Abstract
Cell contacts provide spatial cues that polarize early embryos and epithelial cells. The homophilic adhesion protein E-cadherin is required for contact-induced polarity in many cells. However, it is debated whether E-cadherin functions instructively as a spatial cue, or permissively by ensuring adequate adhesion so that cells can sense other contact signals. In C. elegans, contacts polarize early embryonic cells by recruiting the RhoGAP PAC-1 to the adjacent cortex, inducing PAR protein asymmetry. Here we show that HMR-1/E-cadherin, which is dispensable for adhesion, functions together with HMP-1/α-catenin, JAC-1/p120 catenin, and the previously uncharacterized linker PICC-1/CCDC85/DIPA to bind PAC-1 and recruit it to contacts. Mislocalizing the HMR-1 intracellular domain to contact-free surfaces draws PAC-1 to these sites and depolarizes cells, demonstrating an instructive role for HMR-1 in polarization. Our findings identify an E-cadherin-mediated pathway that translates cell contacts into cortical polarity by directly recruiting a symmetry-breaking factor to the adjacent cortex.
Keywords: polarity, cell contact, adhesion, E-cadherin, RhoGAP, PAR-6
Cell polarity is essential for developmental events such as asymmetric division, formation of epithelial tissues, and morphogenesis. Embryonic blastomeres and mesenchymal cells polarize when cell-cell contacts break surface symmetry, triggering the asymmetric redistribution of cortical PAR proteins1–4. Here the PAR proteins PAR-3 (multi-PDZ domain protein), PAR-6 (PDZ and CRIB domain protein), and atypical protein kinase C (aPKC), establish a signaling center that elaborates contact/contact-free (apicobasal) polarity within the cell2, 5, 6. How symmetry-breaking cell contacts translate into intracellular PAR asymmetry and polarization is poorly understood.
In epithelial cells grown in culture, the homophilic adhesion protein E-cadherin is among the earliest proteins known to accumulate at nascent contacts between cells as they polarize 7–10. E-cadherin is required for apicobasal polarity in many epithelial cell types, and introducing E-cadherin into some mesenchymal cell types is sufficient to induce polarity11–16. These findings have led to the view that E-cadherin could perform an instructive role in polarization by recruiting polarity regulators to cell contacts17. Candidate E-cadherin polarity effectors include regulators of trafficking, such as the exocyst complex, which co-localizes with E-cadherin at nascent cell contacts10, 18. However, an instructive role in polarization for the exocyst or other E-cadherin effectors has not been demonstrated.
Determining the molecular role of E-cadherin in cell polarization is complicated by its requirement for cell adhesion. For example, depleting E-cadherin from early mouse embryos prevents apicobasal PAR protein asymmetry but also severely disrupts blastomere adhesion16, leaving it unclear as to whether loss of polarity is secondary to loss of adhesion. Therefore, it remains possible that E-cadherin is required permissively for contact-induced polarity – i.e. enabling cells to adhere sufficiently such that a cadherin-independent cue can break symmetry19, 20. Some invertebrate and mammalian epithelial cells can polarize in the absence of detectable surface E-cadherin19, 21–23, suggesting that cadherin-independent pathways contribute to polarization in at least some cell types.
In C. elegans early embryos24, 25 (as in mammalian embryos26–28), cell contacts polarize blastomeres by restricting PAR-3, PAR-6, and PKC-3/aPKC to contact-free surfaces. Contacts break symmetry by recruiting the Rho GTPase activating protein (RhoGAP) PAC-1/ARHGAP21 to the adjacent cortex. In turn, PAC-1 locally inhibits the Rho GTPase CDC-42, leaving CDC-42 active at contact-free surfaces where it recruits PAR proteins29. How cell contacts recruit PAC-1 to polarize cells is unknown. The sole C. elegans classic cadherin, E-cadherin homolog HMR-1, also localizes to blastomere cell contacts, although in contrast to E-cadherin in other species HMR-1 is not required for adhesion at this stage 21, 30. Here, we investigate the mechanisms responsible for PAC-1 asymmetry. We show that HMR-1/E-cadherin performs an instructive role in polarization by recruiting PAC-1 to contact sites.
RESULTS
The PAC-1 N-terminal domain mediates cell contact localization
As a first step in determining how PAC-1 is recruited to cell contacts, we performed structure-function experiments to define the domains within PAC-1 responsible for its localization. We detected two distinct isoforms of pac-1 mRNA in embryos – a full-length isoform predicted to encode a protein with central pleckstrin homology (PH) and RhoGAP domains, and a short isoform whose predicted product lacks the N-terminal region and PH domain but retains the RhoGAP domain (Figure 1a). Existing pac-1 mutations affect both full-length and short isoforms (Figure 1a)29. However, an RNAi probe specific to the full-length isoform caused polarity defects identical to those of pac-1 mutants: PAR-6, which in wild type is restricted to contact-free surfaces (Figure 1b, 17/17 embryos), instead localized to both contact-free and contacted surfaces (Figure 1c, 34/34 embryos). Additionally, full-length PAC-1 tagged N-terminally with mCherry (Figure 1a) localized to cell contacts (Figure 1d, 18/18 embryos) and rescued the PAR-6 polarity defects of pac-1 mutants (30/30 embryos). These findings indicate that the full-length PAC-1 isoform, which we refer to hereafter as PAC-1, mediates blastomere polarization.
Figure 1. pac-1 structure-function analysis.
(a) The pac-1 locus; exons are rectangles, introns are chevrons, and transcription start sites are right-angled arrows. Regions of pac-1 encoding the PH (yellow) and GAP (red) domains, the position of the xn6 nonsense mutation, and the site of mCherry insertion within the mCherry-pac-1 transgene are indicated. (b–c) Wild-type and pac-1(5′ RNAi) 7–8 cell embryos stained for PAR-6 (arrows); pac-1(5′ RNAi) targets full-length pac-1 but not the pac-1 short isoform. (d) mCherry-PAC-1 (arrow) at cell contacts in a live 8-cell embryo. (e) Schematic of full-length PAC-1 protein and protein fragments tested for localization; amino acid positions are numbered, position of the PH and GAP domains are shown, and localization pattern is indicated. See Supplementary Figure 1a for transgene expression level quantification. (f–i) Four-cell embryos expressing the indicated GFP-PAC-1 fragments in otherwise wild-type embryos; arrows indicate contact localization. (j) Embryo expressing GFP-PAC-11-574 in which endogenous pac-1 is depleted by RNAi against the 3′ end of pac-1 (see Supplementary Figure 1b,c for controls). Schematized in (e) but not shown: GFP-PAC-1392-838 (localized strongly to cell contacts in 0/54 embryos, although very weak contact localization was evident) and GFP-PAC-12-610 (localized to cell contacts in 48/51 embryos). (k–l) Full-length (FL) mCherry-PAC-1 at cell contacts in control and hmr-1 four-cell embryos. (m) Contact enrichment of mCherry-PAC-1FL in control (n = 18 embryos) and hmr-1 (n = 16 embryos) four-cell embryos (**p = 0.007, Mann-Whitney U test). Samples pooled from three independent experiments. (n–o) GFP-PAC-1N at cell contacts in a control four-cell embryo (n) and in the cytoplasm of a hmr-1 four-cell embryo (o). See Figure 3b for quantification. (p) GFP-PAC-1ΔPH in the cytoplasm of a hmr-1 four-cell embryo. Control embryos are wild-type embryos fed on bacteria containing empty RNAi vector. Embryos are shown live; control and experimental embryos were taken at the same camera exposure. Scale bars, 10μm.
To determine which PAC-1 domains mediate contact localization, we examined PAC-1 fragments fused to green fluorescent protein (GFP) (Figure 1e; transgene expression quantified in Supplementary Figure 1a). Full-length GFP-PAC-1 localized to cell contacts, indistinguishably from mCherry-PAC-1 (Figure 1f, 20/20 embryos). Deleting the PH domain (Figure 1g, 81/84 embryos) or catalytically inactivating the RhoGAP domain29 did not prevent GFP-PAC-1 contact localization. By contrast, removing amino acids 1-574 from the N-terminal domain resulted in cytoplasmic localization (Figure 1h, 25/25 embryos), whereas the N-terminal domain alone fused to GFP localized to cell contacts (Figure 1i, 103/103 embryos). The N-terminal domain still localized to cell contacts in embryos lacking endogenous PAC-1 (Figure 1j, 23/23 embryos; see Supplementary Figure 1b,c for RNAi controls), excluding the possibility that the endogenous protein recruits it there. We conclude that a region of the PAC-1 N-terminus contained within amino acids 1-574, hereafter PAC-1N, is both necessary and sufficient for contact localization.
The homophilic adhesion protein HMR-1/E-cadherin contributes to PAC-1 localization
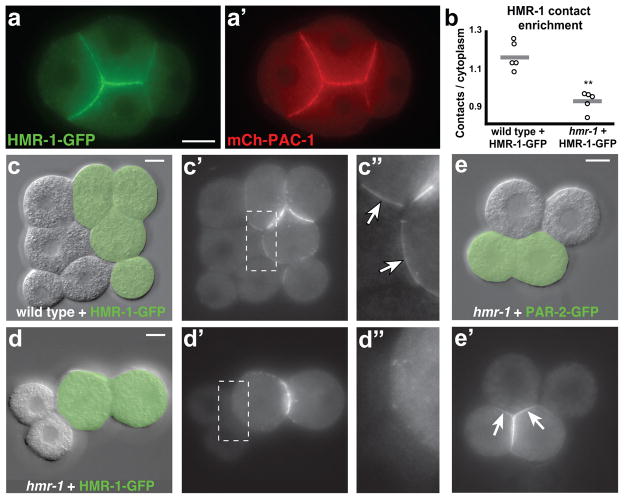
A potential mechanism for localizing PAC-1 is via coupling to a transmembrane protein, such as E-cadherin, that is restricted to cell contacts by homophilic interactions. Because HMR-1/E-cadherin and PAC-1 are both found at cell contacts between blastomeres (Figure 2a,a′), we performed a series of experiments to determine whether HMR-1 has a role in localizing PAC-1. First, we created chimeric cell contacts to test whether HMR-1, like mammalian E-cadherin31, only localizes to contacts when it is present in both touching cells. HMR-1-GFP was enriched at contacts created by combining cells expressing HMR-1-GFP with unmarked wild-type cells (Figure 2b,c-c″, 10/10 embryos). By contrast, HMR-1-GFP was not enriched at chimeric contacts between cells expressing HMR-1-GFP and unmarked cells lacking detectable HMR-1 (Figure 2b,d-d″, 8/8 embryos), which we created by combining a hmr-1 mutant with hmr-1 RNAi as described previously32. To test whether wild-type and hmr-1 cells make effective contacts with each other, we analyzed the localization of GFP-PAR-2, which is recruited to cell contacts independently of HMR-130. GFP-PAR-2 was enriched at chimeric contacts between wild-type and hmr-1 cells (Figure 2e,e′, 10/10 embryos), confirming that HMR-1 is not needed for cell contact formation. We conclude that HMR-1 cannot localize to a cell contact unless it is present in both touching cells, strongly suggesting that HMR-1 localizes to cell contacts through homophilic interactions.
Figure 2. HMR-1-GFP localization in chimeric embryos.
(a-a′) HMR-1-GFP and mCherry-PAC-1 co-expressed and shown live in a four-cell embryo. (b) Quantification of HMR-1-GFP contact enrichment in chimeric embryos, expressed as the ratio between signal at chimeric contacts over cytoplasmic signal within HMR-1-GFP-expressing cells (see Methods). Individual data points are indicated by open circles and the average value is shown as a gray line. Only embryos with chimeric contacts all within one image plane were analyzed (n=5 chimeras for each genotype, **p = 0.009, Mann-Whitney U test). Samples pooled from two independent experiments. (c–d″) Chimeric embryos created by combining an embryo expressing HMR-1-GFP (green shading in c,d) with a wild-type embryo (c-c″; wild-type + HMR-1-GFP chimera) or a hmr-1 embryo (d-d″; hmr-1 + HMR-1-GFP chimera). DIC and GFP channels are shown, and a blowup of the chimeric contact (boxed region) is shown in (c″, d″). HMR-1-GFP can be seen at the chimeric contacts in the wild-type + HMR-1-GFP embryo (arrows in c″). (e-e′) DIC (e) and GFP (e′) images of a chimeric embryo created by combining an embryo expressing PAR-2-GFP (green shading) with a hmr-1 embryo. PAR-2-GFP can be seen accumulating at the chimeric contacts (10/10 embryos; arrows in e′). Scale bars, 10μm.
To determine whether HMR-1 has a role in recruiting PAC-1 to cell contacts, we compared the localization of full-length PAC-1 and a subset of PAC-1 deletion fragments in wild-type and hmr-1 embryos. Full-length mCherry-PAC-1 localized to cell contacts in hmr-1 embryos (Figure 1k,l 16/16 embryos), although the relative amount at cell contacts versus the cytoplasm was slightly but significantly reduced compared to wild type (Figure 1m). By contrast, GFP-PAC-1N, which is found at cell contacts in wild-type embryos (Figure 1n), did not localize to cell contacts in hmr-1 embryos (Figure 1o; quantified in Figure 3b). We draw two conclusions from these observations. First, because HMR-1 is required to localize PAC-1N but only contributes partially to localizing full-length PAC-1, we infer that HMR-1 has a partially redundant role in localizing PAC-1 together with an HMR-1-independent pathway. Second, because PAC-1 fragments that lack the N-terminal domain but retain the remainder of the protein do not localize in wild-type embryos, the PAC-1 N-terminal domain is essential for both the HMR-1-dependent and HMR-1-independent localization mechanisms.
Figure 3. The role of catenins in PAC-1N localization.
(a) Schematic of the C. elegans cadherin-catenin complex at a cell-cell contact. (b) Quantification of GFP-PAC-1N contact enrichment in the indicated genotypes; contact enrichment is expressed as a ratio of the level of GFP-PAC-1N at a cell contact versus the level within the adjacent cytoplasm. Circles represent individual embryo measurements (control n = 21 embryos, hmp-1(RNAi) n = 34, jac-1 n = 23, jac-1 + hmp-1(RNAi) n = 20, hmr-1 n = 16) and gray bars indicate the mean value. Samples pooled from three independent experiments for hmp-1(RNAi); all other samples pooled from two independent experiments. The mean values of jac-1, hmp-1(RNAi), jac-1 + hmp-1(RNAi), and hmr-1 all differ significantly from control (p < 10−8). The mean of jac-1 + hmp-1(RNAi) differs significantly from both jac-1 and hmp-1(RNAi) (p < 10−8) but is not significantly different from hmr-1. See Supplementary Table 1 for Mann-Whitney U test statistical comparisons. (c–f) Images of GFP-PAC-1N in live four-cell embryos of the indicated genotype (see Figure 1o for comparison to hmr-1). Control embryos are wild-type embryos fed on bacteria containing RNAi empty vector. jac-1 mutant embryos were also fed empty vector RNAi bacteria. Scale bars, 10μm.
We next examined GFP-PAC-1ΔPH in hmr-1 mutants. In contrast to wild-type embryos, in which GFP-PAC-1ΔPH localizes to contacts (see Figure 1g), GFP-PAC-1ΔPH was present in the cytoplasm of hmr-1 embryos (Figure 1p, 53/55 embryos). This finding indicates that the redundant HMR-1-independent localization mechanism requires the PH domain, and that HMR-1 becomes essential for localizing PAC-1 when the redundant pathway is compromised. Despite the importance of the PH domain in the hmr-1 mutant background, its function is dispensable in wild-type embryos since GFP-PAC-1ΔPH localized to contacts and rescued the polarity defects of pac-1 mutant embryos (Supplementary Figure 2). This finding suggests that the HMR-1 pathway is sufficient to polarize cells when the redundant PAC-1 localization mechanism is compromised.
JAC-1/p120 catenin and HMP-1/α-catenin recruit PAC-1 to HMR-1
Next, we focused on identifying the molecular links between HMR-1 and the PAC-1 N-terminal domain that help recruit PAC-1 to cell contacts. Like mammalian E-cadherin, the HMR-1 intracellular domain interacts with different catenins through distinct binding domains (summarized in Figure 3a): HMP-2/β-catenin binds to a C-terminal domain and recruits HMP-1/α-catenin, whereas JAC-1/p120 catenin binds to a juxtamembrane domain33, 34. To determine whether catenins provide a link between PAC-1 and HMR-1, we compared the relative level of GFP-PAC-1N at cell contacts in wild-type embryos and in embryos depleted of HMP-2, HMP-1, or JAC-1. hmp-2 and hmp-1 zygotic mutants arrest during mid-embryogenesis21, so homozygous null mutant embryos lacking both maternal and zygotic protein cannot be easily obtained. However, RNAi knockdown of HMP-2 or HMP-1 caused an equivalent partial loss of GFP-PAC-1N from contacts, which we quantified in live embryos (Figure 3b–d, Supplementary Figure 3). Because knockdown of HMP-2 or HMP-1 eliminated HMP-1 immunostaining at cell contacts (Supplementary Figure 4a,b,g), we infer that HMP-1 has a partially redundant role in recruiting PAC-1, and that the contribution of HMP-2 could be limited to localizing HMP-1.
The expression and function of jac-1 had not been examined in early embryos34. To determine if JAC-1 is expressed, we raised an antibody against the conserved Armadillo repeats, which mediate the interaction between JAC-1/p120 catenin and HMR-1/E-cadherin in worms and mammals34, 35. α-JAC-1 stained cell contacts, which we verified by constructing a GFP-JAC-1 fusion protein (Supplementary Figure 4c–e). To assess jac-1 function, we deleted most of the jac-1 gene, including the Armadillo repeat region (Supplementary Figure 4i). jac-1 embryos were viable (wild type: 98% viable, n=555; jac-1: 99% viable, n=641), but showed a partial loss of GFP-PAC-1N from cell contacts, similar to hmp-1(RNAi) embryos (Figure 3b,e; statistical comparisons in Supplementary Table 1). Removing HMP-1 in jac-1 embryos caused a complete loss of GFP-PAC-1N from contacts, as in hmr-1 embryos (Figure 3b,f, Supplementary Table 1), indicating that HMP-1 and JAC-1 each contribute partially to PAC-1 localization.
HMP-1-GFP localized normally in jac-1 embryos, and GFP-JAC-1 localized normally in hmp-1(RNAi) embryos (Supplementary Figure 5a–f), indicating that JAC-1 and HMP-1 are likely to regulate PAC-1 independently rather than through cross-regulation. To test whether JAC-1 or HMP-1 depletion could affect PAC-1 localization indirectly by altering HMR-1 levels, we measured the amount of HMR-1-GFP at cell contacts in live embryos. HMR-1-GFP levels at cell contacts were equivalent in wild-type, hmp-1(RNAi), jac-1, and jac-1 hmp-1(RNAi) embryos (Supplementary Figure 5g–k). Endogenous HMR-1 protein also appeared equivalent in immunostained wild-type and jac-1 hmp-1(RNAi) embryos (Supplementary Figure 5l,m). By contrast, both HMP-1 and GFP-JAC-1 localization was lost from cell contacts in hmr-1 embryos (Supplementary Figure 4f,h). All together, these results indicate that JAC-1 and HMP-1 function independently and in parallel, downstream of HMR-1, to help recruit PAC-1 to cell contacts.
The coiled-coil protein PICC-1 links PAC-1 to JAC-1
We performed yeast two-hybrid experiments to search for a physical link between PAC-1 and the catenins. Because direct two-hybrid tests between PAC-1N and HMP-1 or JAC-1 did not reveal an interaction (Supplementary Figure 7a), we searched for linker proteins by screening a library of mixed-stage cDNAs using PAC-12–610 as bait. The most commonly captured prey clone was F29G9.2, an uncharacterized conserved gene homologous to the poorly characterized human genes CCDC85A (coil-coil domain-containing protein 85A), CCDC85B/DIPA, and CCDC85C (Supplementary Figure 6b). Like its human homologues, F29G9.2 is predicted to encode a protein with multiple coiled-coil domains. Accordingly, we renamed it picc-1 (PAC-1-interacting coiled-coil protein 1).
CCDC85B/DIPA was recently shown to bind p120 isoform 1A36, 37. To determine whether PICC-1 functions analogously by recruiting PAC-1 to JAC-1/p120 and therefore to HMR-1, we first created transgenes that express GFP-PICC-1 from either endogenous or heterologous regulatory sequences. In wild-type (80/80 embryos) and pac-1(RNAi) (35/35 embryos) early embryos, GFP-PICC-1 localized to cell contacts (Figure 4a,b). However, GFP-PICC-1 was absent from contacts in hmr-1 (50/50 embryos) and jac-1 (44/44 embryos) mutants (Figure 4c, Supplementary Figure 6c). By contrast, GFP-PICC-1 remained at cell contacts in hmp-1(RNAi) embryos (31/31 embryos, Supplementary Figure 6d), suggesting that PICC-1 functions downstream of JAC-1 but independently of HMP-1. To test whether PICC-1 has a role in recruiting PAC-1 downstream of JAC-1, we deleted the picc-1 gene (see Supplementary Figure 6a) and examined the localization of GFP-PAC-1N in single and double mutant combinations. Like jac-1 mutants, picc-1 mutants were viable (97% viable, n=437), allowing us to obtain early embryos lacking maternal and zygotic PICC-1 protein. As in jac-1 and hmp-1(RNAi) embryos, GFP-PAC-1N was partially lost from cell contacts in picc-1 embryos (Figure 4d,e,h, Supplementary Table 1). In addition, GFP-PAC-1N was partially lost from contacts in jac-1; picc-1 double mutant embryos, similar to each single mutant (Figure 4f,h, Supplementary Table 1). By contrast, GFP-PAC-1N was absent from cell contacts in picc-1 hmp-1(RNAi) embryos (Figure 4g,h), similar to jac-1 hmp-1(RNAi) embryos (Supplementary Table 1). All together, these observations indicate that PICC-1 functions downstream of HMR-1 and JAC-1, and in parallel to HMP-1, to help recruit PAC-1 to cell contacts.
Figure 4. PICC-1 localization and role in PAC-1N localization.
(a–c) GFP-PICC-1 localization in four-cell embryos of the indicated genotype. (d–g) GFP-PAC-1N localization in four-cell embryos of the indicated genotype. Control embryos are wild-type embryos fed on bacteria containing empty RNAi vector. picc-1 and jac-1; picc-1 mutant embryos were also fed empty vector RNAi bacteria. (h) Quantification of GFP-PAC-1N contact enrichment in the indicated genotypes; contact enrichment is expressed as a ratio of the level of GFP-PAC-1N at a cell contact versus the level within the adjacent cytoplasm. Circles represent individual embryo measurements (control n = 22 embryos, picc-1 n = 19, jac-1; picc-1 n = 23, picc-1 + hmp-1(RNAi) n = 18, jac-1; picc-1 + hmp-1(RNAi) n = 23) and gray bars indicate the mean value. The mean values of picc-1, jac-1; picc-1, picc-1 + hmp-1(RNAi) and jac-1; picc-1 + hmp-1(RNAi) all differ significantly from control (p < 10−6). Mean values for picc-1 are marginally significantly different from those for jac-1; picc-1 (p < 0.05) and significantly different from those for picc-1 + hmp-1(RNAi) (p < 10−4). See Supplementary Table 1 for Mann-Whitney U test statistical comparisons. Samples were pooled from two independent experiments. Scale bars, 10μm.
To determine if PICC-1 recruits PAC-1 by physically linking it to JAC-1 and therefore indirectly to the HMR-1 intracellular domain, we performed a combination of yeast two-hybrid tests and immunoprecipitations (IPs). In direct two-hybrid tests, PICC-1 interacted with both PAC-1N and with JAC-1 (Figure 5a, controls in Supplementary Figure 7a), and we recapitulated the previously reported interaction between JAC-1 and the HMR-1 intracellular domain (Supplementary Figure 7a)34. To examine interactions in vivo, we performed co-IP experiments using lysates from embryos expressing GFP-PICC-1. α-JAC-1 antiserum detected four distinct JAC-1 species in embryonic extracts (apparent molecular weight: 121, 137, 158, and 170 kD), which we verified were specific by analyzing jac-1(xn15) mutant extracts (Figure 5b). IP with α-JAC-1 pulled down all four JAC-1 species and co-immunoprecipitated PICC-1-GFP from otherwise wild-type embryos, but not from jac-1 mutant embryos expressing PICC-1-GFP (Figure 5b). Reciprocal IP experiments using α-GFP antibodies to pull down PICC-1-GFP co-immunoprecipitated JAC-1 (Supplementary Figure 7b). We next searched for an in vivo interaction between PICC-1 and PAC-1 by performing IP experiments in a strain expressing both PICC-1-GFP and mCherry-HA-PAC-1. IP with α-GFP pulled down PICC-1-GFP and co-immunoprecipitated mCherry-HA-PAC-1 (Figure 5c). Reciprocal IP experiments using α-HA antibodies pulled down mCherry-HA-PAC-1 and co-immunoprecipitated PICC-1-GFP (Supplementary Figure 7c). We conclude that PICC-1 can interact with JAC-1 and PAC-1 in both yeast and C. elegans embryonic extracts.
Figure 5. PICC-1 physical interactions with PAC-1 and JAC-1.
(a) Yeast two-hybrid interactions between PICC-1 fused to the GAL4 DNA-binding domain (DBD-bait) and GAL-4 activation domain fusions with PAC-11-574 or JAC-1 (AD-prey). Yeast was plated on selective medium requiring an interaction for growth (see Methods); strong (‘++’: DBD-Krev1 + AD-RalGDS), medium (‘+’: DBD-Krev1 + AD-RalGDSm1), and no interaction (‘−’: DBD-Krev1 + AD-RalGDSm2) controls are shown for comparison. (b) Immunoprecipitation of JAC-1 (four isoforms are visible) and co-immunoprecipitation of PICC-1-GFP from embryonic lysates; jac-1(xn15) mutants (‘xn15’) expressing PICC-1-GFP were used as a control; ‘WT’ = wild-type jac-1 allele. (c) Immunoprecipitation of PICC-1-GFP and co-immunoprecipitation of mCherry-HA-PAC-1 from embryonic lysates; wild-type embryos, and embryos expressing mCherry-HA-PAC-1 but not PICC-1-GFP were used as controls. Immunoprecipitation experiments shown in (b) and (c) were performed twice, and representative examples are shown. Tubulin levels in the input (total embryonic lysate) are shown as a loading control. ‘IB,’ Immunoblot; ‘IP,’ Immunoprecipitation. Uncropped blots are shown in Supplementary Figure 8.
HMR-1 is sufficient to ectopically recruit PAC-1 and depolarize cells
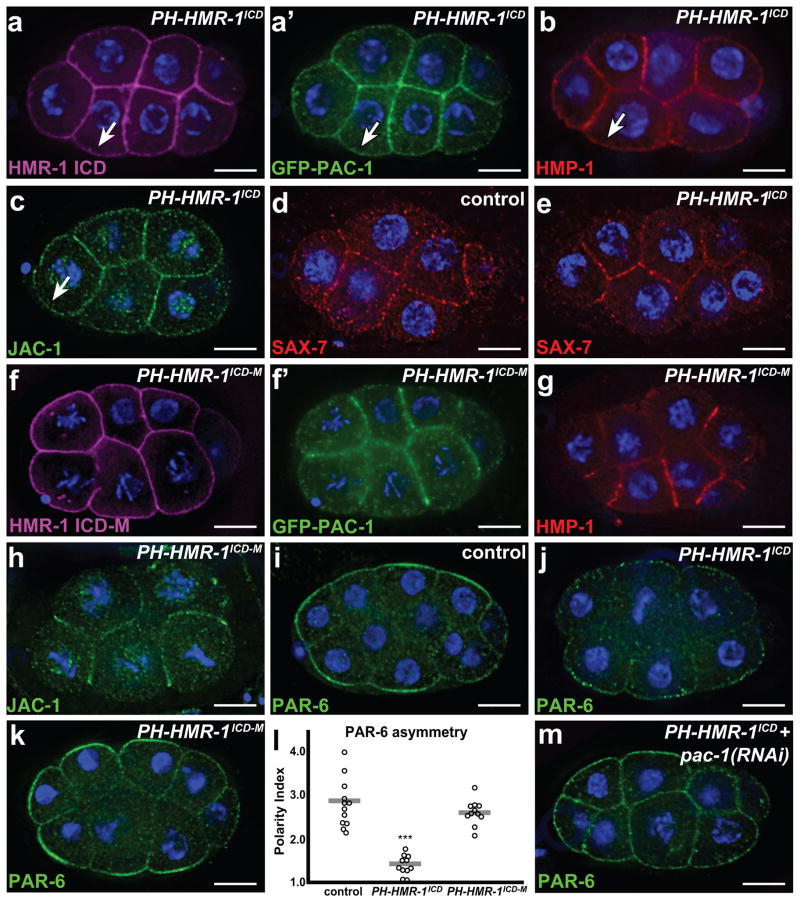
Our analysis of the catenins and PICC-1 suggested that HMR-1 performs an instructive role in polarization by recruiting PAC-1 to cell contact sites. We tested this hypothesis directly by asking whether mislocalizing the HMR-1 intracellular domain (HMR-1ICD) to contact-free surfaces recruited GFP-PAC-1 to these sites. To mislocalize HMR-1ICD, we fused it to the rat PLC1δ1 PH domain, which targets proteins to the plasma membrane of C. elegans blastomeres38, 39. HA-tagged PH-HMR-1ICD expressed from a heat-shock promoter localized uniformly around the plasma membrane (Figure 6a), did not disrupt cell adhesion (as assessed by staining with the cell contact marker SAX-7, Figure 6d,e), and retained its ability to bind HMP-1 and JAC-1 (arrows, Figure 6b,c). PH-HMR-1ICD also recruited GFP-PAC-1 to contact-free surfaces (arrow, Figure 6a′). Preventing catenin interactions by mutating the juxtamembrane domain and deleting the C-terminal domain (PH-HMR-1ICD–M), which prevents binding of JAC-1 and HMP-2 (and therefore HMP-1), respectively33, 34 (Figure 6g,h), blocked the recruitment of GFP-PAC-1 to contact-free surfaces (Figure 6f′).
Figure 6. HMR-1 sufficiency in PAC-1 recruitment and cell polarization.
Control embryos are heat-shocked sibling embryos lacking the indicated transgenic array. All PH-HMR-1ICD and PH-HMR-1ICD-M constructs include an HA tag, which was used for localization of the fusion protein in immunostained embryos. (a) PH-HMR-1ICD expression; arrow indicates localization to a contact-free surface. (a′) GFP-PAC-1 immunostaining in same embryo expressing PH-HMR-1ICD; arrow denotes ectopic localization of GFP-PAC-1 at contact-free surface (GFP-PAC-1 localized pan-cortically in 22/22 embryos). (b) HMP-1 immunostaining in embryo expressing PH-HMR-1ICD (50/50 embryos with pan-cortical localization; arrow indicates HMP-1 at a contact-free surface). (c) JAC-1 immunostaining in embryo expressing PH-HMR-1ICD (51/51 embryos with pan-cortical localization; arrow indicates JAC-1 at a contact-free surface). (d–e) SAX-7 immunostaining in a control embryo (37/37 embryos with contact localization) and embryo expressing PH-HMR-1ICD (33/33 embryos with contact localization). (f) PH-HMR-1ICD-M localization. (f′) GFP-PAC-1 immunostaining in same embryo expressing PH-HMR-1ICD-M; GFP-PAC-1 is present at contacted but not contact-free surfaces (29/29 embryos). (g) HMP-1 immunostaining in embryo expressing PH-HMR-1ICD-M (50/50 embryos with contact localization). (h) JAC-1 immunostaining in embryo expressing PH-HMR-1ICD-M (48/48 embryos with contact localization). (i–k) PAR-6 immunostaining in embryos of the indicated genotype. (l) Quantification of the PAR-6 polarity index, defined as the ratio of PAR-6 levels at contact-free surfaces versus half of the levels at contacted surfaces. Individual measurements are indicated with circles, and the gray bar indicates mean value. There is no significant difference in PAR-6 polarity index between control embryos (n = 12 embryos) and embryos expressing PH-HMR-1ICD-M (n = 11 embryos), while embryos expressing PH-HMR-1ICD (n = 11 embryos) differ significantly from both control embryos and embryos expressing PH-HMR-1ICD-M (***p < 10−5, Mann-Whitney U test). Samples were pooled from three independent experiments. (m) pac-1(RNAi) embryo expressing PH-HMR-1ICD; PAR-6 is present at high levels at both contacted and contact-free surfaces (15/15 embryos). Scale bars, 10μm.
To determine if blastomeres depolarized when HMR-1ICD recruited PAC-1 to contact-free surfaces, we immunostained embryos for PAR-6. In contrast to control heat-shocked embryos and embryos expressing HMR-1ICD–M, where PAR-6 was markedly enriched at contact-free surfaces (Figure 6i,k), PH-HMR-1ICD -expressing embryos had greatly reduced levels of PAR-6 at the cell cortex (Figure 6j); this is the result expected if PAC-1 recruited to contact-free surfaces by the HMR-1ICD represses CDC-42 at these sites, since CDC-42 is required for cortical PAR-6 recruitment 29. We quantified these results by measuring the ‘polarity index’ – defined as the ratio of PAR-6 levels at a contact-free surface divided by half the PAR-6 levels at a cell-cell contact (Figure 6l). Depleting PAC-1 in embryos expressing PH-HMR-1ICD resulted in high levels of PAR-6 at all cell surfaces (Figure 6m, 15/15 embryos), demonstrating that the loss of PAR-6 requires PAC-1, and indicating that PAC-1 functions downstream of the HMR-1 intracellular domain to mediate polarization. Together, these results demonstrate that HMR-1 performs an instructive role in contact-induced cell polarization by recruiting the symmetry-breaking polarity regulator PAC-1 to the adjacent cortex, and that PAC-1 recruitment depends on the ability of catenins to bind the HMR-1 intracellular domain.
DISCUSSION
Our findings identify a pathway that translates cell contact cues into polarity within the cell. We showed previously that the RhoGAP PAC-1 breaks cortical symmetry in C. elegans blastomeres when it is recruited to cell contacts, thereby inducing PAR protein asymmetry29. Here, we identify HMR-1/E-cadherin as a contact-directed cell surface cue that recruits PAC-1, translating polarity information from the cell surface to the adjacent cortex. We have identified physical interactions between JAC-1/p120 catenin, which binds the HMR-1 intracellular domain, and a conserved linker protein (PICC-1) that couples PAC-1 to the cadherin-catenin complex. Our experiments suggest a simple model for polarization (Figure 7), wherein homophilic interactions between HMR-1 proteins on adjacent cells stabilize HMR-1 at contacts, resulting in recruitment of catenins, PICC-1, and PAC-1. JAC-1 functions by bringing PICC-1 to the cadherin-catenin complex, while HMP-1/α-catenin contributes to PAC-1 localization through an unknown, independent mechanism. Once at the cortex, PAC-1 locally inactivates CDC-42 to exclude PAR proteins from contacts29, inducing a contact/contact-free PAR protein asymmetry that polarizes cells. HMR-1/E-cadherin provides an ideal cue for inducing contact-induced polarization, as it is able to self-assemble at contacts through homophilic interactions, and to translate polarity information to the cytoplasm via catenins and their effectors. Because full-length PAC-1 is still present at cell contacts when HMR-1 is depleted, our findings also point to the existence of a redundant E-cadherin-independent polarization cue. It is tempting to speculate that the redundant cue involves another self-assembling adhesion protein. Alternatively, given that the redundant pathway requires the PAC-1 PH domain, the cue could be a lipid that is preferentially enriched at cell contacts.
Figure 7. Model for HMR-1-instructed cell polarization.
See Discussion for details. Dashed lines indicate regulation that may not be direct. ‘X’ represents a redundant, HMR-1-independent polarization cue that requires the PAC-1 PH domain. The GAP domain of PAC-1 negatively regulates CDC-42 at cell contacts to inhibit its recruitment of PAR-6 29.
Because E-cadherin is crucial for epithelial cell adhesion in most species 4, 40, its molecular role in contact-induced polarization has been difficult to ascertain. In particular, it is debated whether E-cadherin directly recruits symmetry-breaking factors to cell contacts (functioning instructively), or if cadherin-mediated cell adhesion is a prerequisite for polarity triggered by a cadherin-independent cue (functioning permissively) 19, 20. Complicating this debate, some cell types that normally polarize in the context of neighboring cells, such as Xenopus blastomeres41–43 or mammalian intestinal epithelial cells44, can do so in the absence of cell-cell contact upon specific pharmacological or genetic perturbations. Because C. elegans early embryos do not require HMR-1 for cell adhesion, we have been able to separate its roles in adhesion and polarization. Our findings indicate that HMR-1 functions instructively in polarization by recruiting the symmetry-breaking polarity regulator PAC-1 to cell contacts. This conclusion is based on our observations that (1) HMR-1 has a partially redundant role in localizing PAC-1; (2) PAC-1, PICC-1, JAC-1/p120 catenin, and the HMR-1 intracellular domain exhibit protein-protein interactions; and (3) the HMR-1 intracellular domain, when targeted to contact-free surfaces, recruits PAC-1 to these sites and depolarizes cells. Redundancy with other polarization pathways (such as the one revealed in this study), rather than lack of an instructive function, could explain why E-cadherin is dispensable for the polarization of some epithelial cells19, 21, 23.
The highly conserved polarization pathway we describe here could perform a similar function in contact-induced polarity within mammals. In particular, given the requirement for E-cadherin in polarizing mammalian blastomeres16, it will be important to establish whether E-cadherin functions in this context by recruiting PICC-1 and PAC-1 homologues. Little is known about the in vivo functions of the three picc-1 homologues (ccdc85a, ccdc85b/DIPA, ccdc85c) and two pac-1 homologs (arhgap21 and arhgap23). However, experiments utilizing cultured cell lines have identified some of the same interactions we describe here. For example, ARHGAP21 associates with E-cadherin, binds directly to α-catenin, and localizes to adherens junctions45, 46. Moreover, CCDC85B/DIPA binds p12036, and both CCDC85B/DIPA and CCDC85C localize to junctions36, 47. Thus PAC-1 and PICC-1 homologs are likely to have conserved roles in signaling downstream of E-cadherin, and perhaps in the contact-mediated polarization of blastomeres and epithelial cells.
METHODS
Strains
hmr-1 embryos were of genotype hmr-1(zu389); xnEx42 [hmr-1(+), Pdpy-7::rfp, Pend-1::gfp] + hmr-1(RNAi), as described32. All unc-119 mutants were unc-119(ed3). The following strains were used: N2 (wild type), FT93: pac-1(xn6) FT99: pac-1(xn6) unc-119; xnIs27 [Ppie-1::gfp-pac-1, unc-119(+)], FT228: hmr-1(zu389); xnEx42, FT413: unc-119; xnIs171 [Ppie-1::gfp-pac-1392-838, unc-119(+)], FT422: unc-119 xnIs178 [Ppac-1::mCh-pac-1, unc-119(+)], FT555: pac-1(xn6) unc-119; xnIs234 [Ppie-1p::gfp-pac-1ΔPH, unc-119(+)], FT606: unc-119; picc-1(xn14), FT741: xnSi6 [Pmex-5::hmr-1-gfp::hmr-13′UTR, unc-119(+)]; unc-119, FT816: xnIs178; xnIs243 [Ppicc-1::picc-1-gfp, unc-119(+)], FT973: unc-119; jac-1(xn15), FT1000: unc-119; xnIs371 [Ppie-1::gfp-PAC-11-574, unc-119(+)], FT1001: unc-119; jac-1(xn15); xnIs371, FT1247: hmr-1; xnIs178, FT1380: unc-119; xnIs499 [Ppie-1::gfp-picc-1, unc-119(+)], FT1388: unc-119; xnIs501 [Ppie-1::gfp-pac-1ΔPH, unc-119 (+)], FT1394: unc-119; xnIs502 [Ppie-1::gfp-jac-1, unc-119 (+)], FT1395: jac-1(xn15); xnIs499, FT1405: hmr-1(zu389); xnIs501, FT1466: xnIs507 [Ppac-1::mCh-ha-pac-1, unc-119(+)]; unc-119, FT1560: xnSi6; jac-1(xn15), FT1526: jac1(xn15); xnIs243, FT1557: xnIs507; xnIs499, FT1568: xnIs371; xnEx384 [Phsp16::ha-phplc1∂1-hmr-1ICD, pRF4], FT1569: xnIs371; xnEx385 [Phsp16::ha-phplc1∂1-hmr-1ICD-M, pRF4], FT1570: xnEx386 [Ppie-1::gfp-pac-1575-1604, unc-119(+)], FT1572: hmr-1(zu389); xnIs371, FT1573: picc-1(xn14); xnIs371, FT1574: jac-1(xn15); picc-1(xn14); xnIs371, FT1578: xnSi6; xnIs178, FT1583: xnIs27; xnEx384, FT1584: xnIs27; xnEx385, FT1609: unc-119; xnIs528 (Phmp-1::hmp-1-gfp, unc-119(+)], FT1617: xnIs528 (Phmp-1::hmp-1-gfp, unc-119(+)]; jac-1(xn15), FT1624: unc-119; xnIs534 [Ppie-1::gfp-pac-12-610, unc-119(+)], FT1639: hmr-1(zu389); xnIs499 [Ppie-1:gfp:picc-1, unc-119(+)], KK866: itIs153 (Ppie-1::par-2-gfp; pRF4).
Identification of pac-1 transcripts
Putative transcriptional start sites were identified through analysis of annotated SL1 splice leader sequences (Wormbase, WS241). Two distinct cDNAs that differed in their 5′ end (full-length pac-1 and short pac-1; see Figure 1a) were amplified from embryonic mRNA that was reverse-transcribed (additional pac-1 isoforms are predicted on Wormbase but these were not identified). cDNAs were cloned into Gateway entry clone pDONR221 and sequenced. Full-length pac-1 corresponds to C04D8.1b and short pac-1 corresponds to C04D8.1d (Wormbase, WS241).
jac-1 and picc-1 deletions
jac-1 and picc-1 deletion alleles were created using MosDEL48. For jac-1, Mos insertion ttTi3732, which resides within the fifth jac-1 intron, was excised. Sequence from the ttTi3732 insertion site to the stop codon of jac-1 was replaced with the unc-119 gene from C. briggsae, creating jac-1(xn15). For picc-1, Mos insertion ttTi25649, which resides within the first picc-1 intron, was excised. Sequence from the ttTi25649 insertion site to the picc-1 3′ UTR was replaced with the unc-119 gene from C. briggsae, creating picc-1(xn14).
Transgenes
Ppac-1::mCherry-pac-1 was created by recombineering fosmid WRM063aE11, using SW102 cells and galK selection as described49. mCherry containing introns was inserted immediately after the start codon of full-length pac-1. 4061bp upstream of the start codon and 554bp downstream of the stop codon were recombined into plasmid pPUB50 by gap repair.
For Ppac-1::mCh-ha-pac-1, mCherry and tandem ha sequences were recombineered into the identical position of the pac-1 fosmid. The entire recombineered fosmid was transformed into SW106 cells, and unc-119(+) from plasmid pLoxP unc-119 was inserted into the vector LoxP site as described51.
Ppicc-1::picc-1-gfp was created by recombineering fosmid WRM0651cF11 as described above. gfp was inserted just 5′ to the picc-1 stop codon. 3331bp upstream of the start codon and 378bp downstream of the stop codon were recombined into plasmid pPUB.
For pac-1 structure-function constructs, entry clones containing the indicated sequences were produced by PCR-modification of a full-length pac-1 cDNA entry clone29. Entry clones were recombined into destination vector pID3.01B52 to create fusions with gfp.
Ppie-1::gfp-picc-1 was created by recombining a full-length picc-1 cDNA entry clone (corresponding to WormBase transcript F29G9.2a) into destination vector pID3.01B.
Phmp-1::hmp-1-gfp was obtained from the TransgenOme Project53.
Ppie-1::gfp-jac-1 was created by recombining a full-length jac-1 cDNA entry clone (corresponding to WormBase transcript Y105C5B.21a) into destination vector pID3.01B.
For Phsp16::ha-phplc1∂1-hmr-1ICD, hmr-1 cDNA encoding the intracellular domain (corresponding to amino acids 1104-1223 of the hmr-1a transcript) was amplified and cloned into a Gateway entry vector containing sequences encoding the PH domain from rat PLC1δ1 (PHPLC1δ1 was amplified from GFP- PHPLC1δ1)38. The phPLC1δ1-hmr-1ICD entry plasmid was subsequently cloned into a modified version of pCD6.09AP containing tandem ha sequences39 using Gateway recombination.
For Phsp16::ha-phplc1∂1-hmr-1ICD-M, the catenin-binding domain from the Phsp16::ha-phplc1∂1-hmr-1ICD construct (corresponding to HMR-1 amino acids 1124-1223)33 was removed using PCR.
Site-directed mutagenesis was performed on the resulting plasmid to mutate the juxtamembrane domain, as described34.
Primers sequences used in transgene construction can be found in Supplementary Table 2.
Worm transformation
Ppac-1::mCherry-pac-1, Ppac-1::mCh-ha-pac-1, Ppicc-1::picc-1-gfp,, Ppie-1::gfp-picc-1, Ppie-1::gfp-jac-1 transgenes and all transgenes for pac-1 structure-function were integrated into unc-119(ed3) worms using biolistic transformation54. Phsp16::ha-phplc1∂1-hmr-1ICD and Phsp16::ha-phplc1∂1-hmr-1ICD-M were microinjected to produce extrachromosomal arrays55.
RNAi
Feeding RNAi was performed as described56. hmr-1 RNAi was performed using a previously described hmr-1 feeding construct57. For pac-1 RNAi, the C04D8.1 clone from the Ahringer RNAi feeding library was used58. For hmp-1 RNAi, a probe targeting bases 2163 to 2856 (corresponding to WormBase transcript R13H4.4a) was cloned into vector pPD129.3659. For RNAi against the 5′ end of pac-1, a probe targeting bases 4 to 981 of full-length pac-1 was cloned into pPD129.36. For hmp-2 RNAi, dsRNA against bases 1221-2034 (corresponding to WormBase transcript K05C4.6a) was synthesized in vitro and injected at a concentration of 1μg/μl into young adult worms; F1 embryos were harvested 24 hours later and analyzed.
Quantifying expression levels of gfp-pac-1 strains
Total fluorescence values were obtained in a central focal plane of four-celled embryos expressing each GFP fusion. Using equivalent camera exposures, average background fluorescence (wild-type four-cell embryos) was subtracted from the average fluorescence value for each strain. Signals were integrated over the area of the embryo.
Quantifying contact enrichment of fluorescently tagged proteins
For live four-celled embryos expressing fluorescently tagged proteins, the contact enrichment was calculated as the ratio of the average intensity of a forty-pixel line along the ABa and ABp contact over the average intensity of a forty-pixel line within the ABa cytoplasm. Identical exposure settings were used for all genotypes. For HMR-1-GFP chimeras, contact enrichment was determined by measuring the average value at all chimeric contacts between AB lineage cells and taking a ratio with the average cytoplasmic intensity within the HMR-1-GFP-expressing cells. Only embryos with all chimeric contacts visible in one plane were measured. ImageJ was used for intensity measurements.
Antibody production
Rabbit affinity-purified custom polyclonal antibodies were produced at Covance using the following peptides (JAC-1: CESPHLGHHDVVKYVEAERF, HMR-1: CAPYDELRIYDDERDN). Specificity of the antiserum for immunofluorescence was determined by comparing wild-type embryos to jac-1(xn15) embryos or hmr-1 embryos. In addition to specific staining of cell contacts, α-JAC-1 antiserum also showed non-specific nuclear staining. Immunoblotting specificity of the α-JAC-1 antibody was determined by comparing wild-type and jac-1(xn15) embryonic lysates.
Yeast two-hybrid screen and direct interaction tests
Two-hybrid screening was performed using the ProQuest vector system and manufacturer’s protocol (Invitrogen). PAC-12-610 fused to the GAL4 DNA binding domain (created from the pac-12-610 entry clone and the pDEST32 destination vector using Gateway recombination) was used to screen a prey library consisting of commercial mixed-stage C. elegans cDNAs fused to the GAL4 activation domain (Invitrogen #11288-016, discontinued and a gift from J. Hubbard). 2.6 × 106 clones were screened on SC –Leu –Trp –His agar plates containing 35mM 3AT. Colonies growing under these conditions were retested on SC –Leu –Trp –Ura plates, and were screened for β-galactosidase activity using the manufacturer’s protocol. For prey clones positive for all three assays, DNA was isolated and sequenced, then retransformed into yeast containing bait plasmid and retested for growth on SC –Leu –Trp –His containing 35mM 3AT. 15 clones of F29G9.2, at least 5 of which were independent, were identified.
Direct interaction tests were performed using the ProQuest vector system and SC –Leu –Trp –His plates containing 50mM 3AT, following the manufacturer’s protocol. The following cDNAs were cloned into the GAL4 DNA binding domain destination vector, pDEST32: pac-11-574, hmp-1 (corresponding to WormBase transcript R13H4.4A), and full-length picc-1 (corresponding to WormBase transcript F29G9.2a). The following cDNAs were cloned into the GAL4 activation domain destination vector, pDEST22: pac-11-574, full-length jac-1 (corresponding to WormBase transcript Y105C5B.21a), hmp-2 (corresponding to WormBase transcript K05C4.6A), and hmr-11104-1223. Interaction experiments shown in Figure 5a were performed three times, and a representative example is shown. Interaction experiments shown in Supplementary Figure 7a were performed twice, except for the DBD-HMR-1ICD + AD-JAC-1 control experiment (performed once) and the DBD-PICC-1 + AD-empty vector control experiment (performed twice). Representative examples are shown.
Embryo lysis and immunoprecipitation
Approximately 50μl to 100μl frozen embryos were thawed on ice and three to four volumes of lysis buffer (50mM Tris pH 7.5, 100mM KCl, 150mM NaCl, 10% glycerol, 0.5% Triton-X-100, 1.5mM MgCl2, 100mM phenylmethanesulfonyl fluoride, 20mM β-glycerophosphate, 1 Roche Complete protease inhibitor tablet/10mL lysis buffer) was added. NaCl was omitted from the lysis buffer for immunoprecipitations of PICC-1-GFP and mCherry-HA-PAC-1. After homogenization with a plastic pestle, embryo lysates were sonicated (Branson analog microtip sonicator, 30% amplitude) three times for 15 seconds with one minute on ice between rounds. The soluble fraction was recovered following centrifugation at 14,000 rpm at 4°C for 20 min. Protein concentrations were determined using a detergent-compatible Bradford reagent (Bio-Rad Laboratories). Both the protein concentration and total protein content between control and experimental samples were equalized before beginning immunoprecipitation experiments.
For immunoprecipitation, whole-embryo lysates were pre-cleared by adding 30μL of protein A/G-agarose beads (Santa Cruz Biotechnologies) for 1h at 4°C. Pre-cleared supernatants were incubated with monoclonal mouse anti-GFP (Roche Applied Science, no. 11-814-460-001, clones 7.1 and 13.1, 4μg/mL), polyclonal rabbit anti-JAC-1 antibody (6.7μg/mL), or monoclonal mouse anti-HA antibody (Covance, no. MMS-101P, clone 16B12, 10μg/mL) overnight at 4°C. 20μL of protein A/G-agarose beads was added to the samples the following day for 3h at 4°C. Beads were washed 5x for 10 minutes in lysis buffer at 4°C. After removing the final wash, beads were boiled in 1x Laemmli buffer containing 100mM dithiothreitol for 10 min. at 90°C, and following centrifugation the supernatant was loaded onto 4–12% Bis-Tris SDS-PAGE gels (Invitrogen) for immunoblot analysis. For Western analysis, proteins were transferred to nitrocellulose membranes and probed using standard techniques. Primary antibodies (listed below) were used at the following concentrations (α-GFP, 1:1000; α-JAC-1, 1:10,000; α-HA, 1:1000). HRP-conjugated secondary antibodies (sheep anti-mouse IgG, Amersham, no. NA931, 1:10,000; mouse anti-rabbit IgG light chain, Jackson Immunoresearch Laboratories, no. 211-032-171 1:50,000) and the ECL Prime kit (Amersham) were used for detection.
Immunostaining
Embryos were freeze-fractured, fixed in methanol or methanol and paraformaldehyde, and stained as described previously29. The following primary antibodies were used: rabbit α-PAR-6 1:20,000 60, rabbit α-GFP 1:1,000 (AbCam, no. Ab6556.25), mouse α-HMP-1 1:10 21, rabbit α-HMR-1 1:10,000, mouse α-HA 1:1,000 (Covance, no. MMS-101P, clone 16B12), rabbit α-HA 1:1,600 (Cell Signaling Technologies, no. 3724), rabbit α-JAC-1 1:1,000, mouse α-SAX-7 1:5 (Developmental Studies Hybridoma Bank)61.
Blastomere recombination
Embryos were treated with alkaline hypochlorite, placed in Shelton’s growth medium, and the vitelline membrane was removed by trituration using a 30 μm micropipette (FHC) as described previously62. To create chimeric contacts, embryos within a depression slide were pushed together using a mouth pipet.
Ectopic PH-HMR-1ICD expression
Gravid transgenic worms (Rollers) were heat-shocked at 34°C for one hour. Worms were chopped in room temperature M9 buffer to liberate embryos, which were fixed, co-stained for HA and either PAR-6, GFP, HMP-1, or JAC-1. Polarity index was quantified in 8–12 cell embryos. Polarity index measurements were obtained by determining the ratio of the average intensity of a forty-pixel line along the contact-free surface of an AB lineage cell versus half of the average intensity of a forty-pixel line at that cell’s contact with a neighboring AB cell. ImageJ was used for intensity measurements. Heat-shocked control embryos were siblings that did not carry the Phsp16::ha-phplc1∂1-hmr-1ICD or Phsp16::ha-phplc1∂1-hmr-1ICD-M transgenic array (identified by absence of HA immunostaining). Embryos with weak or cytoplasmic HA staining, which indicates poor fixation, were excluded from the analysis.
Image Acquisition
Images were captured using a Zeiss AxioImager, 40x, 1.3NA objective, and Hammamatsu Orca-R2 camera. Images of fixed embryos were deconvolved using AxioVision software, and are shown as maximum intensity projections of 3–5 adjacent planes spaced 0.3 μm apart. For experiments where images of live embryos were quantified, images were captured using equal exposures. Exposures were measured on the day of the experiment by imaging at least six control embryos for a given experiment and taking the average exposure across all embryos. Images were cropped, rotated, and levels were adjusted in ImageJ and Adobe Photoshop.
Statistical analysis
Statistical tests used and sample sizes for each group are indicated in the figure legends. No statistical method was used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
Supplementary Material
Acknowledgments
We thank Alfred Fisher, Kathrin Gieseler, Greg Hermann, Oliver Hobert, Jane Hubbard, Tony Hyman, and Eric Jorgensen for their generous gifts of strains, plasmids or antibodies. Special thanks to Michael Burel for help in examining GFP-PAC-1ΔPH localization. Thanks to Lionel Christiaen, Thomas Hurd, and members of the Nance lab for comments on the manuscript. This study was funded by a National Science Foundation Graduate Research Fellowship under grant No. 12-A0-00-000165-01 (D.K.), an American Heart Association postdoctoral fellowship (Y.Z.), and NIH grants R01GM098492 (J.N.), R01GM078341 (J.N.), and T32HD007520 (D.C.A).
Footnotes
AUTHOR CONTRIBUTIONS
D.K., D.C.A., and J.N. designed, executed, and analyzed the experiments. J.Y.Y., Y.Z., and J.N. designed, executed and analyzed the co-immunoprecipitation experiments. D.K. and J.N. wrote the manuscript with input from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Roignot J, Peng X, Mostov K. Polarity in mammalian epithelial morphogenesis. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.St Johnston D, Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Johnson MH. From mouse egg to mouse embryo: polarities, axes, and tissues. Annu Rev Cell Dev Biol. 2009;25:483–512. doi: 10.1146/annurev.cellbio.042308.113348. [DOI] [PubMed] [Google Scholar]
- 4.Nelson WJ, Dickinson DJ, Weis WI. Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog Mol Biol Transl Sci. 2013;116:3–23. doi: 10.1016/B978-0-12-394311-8.00001-7. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Zhang M. The Par3/Par6/aPKC complex and epithelial cell polarity. Exp Cell Res. 2013;319:1357–1364. doi: 10.1016/j.yexcr.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Nance J, Zallen JA. Elaborating polarity: PAR proteins and the cytoskeleton. Development. 2011;138:799–809. doi: 10.1242/dev.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams CL, Nelson WJ, Smith SJ. Quantitative analysis of cadherin-catenin-actin reorganization during development of cell-cell adhesion. J Cell Biol. 1996;135:1899–1911. doi: 10.1083/jcb.135.6.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams CL, Chen YT, Smith SJ, Nelson WJ. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 10.Nejsum LN, Nelson WJ. A molecular mechanism directly linking E-cadherin adhesion to initiation of epithelial cell surface polarity. J Cell Biol. 2007;178:323–335. doi: 10.1083/jcb.200705094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson MH, Maro B, Takeichi M. The role of cell adhesion in the synchronization and orientation of polarization in 8-cell mouse blastomeres. J Embryol Exp Morphol. 1986;93:239–255. [PubMed] [Google Scholar]
- 12.Gumbiner B, Stevenson B, Grimaldi A. The role of the cell adhesion molecule uvomorulin in the formation and maintenance of the epithelial junctional complex. J Cell Biol. 1988;107:1575–1587. doi: 10.1083/jcb.107.4.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNeill H, Ozawa M, Kemler R, Nelson WJ. Novel function of the cell adhesion molecule uvomorulin as an inducer of cell surface polarity. Cell. 1990;62:309–316. doi: 10.1016/0092-8674(90)90368-o. [DOI] [PubMed] [Google Scholar]
- 14.Watabe M, Nagafuchi A, Tsukita S, Takeichi M. Induction of polarized cell-cell association and retardation of growth by activation of the E-cadherin-catenin adhesion system in a dispersed carcinoma line. J Cell Biol. 1994;127:247–256. doi: 10.1083/jcb.127.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capaldo CT, Macara IG. Depletion of E-cadherin disrupts establishment but not maintenance of cell junctions in Madin-Darby canine kidney epithelial cells. Mol Biol Cell. 2007;18:189–200. doi: 10.1091/mbc.E06-05-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephenson RO, Yamanaka Y, Rossant J. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development. 2010;137:3383–3391. doi: 10.1242/dev.050195. [DOI] [PubMed] [Google Scholar]
- 17.Nejsum LN, Nelson WJ. Epithelial cell surface polarity: the early steps. Front Biosci. 2009;14:1088–1098. doi: 10.2741/3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeaman C, Grindstaff KK, Nelson WJ. Mechanism of recruiting Sec6/8 (exocyst) complex to the apical junctional complex during polarization of epithelial cells. J Cell Sci. 2004;117:559–570. doi: 10.1242/jcs.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris TJ, Peifer M. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J Cell Biol. 2004;167:135–147. doi: 10.1083/jcb.200406024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Bivic A. E-cadherin-mediated adhesion is not the founding event of epithelial cell polarity in Drosophila. Trends Cell Biol. 2005;15:237–240. doi: 10.1016/j.tcb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Costa M, et al. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanentzapf G, Smith C, McGlade J, Tepass U. Apical, lateral, and basal polarization cues contribute to the development of the follicular epithelium during Drosophila oogenesis. J Cell Biol. 2000;151:891–904. doi: 10.1083/jcb.151.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theard D, Steiner M, Kalicharan D, Hoekstra D, van Ijzendoorn SC. Cell polarity development and protein trafficking in hepatocytes lacking E-cadherin/β-catenin-based adherens junctions. Mol Biol Cell. 2007;18:2313–2321. doi: 10.1091/mbc.E06-11-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nance J, Priess JR. Cell polarity and gastrulation in C. elegans. Development. 2002;129:387–397. doi: 10.1242/dev.129.2.387. [DOI] [PubMed] [Google Scholar]
- 25.Nance J, Munro EM, Priess JR. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 2003;130:5339–5350. doi: 10.1242/dev.00735. [DOI] [PubMed] [Google Scholar]
- 26.Vinot S, et al. Asymmetric distribution of PAR proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev Biol. 2005;282:307–319. doi: 10.1016/j.ydbio.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Thomas FC, et al. Contribution of JAM-1 to epithelial differentiation and tight-junction biogenesis in the mouse preimplantation embryo. J Cell Sci. 2004;117:5599–5608. doi: 10.1242/jcs.01424. [DOI] [PubMed] [Google Scholar]
- 28.Plusa B, et al. Downregulation of Par3 and aPKC function directs cells towards the ICM in the preimplantation mouse embryo. J Cell Sci. 2005;118:505–515. doi: 10.1242/jcs.01666. [DOI] [PubMed] [Google Scholar]
- 29.Anderson DC, Gill JS, Cinalli RM, Nance J. Polarization of the C elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science. 2008;320:1771–1774. doi: 10.1126/science.1156063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grana TM, Cox EA, Lynch AM, Hardin J. SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Dev Biol. 2010;344:731–744. doi: 10.1016/j.ydbio.2010.05.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirano S, Nose A, Hatta K, Kawakami A, Takeichi M. Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J Cell Biol. 1987;105:2501–2510. doi: 10.1083/jcb.105.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chihara D, Nance J. An E-cadherin-mediated hitchhiking mechanism for C elegans germ cell internalization during gastrulation. Development. 2012;139:2547–2556. doi: 10.1242/dev.079863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korswagen HC, Herman MA, Clevers HC. Distinct β-catenins mediate adhesion and signalling functions in C. elegans. Nature. 2000;406:527–532. doi: 10.1038/35020099. [DOI] [PubMed] [Google Scholar]
- 34.Pettitt J, Cox EA, Broadbent ID, Flett A, Hardin J. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J Cell Biol. 2003;162:15–22. doi: 10.1083/jcb.200212136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel JM, Reynolds AB. The tyrosine kinase substrate p120cas binds directly to E-cadherin but not to the adenomatous polyposis coli protein or alpha-catenin. Mol Cell Biol. 1995;15:4819–4824. doi: 10.1128/mcb.15.9.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markham NO, et al. Monoclonal antibodies to DIPA: a novel binding partner of p120-catenin isoform 1. Hybridoma. 2012;31:246–254. doi: 10.1089/hyb.2012.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markham NO, et al. DIPA-family coiled-coils bind conserved isoform-specific head domain of p120-catenin family: potential roles in hydrocephalus and heterotopia. Mol Biol Cell. 2014;25:2592–2603. doi: 10.1091/mbc.E13-08-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Audhya A, et al. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan E, Nance J. Mechanisms of CDC-42 activation during contact-induced cell polarization. J Cell Sci. 2013;126:1692–1702. doi: 10.1242/jcs.124594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stepniak E, Radice GL, Vasioukhin V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb Perspect Biol. 2009;1:a002949. doi: 10.1101/cshperspect.a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardellini P, Davanzo G, Citi S. Tight junctions in early amphibian development: detection of junctional cingulin from the 2-cell stage and its localization at the boundary of distinct membrane domains in dividing blastomeres in low calcium. Dev Dyn. 1996;207:104–113. doi: 10.1002/(SICI)1097-0177(199609)207:1<104::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 42.Muller HA, Hausen P. Epithelial cell polarity in early Xenopus development. Dev Dyn. 1995;202:405–420. doi: 10.1002/aja.1002020410. [DOI] [PubMed] [Google Scholar]
- 43.Fesenko I, et al. Tight junction biogenesis in the early Xenopus embryo. Mech Dev. 2000;96:51–65. doi: 10.1016/s0925-4773(00)00368-3. [DOI] [PubMed] [Google Scholar]
- 44.Baas AF, et al. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell. 2004;116:457–466. doi: 10.1016/s0092-8674(04)00114-x. [DOI] [PubMed] [Google Scholar]
- 45.Sousa S, et al. ARHGAP10 is necessary for α-catenin recruitment at adherens junctions and for Listeria invasion. Nat Cell Biol. 2005;7:954–960. doi: 10.1038/ncb1308. [DOI] [PubMed] [Google Scholar]
- 46.Van Itallie CM, et al. Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J Cell Sci. 2014;127:885–895. doi: 10.1242/jcs.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori N, et al. Ccdc85c encoding a protein at apical junctions of radial glia is disrupted in hemorrhagic hydrocephalus (hhy) mice. Am J Pathol. 2012;180:314–327. doi: 10.1016/j.ajpath.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Frokjaer-Jensen C, et al. Targeted gene deletions in C. elegans using transposon excision. Nat Methods. 2010;7:451–453. doi: 10.1038/nmeth.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarov M, et al. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Nash L, Fisher AL. A simplified, robust, and streamlined procedure for the production of C. elegans transgenes via recombineering. BMC Dev Biol. 2008;8:119. doi: 10.1186/1471-213X-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D’Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 53.Sarov M, et al. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell. 2012;150:855–866. doi: 10.1016/j.cell.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Totong R, Achilleos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134:1259–1268. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- 58.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 59.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 60.Schonegg S, Hyman AA. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development. 2006;133:3507–3516. doi: 10.1242/dev.02527. [DOI] [PubMed] [Google Scholar]
- 61.Hadwiger G, Dour S, Arur S, Fox P, Nonet ML. A monoclonal antibody toolkit for C. elegans. PLoS One. 2010;5:e10161. doi: 10.1371/journal.pone.0010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shelton CA, Bowerman B. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development. 1996;122:2043–2050. doi: 10.1242/dev.122.7.2043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.