia). The genotyping was performed with a PCR-CTPP (polymerase chain reaction with confronting two-pair primers)32. Amplification was carried out in an ABI thermal cycler using two external and two internal sequence-specific primers (Supporting information, Table S1) and tubes of PCR MasterMix (Isogene Laboratory, Moscow, Russia). The PCR products were analysed in 2% agarose gel stained with ethidium bromide. The CFX96 real-time PCR detection system with SYBR Green fluorescent dye was employed to genotype 10% of randomly taken DNA samples for each SNP once more. There was 100% concordance in genotype calling of duplicate samples.
Statistical analysis
Deviation from Hardy–Weinberg equilibrium was assessed by χ2 analysis. As all studied continuous variables did not assume a normal distribution, the Mann–Whitney U-non-parametric test was used to compare such variables. To evaluate associations between gene polymorphisms and studied end-points we performed logistic regression analysis using the SNPStats package33, a free web-based tool designed specifically for genetic association studies. The association with disease is modelled depending on the response variable. For binary variables, unconditional logistic regression models are used. For quantitative response, linear regression models are used to examine the proportion of variation in the response explained by the SNPs. In analysis of the SNPs in relation to the response, SNPStats provides odds ratios (OR), confidence intervals (CI) and the P-values for multiple inheritance models (dominant, recessive, over-dominant, co-dominant and additive), as well as Akaike's information criterion (AIC) indicating the best inheritance genetic model for each specific polymorphism. Other quantitative or categorical variables may be added to the regression models for analysis as covariates and treated as potential confounders. In multivariate models, we adjusted for age, sex, APACHE II score and using (more than 24 h) mechanical ventilation. For the regression models we present OR for the minor allele. Therefore, OR < 1·0 shows a protective effect of the minor allele, and OR > 1·0 shows that the minor allele is a risk allele. The lowest AIC value was considered the best-fitting model for the fitted variant. For genotypes with minor allele frequencies, <10% only dominant and additive genetic models were evaluated.
The influence of multiple testing in the genotypical-dependent outcome (sepsis, mortality, ARDS) was evaluated applying Benjamini–Hochberg step-up false discovery rate (FDR) corrections (implemented in the winpepi computer programs34). The winpepi is a free and user-friendly resource comprising computer programs for a wide spectrum of statistical tools designed for epidemiologists and biomedical researchers. Procedures for adjusting P-values derived from multiple significance tests are supplied by one of the modules of the etcetera program of the winpepi package.
The association study between genetic and biochemical variables was considered exploratory, not requiring corrections for multiple comparisons35.
The winpepi test power calculator (in the compare2 program) was used to evaluate the test power. Post-hoc power calculations were generated using a detected OR assuming the specified model of risk, 5% type 1 error rate and 80% power. In the sepsis study, statistical power was 66·75% (IL1B rs16944; OR = 0·27) and 86·11% (IL8 rs4073; OR = 0·44); in the hospital mortality study, statistical power was 94·96% (IL1B rs16944; OR = 0·41); in the ARDS study, statistical power was 71·45% (IL4 rs2243250; OR = 0·41) and 41·50% (IL13 rs20541; OR = 0·55).
Results
Characteristics of the study population
Disease progression in 430 Caucasian patients from the European region of Russian Federation was evaluated in association with genotype and laboratory data. The baseline characteristics of the study subjects are shown in Table1. Within the first 24 h after ICU admission, the APACHE II and SOFA scores did not differ between patients with and without trauma (APACHE II: 17·98 ± 4·38 and 18·20 ± 4·14, respectively, P = 0·82; SOFA: 11·55 ± 3·64 and 11·59 ± 4·03, respectively, P = 0·92). We calculated pairwise associations for binary variables and found the following Pearson's correlation coefficients (rφ) between clinical end-points: sepsis and mortality, rφ = 0·34; sepsis and ARDS, rφ = 0·21; mortality and ARDS, rφ = 0·30.
Table 1.
Characteristics of the patients included in the study
| Characteristics | All patients, n (portion) mean ± s.d. |
|---|---|
| Total number | 430 |
| Age (years) | 42·12 ± 18·56 |
| Male sex (n, %) | 349 (0·81) |
| Pre-existing conditions | |
| • Cardiovascular diseases | 45 (0·11) |
| • Diabetes | 25 (0·06) |
| • Obesity | 24 (0·06) |
| ICU admission | 402 (0·94) |
| ICU length of stay (d) | 14·82 ± 23·46 |
| Patients on mechanical ventilator | 153 (0·36) |
| APACHE II scorea | 18·08 ± 4·27 |
| SOFA scorea | 11·57 ± 3·83 |
| Diagnosis at admission | |
| • Severe combined trauma/wounding | 236 (0·55) |
| • Bowel obstruction | 28 (0·07) |
| • Inflammatory diseases of the abdominal cavity and retroperitoneal space complicated by destruction | 106 (0·25) |
| • Purulent-inflammatory diseases of the skin, subcutaneous tissue | 44 (0.10) |
| • Other | 19 (0·04) |
| Critical conditions | |
| • Sepsis | 80 (0.19) |
| • ARDS | 75 (0·17) |
| Hospital mortality | 95 (0·22) |
Within the first 24 h after admission. ARDS = adult respiratory distress syndrome; ICU = intensive care unit; APACHE = Acute Physiology and Chronic Health Evaluation; SOFA = Sequential Organ Failure Assessment; s.d. = standard deviation.
The dynamics of changes in SOFA score and laboratory parameters
The dynamics of changes in SOFA score and biochemical blood plasma values in relation to all studied clinical phenotypes is given in Supporting information, Table S2. The mean SOFA score on ICU admission and during the ICU stay was, in general, higher in all studied cases (sepsis, mortality, ARDS, renal and hepatic insufficiency) compared with matched controls. As expected, the maximum differences were found in the mortality set. Normal levels of studied biochemical parameters are considered to be in the range of 3·4–17·1 mmol/L for total bilirubin, 1.7–8.3 mmol/L for BUN, 26·0–174·0 U/L for CK, 53·0–124·0 mmol/L for creatinine and 89·0–221·0 U/L for LDH. In non-survivors, all studied biochemical parameters increased gradually within 2 weeks. In comparison with survivors, the maximum differences were found on the 14th day. Patients with ARDS periodically had higher levels of BUN, creatinine and bilirubin. In patients with renal insufficiency, BUN and creatinine were elevated from the 3rd and 5th to the 14th days, respectively. With few exceptions in patients with liver insufficiency, levels of bilirubin, BUN and creatinine tended to increase over time. In the group of patients with liver insufficiency a very high level of LDH was found by the end of the second week.
We also evaluated changes in biochemical parameters in sepsis patients with and without acute trauma (Supporting information, Table S3). In general, the results seem to be consistent with the expectations. CK and LDH are expressed in many different organs and tissues and their levels were higher in trauma patients. After acute trauma, the levels of CK were elevated on the first and seventh days after admission to the ICU, and the levels of LDH were higher on the seventh day, compared to non-trauma patients with sepsis. BUN, creatinine and bilirubin are key biomarkers of renal and liver failure and their concentrations increased gradually in non-trauma subjects, but the only significant increment was observed for serum bilirubin on the 7th day. SOFA scores did not differ between trauma and non-trauma patients with sepsis.
Available data concerning changes in other biochemical parameters linked with renal and liver insufficiency are presented in Supporting information, Table S4. The most profound effect was found for AST concentrations in patients with liver insufficiency on the 14th day.
Genetic association study of selected candidate genes in sepsis, ARDS and mortality sets
All genotypes were in Hardy–Weinberg equilibrium (Supporting information, Table S5). Within the first day of ICU admission, APACHE II scores were significantly higher for the carriers of the IL6 −174 C/C genotype. Higher SOFA scores correlated with the IL6 −174 C/C genotype and with the IL8 −251 A allele (additive model) (Supporting information, Table S6).
Genotype association results significant after correction for multiple testing are presented in Table2; non-significant data are given in Supporting information, Table S7. Alleles of proinflammatory cytokines IL1B −511A and IL8 −251A were protective against the development of sepsis. The protective effect of the same IL1B allele was found in relation to hospital mortality. In the ARDS set alleles of anti-inflammatory cytokines IL4 −589T and IL13 431A were less frequent in the cases than in the controls.
Table 2.
The distribution of genotypes among patients with/without sepsis, ARDS and survivors/non-survivors
| Genes and genotypes | Controls | Cases | P-value (genetic model), OR (95% CI) | ||
|---|---|---|---|---|---|
| Number (%) | |||||
| Sepsis | |||||
| IL1B | n=334 | n=78 | |||
| G/G | 146 (43·7) | 42 (53·9) | 0·014 (rec)a | ||
| −511 G>A | G/A | 145 (43·4) | 33 (42·3) | 0·27 (0·08–0·92) | |
| A/A | 43 (12·9) | 3 (3·8) | |||
| IL8 | n=342 | n=79 | |||
| T/T | 98 (28·6) | 37 (46·8) | 0·0023 (dom)b | ||
| −251 T>A | T/A | 183 (53·5) | 33 (41·8) | 0·44 (0·26–0·75) | |
| A/A | 61 (17·8) | 9 (11·4) | |||
| Hospital mortality | |||||
| IL1B | n=321 | n=91 | |||
| G/G | 136 (42·4) | 52 (57·1) | 0·0019 (dom)c | ||
| −511 G>A | G/A | 147 (45·8) | 31 (34·1) | 0·41 (0·23–0·73) | |
| A/A | 38 (11·8) | 8 (8·8) | |||
| ARDS | |||||
| IL4 | n=345 | n=72 | |||
| C/C | 198 (57·4) | 51 (70·8) | 0·01 (dom)d | ||
| −589 C>T | C/T | 134 (38·8) | 20 (27·8) | 0·48 (0·27–0·86) | |
| T/T | 13 (3·8) | 1 (1·4) | |||
| IL13 | n=347 | n=74 | |||
| C/C | 179 (51·6) | 51 (68·9) | 0·008 (add)e | ||
| 431 A>Gr* | C/T | 141 (40·6) | 20 (27·0) | 0·55 (0·35–0·88) | |
| T/T | 27 (7·8) | 3 (4·0) | |||
The analysis is adjusted for age, sex, Acute Physiology and Chronic Health Evaluation II (APACHE II) score and using (more than 24 h) of mechanical ventilation. ARDS = adult respiratory distress syndrome; OR = odds ratio; CI = confidence interval. The choice of each genetic model was based on Akaike information criterion (AIC) value. The genetic model: rec = recessive: dom = dominant; add = additive.
IL13 rs20541 (431 A>G) alleles are reported in reverse orientation to genome (A allele is T); http://www·ncbi·nlm·nih·gov/projects/SNP/snp_ref·cgi?rs=20541. Benjamini–Hochberg step-up false discovery rate (FDR)-adjusted P-values:
P = 0·042
P = 0·014
P = 0·011
P = 0·030
P = 0·030.
Correlation between genotypes and biochemical parameters
Dynamic changes in SOFA scores and biochemical blood plasma values were assessed in relation to the studied SNPs. Addressing the SOFA score, it should be noted that within the first day on ICU admission, similar associations were found in the whole sample (n = 430) and in the observational group (n = 88) for the IL6 and IL8 SNPs (Supporting information, Tables S6 and S8, Fig. 1). The majority of the dynamic associations was not stable and might have arisen by chance (Supporting information, Table S8). The most prominent associations were found for SNPs IL8 −251 T>A and IL6 −174 G>C (Figs 1, 2). The IL8 −251 A allele correlated with higher SOFA score during the entire observation time (Fig. 1). The carriers of the IL6 C/C genotype appeared to have elevated bilirubin levels from the first to the 14th day. LDH, CK and BUN levels were initially higher in patients with IL6 G allele. However, over time, these associations inverted and the carriers of the IL6 C/C genotype had significantly elevated concentrations of LDH, CK and BUN by the end of the second week (Fig. 2). Creatinine level also tended to increase in patients with the IL6 C/C genotype, but the differences did not reach statistical significance (Supporting information, Table S8).
Fig 1.

Dynamic changes in sequential organ failure assessment (SOFA) score in the carriers of different IL8 genotypes. Mean values and standard errors (s.e.) of SOFA scores are plotted by IL8 —251 T>A genotypes in five points of measurements between 1 and 14 days after admission to the intensive care unit. Bars represent mean SOFA values for patients carrying specified genotype. P-values are given for recessive model (A/A versus T/T-T/A).
Fig 2.
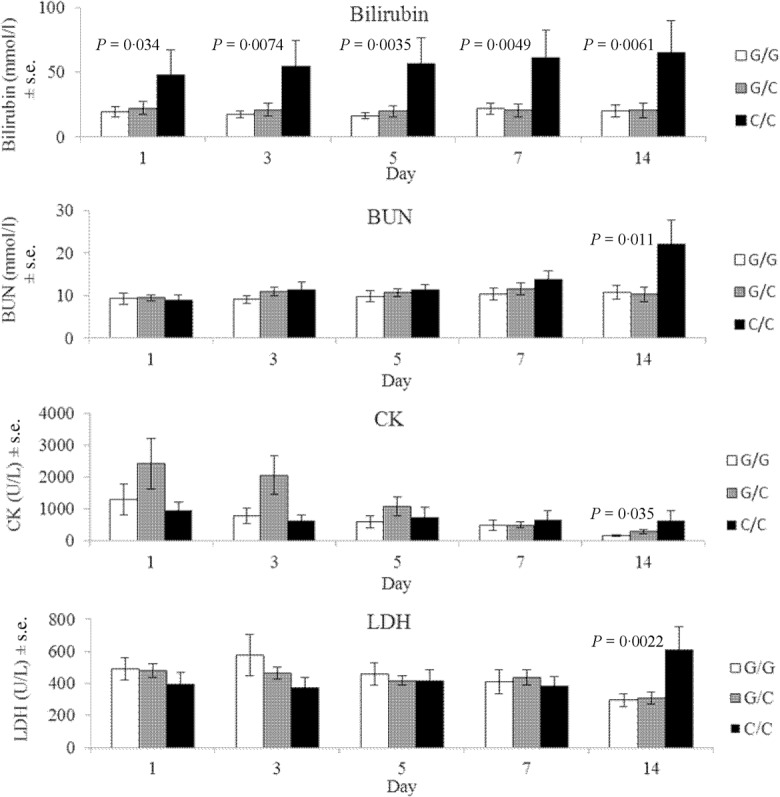
Dynamic changes in biochemical indicators in the carriers of different IL6 genotypes. Mean values and standard errors (s.e.) of bilirubin, blood urea nitrogen (BUN), creatine kinase (CK) and lactate dehydrogenase (LDH) values are plotted by the IL6 —174 G>C genotypes in five points of measurements between 1 and 14 days after admission to the intensive care unit. Bars represent mean indicator values for patients carrying specified genotype. Significant P-values are given for recessive model (C/C versus G/G-G/C). The analysis is adjusted for age, sex, APACHE II score, and using (more than 24 h) of mechanical ventilation.
Biochemical characteristics and genotype–phenotype correlation
The revealed genotypical associations with biochemical traits were used to analyse additional clinical phenotypes. The groups with renal and liver insufficiency were small. For this reason, they were not included in the initial genetic association study. Based on the results of bilirubin and BUN associations with IL6, we explored an interaction between the IL6 polymorphism and liver and renal insufficiency (Table3). IL6 C/C genotype appeared to be associated with liver insufficiency. A marginally significant association was also found for renal insufficiency under the same genetic model.
Table 3.
The distribution of IL6 genotypes among patients with/without renal and liver insufficiency
| Controls | Cases | ||||
|---|---|---|---|---|---|
| Genotypes | Number (%) | P-value (genetic model),c OR (95% CI) | |||
| Renal insufficiency | |||||
| n = 387 | n = 38 | ||||
| IL6 | G/G | 137 (35·4) | 14 (36·8) | 0·05 (rec) | |
| −174 G>C | G/C | 181 (46·8) | 12 (31·6) | 2·21 (1·00–4·78) | |
| C/C | 69 (17·8) | 12 (31·6) | |||
| Liver insufficiency | |||||
| IL6 | n = 400 | n = 25 | |||
| G/G | 141 (35·4) | 10 (40·0) | 0·012 (rec) | ||
| −174 G>C | G/C | 188 (47·0) | 5 (20·0) | 3·10 (1·33–7·23) | |
| C/C | 71 (17·8) | 10 (40·0) | |||
The analysis is adjusted for age, sex, Acute Physiology And Chronic Health Evaluation II (APACHE II) score, and using (more than 24 h) of mechanical ventilation. OR = odds ratio; CI = confidence interval. The genetic model: rec = recessive.
Discussion
In this study we investigated functional polymorphic variants in the genes of cytokines in relation to clinical phenotypes in patients with critical illness.
Alleles of proinflammatory cytokines IL1B −511A and IL8 −251A were associated with protection against sepsis. In active inflammation, interleukin IL-1B is known to serve as triggering cytokine for the cytokine cascades. In-vitro studies have shown that allele A in the promoter (upstream of the transcriptional start site) SNP −511 G>A is related to increased lipopolysaccharide (LPS)-induced IL-1B protein secretion36. In a recent meta-analysis, the SNP −511A allele was less frequent in patients with sepsis compared to controls among Caucasians (one study) and Asians (four studies), although the results failed to reach statistical significance37. The same direction of the effect was observed in our study for the same genetic model. Sepsis is a risk factor for mortality, and the revealed association of the SNP −511A allele with improved survival supports our findings in the group of patients with sepsis. These data are in line with the results of other studies of bacterial infections. The IL1B −511A allele has been protective against human leptospirosis38, Helicobacter pylori eradication failure39 and susceptibility to bacteraemia within the first year after kidney transplantation40.
IL-8 is a member of the chemokine family that initiates and enhances inflammation by activation and chemotaxis of immune cells. The results of the studies of genotype-specific influence on IL8 gene expression are controversial. Higher LPS-stimulated expression of IL8 mRNA was shown for the IL8 SNP −251A allele in Caucasians41,42, but for the allele T in Chinese43,44. The IL8 transcription is also influenced by haplotype and is tissue-specific45,46. The protective effect of the IL8 −251A allele in acute infections has been observed in acute suppurative apical periodontitis, but not chronic non-suppurative apical periodontitis47 and respiratory syncytial virus (RSV) bronchiolitis42.
Sepsis is a condition caused by a hyperinflammatory immune response to infection. In the 1990s, sepsis was believed to be associated with an exacerbated production of mainly proinflammatory cytokines. Later, it became evident that a tightly regulated balance in the cytokine network, which comprises proinflammatory cytokines, anti-inflammatory cytokines and soluble inhibitors of proinflammatory cytokines, is crucial to protect from sepsis48. The data today have shown that deficiencies in the production of proinflammatory cytokines result in defective activation of the host defence against infectious pathogens. Innate deficiency in cytokine release leads to a rapid accumulation of the microorganisms and may be followed by secondary systemic inflammatory and anti-inflammatory reactions49. The results of the current study support this assumption, as high-producing alleles of proinflammatory cytokines IL1B −511A and IL8 −251A protected against sepsis development.
IL-8 is considered one of the most important cytokines in the pathogenesis of ARDS, with the −251A allele being associated with the acute lung injury (ALI) susceptibility and related outcomes (references in50). The opposite direction of the IL8 genetic associations in acute infections (including sepsis in the current study) and ALI/ARDS may be linked to pathophysiological differences between sepsis-related ARDS and non-sepsis-related ARDS51. In our sample, only 12 people (∼3%) developed ARDS as a sequela from sepsis and, in line with literature data, the risk allele A (−251 T>A) was found more frequently in patients with ARDS than in non-ARDS controls, although the results were non-significant (P = 0·15, OR = 1·54, 95% CI = 0·84–2·80, dominant model) due to the insufficient statistical power of the test (26·67% at the two-sided significance level 0·05). IL-8 may be produced early in the inflammatory response, and may persist for a relatively long period of time52. A profound proinflammatory response might have a beneficial role in activating host defence, but a prolonged proinflammatory state can promote organ damage1. We found that a high-producing IL8 −251A allele was associated with higher SOFA scores, and this association is consistent with the above statement.
Variant alleles of anti-inflammatory cytokines IL4 −589T and IL13 431A were associated with protection against ARDS. Type 2 cytokines IL-4 and IL-13 are two closely related cytokines that play simultaneously overlapping and distinct roles in type 2 immunity53. Although these cytokines are usually considered as inducers of humoral response through the immunoglobulin (Ig)-mediated allergic/inflammatory pathway, they can also influence humoral responses during infections with extracellular pathogens54,55 and inhibit secretion of proinflammatory cytokines in response to bacterial toxins56,57. The genes for IL4 and IL13 are located in a cluster of cytokine genes in the region 5q23-31 (also including IL3, IL5, IL9, IL15, GMCSF and interferon regulatory factor)23. The IL4 promoter variant −589C>T allele T is considered a high-producer allele58. −589C>T SNP is linked tightly to 18 other SNPs, among which is −33C>T (rs2070874). Haplotype TT comprising both variant alleles in linkage disequilibrium was associated with threefold higher transcriptional activity in vitro and in vivo59.
The IL13 missense SNP (431 A>G) is also described as functional, with allele A being associated with higher IL-13 production60. This allele is in linkage disequilibrium with variant allele T of promoter SNP −1055C>T (rs1800925), which is also associated with higher IL-13 expression in patients with allergic asthma and non-atopic controls61.
The balance between proinflammatory and anti-inflammatory mediators may determine the extent of lung injury. Anti-inflammatory cytokines may protect against lung injury62. Although Th2 cytokines have been characterized not only as major contributors to allergy but also as mediators of response to pathogens, there have been very few studies conducted on the role of IL4 and IL13 SNPs in infectious and non-allergic inflammation diseases. The IL4 −589T allele protected against HIV-1 disease progression by reducing virus load63, and was less frequent in patients with tuberculous infection than in controls64. The carriers of the IL13 431A allele had a reduced risk of severe malaria65. For the first time, we observed the protective effect of high-producing alleles of anti-inflammatory cytokines IL4 −589T and IL13 431A in ARDS, and these results seem to have a biological rationale.
To understand further the genetic susceptibility to severe complications of trauma/injury, routine biochemical parameters were evaluated and related to the development of critical conditions and genetic variability in the studied genes. The IL6 –174 C/C genotype was linked consistently with the elevated levels of biochemical indicators: bilirubin, BUN, CK and LDH, but the only stable association from the first to the 14th day was found for bilirubin. IL-6 is a multi-functional cytokine with both pro- and anti-inflammatory activities and a central role in host defence66. It is produced by a large quantity of cell types and has many functions, including stimulation of the hepatic acute-phase proteins such as C-reactive protein (CRP) and fibrinogen. This function is reflected in one of the alternative names for IL-6: hepatocyte stimulatory factor (HSF) (http://omim.org/entry/147620). The liver is a crucial organ in the first line of host defence67, and IL-6 is the critical cytokine during the hepatic acute-phase reaction68. It is secreted by hepatic Kupffer cells (liver macrophages) in response to liposaccharide or tumour necrosis factor (TNF)69. IL-6 is known to inhibit an endotoxin-associated increase in TNF-α70 and to protect hepatocytes from TNF-α-induced hepatic injury71,72. Moreover, IL-6 increases the production of anti-inflammatory cytokines IL-1RA and IL-10 and stress-related anti-inflammatory hormone cortisol73. Many of the hepatoprotective effects of IL-6 are mediated through the activation of signal transducer and activator of transcription-3 (STAT-3) and mitogen-activated protein kinase (MAPK) signalling pathways69,74. Apart from the acute-phase response reaction, peaking within 2–3 days, IL-6 exerts anti-oxidant capacity in hepatocytes and protects liver from drug-induced hepatoxicity75–77. SNPs in the promoter region of the IL6 gene may be responsible for variations in transcription that subsequently affect serum levels. The best-characterized of these polymorphisms is a promoter SNP rs1800795 at position −174, upstream of the transcription start site, involving transversion of guanine for cytosine. Several studies of the associations of this SNP with severe acute conditions have shown that G allele is favourable for the disease outcome. Patients with the −174 G/G genotype demonstrated an improved survival in sepsis78,79 and had a reduced risk of septic shock80,81 and extrapulmonary pneumococcal dissemination82. In subjects with pneumococcal community-acquired pneumonia the −174 G/G genotype was protective against septic shock, ARDS, MODS and mortality25. By contrast, non-significant associations with severity or outcome have been observed in two other studies of critical conditions83,84. Although certain discrepancies also exist in relation to an assessment of genotype influence on gene expression, the IL6 −174 G allele is usually considered a high-producing allele80,83,85–88. The evidence for polymorphisms in the IL6 gene affecting the risk of organ failure is scarce. In addressing this theme, additional to the above-mentioned study by Martín-Loeches et al.25, investigation by Verduijn et al.89 has shown that the IL6 −174 C/C genotype is associated with mortality and technique failure in European peritoneal dialysis patients. In our study, we also found correlations between the IL6 –174 C/C genotype and liver and kidney insufficiency, the conditions connected closely to the levels of bilirubin, BUN and creatinine. Other clinical phenotypes and SOFA scores after 1 week of ICU stay were not associated with the IL6 polymorphic variant, although initially the carriers of the IL6 –174 C/C genotype had higher APACHE II and SOFA scores. While the severity of the state in the early period after injury depends upon the severity of the damage and the intensity of the acute-phase response, the development of different complications and critical conditions over time may be affected by genetically mediated associations of certain metabolic pathways. In our sample, it is the IL6 association with the level of biochemical parameters, notably bilirubin, and liver and kidney insufficiency in patients without the initially compromised liver and kidney functions.
The study has several limitations. Our sample size was modest, and the study is powered to detect only relatively large effect sizes (minimum detectable OR ∼1·6–1·9; reciprocal OR∼ 0·52–0·63). Although having an exploratory character, an association study between genetic and biochemical variables is limited by the multiple comparison issue. Our patients were mainly men, and the results may not be generalizable to women. The results require replication, but the revealed associations are important to understand the pathways and mechanisms of the development and progression of acute severe complications in patients with trauma/injury.
In summary, first, we present new results about the protective role of high-producing alleles of anti-inflammatory cytokines IL4 −589T and IL13 431A in patients with ARDS.
Secondly, for the first time, we tested correlations between SNPs in the genes of cytokines and the dynamics of routine biochemical parameters. Our data brought novel observations that higher concentrations of bilirubin are associated routinely with the IL6 −174C/C genotype. This genotype also appeared to be linked with the increased levels of BUN, CK and LDH, but only up to the end of the second week. It is an interesting challenge for future research directions to determine whether stronger hepatotoxic effects in the carriers of the −174C/C genotype were causal and/or synergistic for these last associations. The tested approach seems to be useful in genetic association studies, as biochemical indicators, being the intermediate metabolites, may have greater reliability and demonstrate larger effect sizes than those reported to disease itself90.
Thirdly, we observed the protective effects of high-producing alleles −511A IL1B and −251A IL8 in patients with sepsis. At the same time, −251A allele of the IL8 gene was associated consistently with higher SOFA score. The associations of polymorphic variations in the genes of proinflammatory cytokines IL1B and IL8 in patients with acute infectious, including sepsis, have been studied previously with inconsistent results. Our data are in line with the premise that a powerful proinflammatory response can have beneficial effects on sepsis development, but may increase the risk of subsequent organ damage1,91.
It is assumed that, for infectious diseases, genetics is more important than for cancer or cardiovascular diseases92. The PIRO concept92, which attempts to characterize sepsis across four components (predisposition, infection, response, organ dysfunction) is designed to classify states of sepsis. PIRO recognizes that the incidence and outcome of sepsis are influenced by genetic susceptibility, which has been suggested as a risk stratification tool and an inclusion criterion for therapeutic trials.
Acknowledgments
This work was supported by grants from the Presidium of the Russian Academy of Sciences program ‘Fundamental Sciences for Medicine’—2012, 2013, 2014.
Disclosures
There are no conflicts of interest.
References
- Dinarello CA. Proinflammatory and anti-inflammatory cytokines as mediators in the pathogenesis of septic shock. Chest. 1997;112:321S–9S. doi: 10.1378/chest.112.6_supplement.321s. [DOI] [PubMed] [Google Scholar]
- Remick DG. Cytokine therapeutics for the treatment of sepsis: why has nothing worked? Curr Pharm Des. 2003;9:75–82. doi: 10.2174/1381612033392567. [DOI] [PubMed] [Google Scholar]
- Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 2008;8:776–87. doi: 10.1038/nri2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar J, Maca-Meyer N, Pérez-Méndez L, Flores C. Bench-to-bedside review: understanding genetic predisposition to sepsis. Crit Care. 2004;8:180–9. doi: 10.1186/cc2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namath A, Patterson AJ. Genetic polymorphisms in sepsis. Crit Care Clin. 2009;25:835–56. doi: 10.1016/j.ccc.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Wong HR. Genetics and genomics in pediatric septic shock. Crit Care Med. 2012;40:1618–26. doi: 10.1097/CCM.0b013e318246b546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrakos C, Vincent JL. Sepsis biomarkers: a review. Crit Care. 2010;14:R15. doi: 10.1186/cc8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar D, Moore FA, Moore EE, Balogh Z. Postinjury multiple organ failure. Injury. 2009;40:912–18. doi: 10.1016/j.injury.2009.05.024. [DOI] [PubMed] [Google Scholar]
- Ratzinger F, Schuardt M, Eichbichler K, et al. Utility of sepsis biomarkers and the infection probability score to discriminate sepsis and systemic inflammatory response syndrome in standard care patients. PLOS ONE. 2013;8:e82946. doi: 10.1371/journal.pone.0082946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field E, Horst HM, Rubinfeld IS, et al. Hyperbilirubinemia: a risk factor for infection in the surgical intensive care unit. Am J Surg. 2008;195:304–7. doi: 10.1016/j.amjsurg.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Zhai R, Sheu CC, Su L, et al. Serum bilirubin levels on ICU admission are associated with ARDS development and mortality in sepsis. Thorax. 2009;64:784–90. doi: 10.1136/thx.2009.113464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LJ, McKinley BA, Turner KL, et al. The epidemiology of sepsis in general surgery patients. J Trauma. 2011;70:672–80. doi: 10.1097/TA.0b013e31820e7803. [DOI] [PubMed] [Google Scholar]
- Pate JJ, Taneja A, Niccum D, et al. The association of serum bilirubin levels on the outcomes of severe sepsis. J Intens Care Med. 2013 doi: 10.1177/0885066613488739. doi: 10.1177/0885066613488739. [DOI] [PubMed] [Google Scholar]
- Zilberberg MD, Carter C, Lefebvre P, et al. Red blood cell transfusions and the risk of acute respiratory distress syndrome among the critically ill: a cohort study. Crit Care. 2007;11:R63. doi: 10.1186/cc5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Xu MS, Ching JC, et al. CD209 (DC-SIGN) -336A>G promoter polymorphism and severe acute respiratory syndrome in Hong Kong Chinese. Hum Immunol. 2010;71:702–7. doi: 10.1016/j.humimm.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez A, Shental O, Tchebiner JZ, et al. Diagnostic and prognostic value of very high serum lactate dehydrogenase in admitted medical patients. Isr Med Assoc J. 2014;16:439–43. [PubMed] [Google Scholar]
- Drent M, Cobben NA, Henderson RF, et al. Usefulness of lactate dehydrogenase and its isoenzymes as indicators of lung damage or inflammation. Eur Respir J. 1996;9:1736–42. doi: 10.1183/09031936.96.09081736. [DOI] [PubMed] [Google Scholar]
- Borgatta B, Pérez M, Rello J, et al. Elevation of creatine kinase is associated with worse outcomes in 2009 pH1N1 influenza A infection. Intens Care Med. 2012;38:1152–61. doi: 10.1007/s00134-012-2565-5. [DOI] [PubMed] [Google Scholar]
- Teng L, Yu M, Li JM, et al. Matrix metalloproteinase-9 as new biomarkers of severity in multiple organ dysfunction syndrome caused by trauma and infection. Mol Cell Biochem. 2012;360:271–7. doi: 10.1007/s11010-011-1066-0. [DOI] [PubMed] [Google Scholar]
- Gao M, Zhang L, Liu Y, et al. Use of blood urea nitrogen, creatinine, interleukin-6, granulocyte-macrophage colony stimulating factor in combination to predict the severity and outcome of abdominal sepsis in rats. Inflamm Res. 2012;61:889–97. doi: 10.1007/s00011-012-0481-3. [DOI] [PubMed] [Google Scholar]
- Chen M, Wang B, Xu Y, et al. Diagnostic value of serum leptin and a promising novel diagnostic model for sepsis. Exp Ther Med. 2014;7:881–6. doi: 10.3892/etm.2014.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zeng L, Zhou J, et al. Clinical relevance of 13 cytokine gene polymorphisms in Chinese major trauma patients. Intens Care Med. 2010;36:1261–5. doi: 10.1007/s00134-010-1797-5. [DOI] [PubMed] [Google Scholar]
- Paffen E, Medina P, de Visser MC, et al. The -589C>T polymorphism in the interleukin-4 gene (IL-4) is associated with a reduced risk of myocardial infarction in young individuals. J Thromb Haemost. 2008;6:1633–8. doi: 10.1111/j.1538-7836.2008.03096.x. [DOI] [PubMed] [Google Scholar]
- Wacharasint P, Nakada TA, Boyd JH, et al. AA genotype of IL-8 -251A/T is associated with low PaO(2)/FiO(2) in critically ill patients and with increased IL-8 expression. Respirology. 2012;17:1253–60. doi: 10.1111/j.1440-1843.2012.02244.x. [DOI] [PubMed] [Google Scholar]
- Martín-Loeches I, Solé-Violán J, Rodríguez de Castro F, et al. Variants at the promoter of the interleukin-6 gene are associated with severity and outcome of pneumococcal community-acquired pneumonia. Intens Care Med. 2012;38:256–62. doi: 10.1007/s00134-011-2406-y. [DOI] [PubMed] [Google Scholar]
- Stanilova SA, Miteva LD, Karakolev ZT, Stefanov CS. Interleukin-10-1082 promoter polymorphism in association with cytokine production and sepsis susceptibility. Intens Care Med. 2006;32:260–6. doi: 10.1007/s00134-005-0022-4. [DOI] [PubMed] [Google Scholar]
- Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818–29. [PubMed] [Google Scholar]
- Vincent JL, Moreno R, Takala J, et al. The SOFA (sepsis-related organ failure assessment) score to describe organ dysfunction/failure. Intens Care Med. 1996;22:707–10. doi: 10.1007/BF01709751. [DOI] [PubMed] [Google Scholar]
- Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–55. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- Levy MM, Fink MP, Marshall JC, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250–6. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- Ranieri VM, Rubenfeld GD, Thompson BT, et al. Acute respiratory distress syndrome: the Berlin definition. JAMA. 2012;307:2526–33. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- Hamajima N. PCR–CTPP. A new genotyping technique in the era of genetic epidemiology. Expert Rev Mol Diagn. 2001;1:119–23. doi: 10.1586/14737159.1.1.119. [DOI] [PubMed] [Google Scholar]
- Sole X, Guino E, Valls J, et al. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–9. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Abramson JH. WINPEPI updated: computer programs for epidemiologists, and their teaching potential. Epidemiol Perspect Innov. 2011;8:1. doi: 10.1186/1742-5573-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–6. [PubMed] [Google Scholar]
- Pociot F, Molvig J, Wogensen L, et al. A TaqI polymorphism in the human interleukin-1 beta (IL-1 beta) gene correlates with IL-1 beta secretion in vitro. Eur J Clin Invest. 1992;15:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]
- Zhang AQ, Pan W, Gao JW, et al. Associations between interleukin-1 gene polymorphisms and sepsis risk: a meta-analysis. BMC Med Genet. 2014;15:8. doi: 10.1186/1471-2350-15-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves LM, Bulhões SM, Branco CC, et al. Human leptospirosis: seroreactivity and genetic susceptibility in the population of São Miguel Island (Azores, Portugal) PLOS ONE. 2014;9:e108534. doi: 10.1371/journal.pone.0108534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Furuta T, Yamaoka Y. Influence of inflammatory cytokine polymorphisms on eradication rates of Helicobacter pylori. J Gastroenterol Hepatol. 2009;24:1725–32. doi: 10.1111/j.1440-1746.2009.06047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan QQ, Ye QF, Ma Y, Zhou JD. Genetic association of interleukin-1β (−511C/T) and its receptor antagonist (86-bpVNTR) gene polymorphism with susceptibility to bacteremia in kidney transplant recipients. Transplant Proc. 2012;44:3026–8. doi: 10.1016/j.transproceed.2012.05.081. [DOI] [PubMed] [Google Scholar]
- Wacharasint P, Nakada TA, Boyd JH, et al. AA genotype of IL-8 −251A/T is associated with low PaO(2)/FiO(2) in critically ill patients and with increased IL-8 expression. Respirology. 2012;17:1253–60. doi: 10.1111/j.1440-1843.2012.02244.x. [DOI] [PubMed] [Google Scholar]
- Hull J, Thomson A, Kwiatkowski D. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax. 2000;55:1023–7. doi: 10.1136/thorax.55.12.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Zeng L, Zhou J, et al. Clinical relevance of 13 cytokine gene polymorphisms in Chinese major trauma patients. Intens Care Med. 2010;36:1261–5. doi: 10.1007/s00134-010-1797-5. [DOI] [PubMed] [Google Scholar]
- Lee WP, Tai DI, Lan KH, et al. The -251T allele of the interleukin-8 promoter is associated with increased risk of gastric carcinoma featuring diffuse-type histopathology in Chinese population. Clin Cancer Res. 2005;11:6431–41. doi: 10.1158/1078-0432.CCR-05-0942. [DOI] [PubMed] [Google Scholar]
- Hull J, Ackerman H, Isles K, et al. Unusual haplotypic structure of IL-8, a susceptibility locus for a common respiratory virus. Am J Hum Genet. 2001;69:413–19. doi: 10.1086/321291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacking D, Knight JC, Rockett K, et al. Increased in vivo transcription of an IL-8 haplotype associated with respiratory syncytial virus disease-susceptibility. Genes Immun. 2004;5:274–82. doi: 10.1038/sj.gene.6364067. [DOI] [PubMed] [Google Scholar]
- Amaya MP, Criado L, Blanco B, et al. Polymorphisms of pro-inflammatory cytokine genes and the risk for acute suppurative or chronic nonsuppurative apical periodontitis in a Colombian population. Int Endod J. 2013;46:71–8. doi: 10.1111/j.1365-2591.2012.02097.x. [DOI] [PubMed] [Google Scholar]
- Schulte W, Bernhagen J, Bucala R. Cytokines in sepsis: potent immunoregulators and potential therapeutic targets – an updated view. Mediat Inflamm. 2013;2013:165974. doi: 10.1155/2013/165974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, van der Meer JW, van Deuren M, Kullberg BJ. Proinflammatory cytokines and sepsis syndrome: not enough, or too much of a good thing? Trends Immunol. 2003;24:254–8. doi: 10.1016/s1471-4906(03)00079-6. [DOI] [PubMed] [Google Scholar]
- O'Mahony DS, Glavan BJ, Holden TD, et al. Inflammation and immune-related candidate gene associations with acute lung injury susceptibility and severity: a validation study. PLOS ONE. 2012;7:e51104. doi: 10.1371/journal.pone.0051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu CC, Gong MN, Zhai R, et al. Clinical characteristics and outcomes of sepsis-related vs non-sepsis-related ARDS. Chest. 2010;138:559–67. doi: 10.1378/chest.09-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforge LE, FantonE JC, Kenney JS, Remick DJ. Oxygen radical scavengers selectively inhibit interleukin 8 production in human whole blood. J Clin Invest. 1992;90:2123–9. doi: 10.1172/JCI116097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Locksley RM. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol. 2013;31:317–43. doi: 10.1146/annurev-immunol-032712-095906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Nishikawa S, Miura T, et al. Interleukin-4 and interleukin-10 are involved in host resistance to Staphylococcus aureus infection through regulation of gamma interferon. Infect Immun. 2000;68:2424–30. doi: 10.1128/iai.68.5.2424-2430.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AQ, Shen Y, Wu ZQ, et al. Endogenous pro- and anti-inflammatory cytokines differentially regulate an in vivo humoral response to Streptococcus pneumoniae. Infect Immun. 2002;70:749–61. doi: 10.1128/iai.70.2.749-761.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumhofer JM, Beinhauer BG, Wang JE, et al. Gene transfer with IL-4 and IL-13 improves survival in lethal endotoxemia in the mouse and ameliorates peritoneal macrophages immune competence. Eur J Immunol. 1998;28:610–15. doi: 10.1002/(SICI)1521-4141(199802)28:02<610::AID-IMMU610>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Collighan N, Giannoudis PV, Kourgeraki O, et al. Interleukin 13 and inflammatory markers in human sepsis. Br J Surg. 2004;91:762–8. doi: 10.1002/bjs.4521. [DOI] [PubMed] [Google Scholar]
- Rosenwasser LJ, Klemm DJ, Dresback JK, et al. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995;25(Suppl 2):74–8. doi: 10.1111/j.1365-2222.1995.tb00428.x. [DOI] [PubMed] [Google Scholar]
- Nakashima H, Miyake K, Inoue Y, et al. Association between IL-4 genotype and IL-4 production in the Japanese population. Genes Immun. 2002;3:107–9. doi: 10.1038/sj.gene.6363830. [DOI] [PubMed] [Google Scholar]
- Vladich FD, Brazille SM, Stern D, et al. IL-13 R130Q, a common variant associated with allergy and asthma, enhances effector mechanisms essential for human allergic inflammation. J Clin Invest. 2005;115:747–54. doi: 10.1172/JCI22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pouw Kraan TC, van Veen A, Boeije LC, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun. 1999;1:61–5. doi: 10.1038/sj.gene.6363630. [DOI] [PubMed] [Google Scholar]
- Parsons PE. Interleukin-10: the ambiguity in sepsis continues. Crit Care Med. 1998;26:818–19. doi: 10.1097/00003246-199805000-00007. [DOI] [PubMed] [Google Scholar]
- Nakayama EE, Meyer L, Iwamoto A, et al. Protective effect of interleukin-4 -589T polymorphism on human immunodeficiency virus type 1 disease progression: relationship with virus load. J Infect Dis. 2002;185:1183–6. doi: 10.1086/339825. [DOI] [PubMed] [Google Scholar]
- Naslednikova IO, Urazova OI, Voronkova OV, et al. Allelic polymorphism of cytokine genes during pulmonary tuberculosis. Bull Exp Biol Med. 2009;148:175–80. doi: 10.1007/s10517-009-0674-0. [DOI] [PubMed] [Google Scholar]
- Manjurano A, Clark TG, Nadjm B, et al. Candidate human genetic polymorphisms and severe malaria in a Tanzanian population. PLOS ONE. 2012;7:e47463. doi: 10.1371/journal.pone.0047463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Taga T, Kishimoto T. Interleukin-6 in biology and medicine. Adv Immunol. 1993;54:1–78. doi: 10.1016/s0065-2776(08)60532-5. [DOI] [PubMed] [Google Scholar]
- Seki S, Habu Y, Kawamura T, et al. The liver as a crucial organ in the first line of host defense: the roles of Kupffer cells, natural killer (NK) cells and NK1.1 Ag+ T cells in T helper 1 immune responses. Immunol Rev. 2000;174:35–46. doi: 10.1034/j.1600-0528.2002.017404.x. [DOI] [PubMed] [Google Scholar]
- Yang XP, Schaper F, Teubner A, et al. Interleukin-6 plays a crucial role in the hepatic expression of SOCS3 during acute inflammatory processes in vivo. J Hepatol. 2005;43:704–10. doi: 10.1016/j.jhep.2005.02.048. [DOI] [PubMed] [Google Scholar]
- Taub R. Hepatoprotection via the IL-6/Stat3 pathway. J Clin Invest. 2003;112:978–80. doi: 10.1172/JCI19974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkie R, Ostrowski SR, Jauffred S, et al. Exercise and IL-6 infusion inhibit endotoxin-induced TNF-alpha production in humans. FASEB J. 2003;17:884–6. doi: 10.1096/fj.02-0670fje. [DOI] [PubMed] [Google Scholar]
- Mizuhara H, O'Neill E, Seki N, et al. T cell activation-associated hepatic injury: mediation by tumor necrosis factors and protection by interleukin 6. J Exp Med. 1994;179:1529–37. doi: 10.1084/jem.179.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Gantner F, Bohlinger I, et al. Tumor necrosis factor-induced hepatocyte apoptosis precedes liver failure in experimental murine shock models. Am J Pathol. 1995;146:1220–34. [PMC free article] [PubMed] [Google Scholar]
- Steensberg A, Fischer CP, Keller C, et al. IL-6 enhances plasma IL-1ra, IL-10, and cortisol in humans. Am J Physiol Endocrinol Metab. 2003;285:E433–37. doi: 10.1152/ajpendo.00074.2003. [DOI] [PubMed] [Google Scholar]
- Bansal MB, Kovalovich K, Gupta R, et al. Interleukin-6 protects hepatocytes from CCl4-mediated necrosis and apoptosis in mice by reducing MMP-2 expression. J Hepatol. 2005;42:548–56. doi: 10.1016/j.jhep.2004.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir GA, Mohsin S, Khan M, et al. Mesenchymal stem cells and interleukin-6 attenuate liver fibrosis in mice. J Transl Med. 2013;11:78. doi: 10.1186/1479-5876-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wruck CJ, Streetz K, Pavic G, et al. Nrf2 induces interleukin-6 (IL-6) expression via an antioxidant response element within the IL-6 promoter. J Biol Chem. 2011;286:4493–9. doi: 10.1074/jbc.M110.162008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoku DB. Drug-induced hepatotoxicity: metabolic, genetic and immunological basis. Int J Mol Sci. 2014;15:6990–7003. doi: 10.3390/ijms15046990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter B, Raufhake C, Erren M, et al. Effect of the interleukin-6 promoter polymorphism (-174 G/C) on the incidence and outcome of sepsis. Crit Care Med. 2002;30:32–7. doi: 10.1097/00003246-200201000-00005. [DOI] [PubMed] [Google Scholar]
- Zidan HE, Elbehedy RM, Azab SF. IL6-174 G/C gene polymorphism and its relation to serum IL6 in Egyptian children with community-acquired pneumonia. Cytokine. 2014;67:60–4. doi: 10.1016/j.cyto.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Tischendorf JJ, Yagmur E, Scholten D, et al. The interleukin-6 (IL6)-174 G/C promoter genotype is associated with the presence of septic shock and the ex vivo secretion of IL6. Int J Immunogenet. 2007;34:413–18. doi: 10.1111/j.1744-313X.2007.00712.x. [DOI] [PubMed] [Google Scholar]
- Michalek J, Svetlikova P, Fedora M, et al. Interleukin-6 gene variants and the risk of sepsis development in children. Hum Immunol. 2007;68:756–60. doi: 10.1016/j.humimm.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Schaaf B, Rupp J, Muller-Steinhardt M, et al. The interleukin-6 -174 promoter polymorphism is associated with extrapulmonary bacterial dissemination in Streptococcus pneumoniae infection. Cytokine. 2005;31:324–8. doi: 10.1016/j.cyto.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Burzotta F, Iacoviello L, Di Castelnuovo A, et al. Relation of the -174 G/C polymorphism of interleukin-6 to interleukin-6 plasma levels and to length of hospitalization after surgical coronary revascularization. Am J Cardiol. 2001;88:1125–8. doi: 10.1016/s0002-9149(01)02046-x. [DOI] [PubMed] [Google Scholar]
- Solé-Violán J, de Castro Fv, García-Laorden MI, et al. Genetic variability in the severity and outcome of community-acquired pneumonia. Respir Med. 2010;104:440–7. doi: 10.1016/j.rmed.2009.10.009. [DOI] [PubMed] [Google Scholar]
- Rivera-Chavez FA, Peters-Hybki DL, Barber RC, O'Keefe GE. Interleukin-6 promoter haplotypes and interleukin-6 cytokine responses. Shock. 2003;20:218–23. doi: 10.1097/01.shk.0000079425.52617.db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudorf S, Krabbe KS, Berg RM, et al. Common studied polymorphisms do not affect plasma cytokine levels upon endotoxin exposure in humans. Clin Exp Immunol. 2008;152:147–52. doi: 10.1111/j.1365-2249.2008.03612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wypasek E, Undas A, Sniezek-Maciejewska M, et al. The increased plasma C-reactive protein and interleukin-6 levels in patients undergoing coronary artery bypass grafting surgery are associated with the interleukin-6-174G > C gene polymorphism. Ann Clin Biochem. 2010;47:343–9. doi: 10.1258/acb.2010.090305. [DOI] [PubMed] [Google Scholar]
- Satti HS, Hussain S, Javed Q. Association of interleukin-6 gene promoter polymorphism with coronary artery disease in Pakistani families. ScientificWorldJournal. 2013;2013:53836. doi: 10.1155/2013/538365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduijn M, Maréchal C, Coester AM, et al. The -174G/C variant of IL6 as risk factor for mortality and technique failure in a large cohort of peritoneal dialysis patients. Nephrol Dial Transplant. 2012;27:3516–23. doi: 10.1093/ndt/gfs128. [DOI] [PubMed] [Google Scholar]
- Dharuri H, Henneman P, Demirkan A, et al. Automated workflow-based exploitation of pathway databases provides new insights into genetic associations of metabolite profiles. BMC Genomics. 2013;14:865. doi: 10.1186/1471-2164-14-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, van der Meer JW, van Deuren M, Kullberg BJ. Proinflammatory cytokines and sepsis syndrome: not enough, or too much of a good thing? Trends Immunol. 2003;24:254–8. doi: 10.1016/s1471-4906(03)00079-6. [DOI] [PubMed] [Google Scholar]
- Angus DC, Burgner D, Wunderink R, et al. The PIRO concept: P is for predisposition. Crit Care. 2003;7:248–51. doi: 10.1186/cc2193. [DOI] [PMC free article] [PubMed] [Google Scholar]


