Abstract
Chlamydia trachomatis actively subverts the minus-end directed microtubule motor, dynein, to traffic along microtubule tracks to the Microtubule Organizing Center (MTOC) where it remains within a membrane bound replicative vacuole for the duration of its intracellular development. Unlike most substrates of the dynein motor, disruption of the dynactin cargo-linking complex by over-expression of the p50 dynamitin subunit does not inhibit C. trachomatis transport. A requirement for chlamydial protein synthesis to initiate this process suggests that a chlamydial product supersedes a requirement for p50 dynamitin. A yeast 2-hybrid system was used to screen the chlamydia inclusion membrane protein CT850 against a HeLa cell cDNA library and identified an interaction with the dynein light chain DYNLT1 (Tctex1). This interaction was at least partially dependent upon an (R/K-R/K-X-X-R/K) motif that is characteristic of DYNLT1 binding domains. CT850 expressed ectopically in HeLa cells localized at the MTOC and this localization is similarly dependent upon the predicted DYNLT1 binding domain. Furthermore, DYNLT1 is enriched at focal concentrations of CT850 on the chlamydial inclusion membrane that are known to interact with dynein and microtubules. Depletion of DYNLT1 disrupts the characteristic association of the inclusion membrane with centrosomes. Collectively, the results suggest that CT850 interacts with DYNLT1 to promote appropriate positioning of the inclusion at the MTOC.
Keywords: Chlamydia, microtubules, dynein, vesicle trafficking
Graphical Abstract
1. INTRODUCTION
Chlamydial trachomatis is the causative agent of several significant diseases of humans. Distinct serological variants, or serovars, are responsible for the different diseases such as trachoma, the leading cause of infectious blindness worldwide [1]. Other serovars are the causative agents of a variety of sexually transmitted diseases including urethritis, cervicitis, pelvic inflammatory disease, and a more systemic, granulomatous disease, lymphogranuloma venereum [2].
Chlamydiae are bacterial, obligate intracellular pathogens that undergo a biphasic life cycle consisting of an environmentally resistant extracellular form called the elemementary body (EB) and an intracellular replicative form known as the reticulate body (RB) [3]. EBs actively trigger endocytosis by eukaryotic host cells where they remain within a membrane bound vesicle termed an inclusion for the duration of their intracellular developmental cycle [4]. Soon after internalization, EBs initiate protein synthesis and modify the inclusion membrane by the insertion of a number of type III secreted effector proteins collectively known as Incs [5,6]. Once modified by chlamydial proteins, a number of interactions with the host cell are observed [7]. Among these are trafficking of C. trachomatis in a dynein dependent manner to the microtubule organizing center (MTOC) where the inclusion remains in a perinuclear location as it grows to accommodate the increasing number of bacteria [8,9,10]. Trafficking of C. trachomatis to the MTOC is dependent upon chlamydial protein synthesis and an intact microtubular network. Microinjection of antibodies to the minus-end directed motor protein complex, dynein, inhibits chlamydial trafficking to the MTOC [9].
Microtubule motors play important roles in a number of essential functions in eukaryotic cells including organelle structure and positioning, chromosome segregation during mitosis, and vesicular transport [11]. The microtubule network is organized with a minus end focused at the MTOC and plus end at the cell periphery and serves as a scaffold for the transport of the various cellular cargoes by ATP-dependent microtubule motors. The microtubule motors consist of the dynein and kinesin superfamily proteins which are the major minus-end and plus-end directed motors, respectively. The dynein motor is comprised of two heavy chains and multiple intermediate, light intermediate, and light chains [12,13]. Also required for most, if not all, dynein functions is the cargo-linking and activating complex dynactin. Dynactin is a large, multisubunit complex that consists of at least seven distinct proteins [14]. Overexpression of one of these components, p50 dynamitin, is inhibitory to dynein dependent trafficking of most physiological cargo [15,16]. Surprisingly, overexpression of p50 dynamitin does not inhibit trafficking of C. trachomatis inclusions [9]. Because the dynactin complex is required by all known cargo trafficked to the MTOC, we hypothesized that an unknown chlamydial protein(s) may supersede a requirement for an intact dynactin complex.
Recently, we have identified a microdomain on the C. trachomatis inclusion membrane that is enriched in cholesterol, active Src-family kinases, and at least four Incs (IncB, CT101, CT222, and CT850) [17]. These microdomains are focal points for microtubules and are tightly associated with centrosomes, which organize microtubules at the MTOC. We have speculated that these inclusion microdomains serve as a platform for stable interactions with dynein and consequently centrosomes and the MTOC. One of the microdomain components, CT850, when ectopically expressed in HeLa cells, forms aggregates that associate with centrosomes [17]. Here we show that CT850 encodes a predicted binding domain for a dynein light chain isoform, DYNLT1, and that this domain promotes association of CT850 with dynein.
2. Materials and Methods
2.1 Organism and Cell Culture
Chlamydia trachomatis serovar L2 (LGV 434) was propagated in HeLa 229 cells and purified by Renografin density gradient centrifugation as previously described [18].
2.2 Yeast 2-Hybrid Analysis
Yeast strain Y190 was purchased from Clontech and AH109 was provided by M.A. Scidmore. Yeast were transformed using the Yeast Maker Yeast Transformation System 2 (Clontech) and transformants selected on synthetically defined (SD) plates lacking appropriate nutrients (Clontech). Yeast matings were performed according to the Matchmaker Gold Yeast Two Hybrid System (Clontech) as previously described [19]. Plasmids were recovered from yeast using PrepEase Yeast Plasmid Isolation Kit (Affymetrix) and transformed into E. coli strain XL1-blue (Stratagene).
GAL4 DNA-binding domain (DNA-BD) fusions were constructed in pGBKT7 (Clontech). The carboxy terminus of CT850 was generated by PCR-amplification of CT850 (861bp) from L2 genomic DNA using a forward primer with a 5′ NcoI site (5′-AAACCATGGAGGAAGGACATATTAAG-3′) and reverse primer with a BamHI site (5′-AAAGGATCCTTACCGATTCTGGTT-3′) and ligated into the NcoI and BamHI sites of pGBKT7. GBK-CT850Δtbd was generated in two fragments. The 5′ fragment was generated by PCR amplification using the forward primer from above with the reverse primer containing a XhoI site (5′-AAACTCGAGAAAGGAATCCTTAAGCGTAAA-3) and the 3′ fragment was generated using the reverse primer from above and the forward primer containing a XhoI site (5′-AAACTCGAGCGCTTCTTAGCTCAAGAGCAA-3′). The two fragments were then digested with XhoI and ligated. The ligated fragment was then digested with NcoI and BamHI and ligated into the NcoI and BamHI sites of pGBKT7. GAL4 activation domain (AD) fusions were constructed in pGADT7 (Clontech). Full length dynein light chain DYNLT1 was PCR amplified from HeLa cDNA using the forward primer containing an EcoRI site (5′-AAAGAATTCATGGAAGACTACCAGGCT-3′) and the reverse primer containing a BamHI (5′-AAAGGATCCGTCAAATAGACATGTCCGAA-3′) site and ligated into the EcoRI and BamHI sites of pGADT7. Full length dynein light chain DYNLT1 and CT850Δtbd were PCR amplified as above except containing an Xho1 site for construction of mCherry fusions.
2.3 Immunofluorescent microscopy
Tubulin staining was performed using mouse anti-γ tubulin, clone GTU-88 (Sigma). The anti-DYNLT1 monoclonal antibody was from Millipore. The chlamydia antibodies used were polyclonal rabbit anti-C. trachomatis L2 EB. Rabbit anti-peptide antibodies against Inc850 (TVKDSFLKKARRERFLA) were prepared commercially by Quality Controlled Biochemicals, Inc (Hopkinton, MA). C. trachomatis-infected cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS and stained for immunofluorescence microscopy as previously described [17].
Images were acquired on a Nikon Eclipse 80i microscope using a 60× 1.4 NA oil immersion objective (Nikon, USA) or a Zeiss LSM 510 Meta laser confocal scanning microscope using a 63×, 1.4 NA oil objective (Carl Zeiss MicroImaging, Inc., Maple Grove, MN)
To determine distance from the chlamydial inclusion to centrosomes, C. trachomatis-infected HeLa cells at 18 hr post-infection were fixed and stained with anti-chlamydia sera and anti-γ tubulin for centrosomes. Images were acquired and merged and the closest distance from the inclusion membrane to the nearest centrosome was determined using an NIS-Elements AR software package (Nikon).
2. 4 siRNA knockdown
HeLa cells were plated to approximately 50% confluency on coverslips in 24-well plates (Corning, Lowell, MA). Cells were transfected with ON-TARGETplus siRNA corresponding to DYNLT1 or non-targeting sequence #1 (Thermo Scientific/Dharmacon, Lafayette, CO) according to the manufacturer’s instructions. At 48 hours after transfection, cells were infected with C. trachomatis serovar L2 and 24 hours post-infection cells were fixed with methanol for immunostaining.
2.5 Co-Immunoprecipitation and Immunoblotting
HeLa cells were transfected with CT850-mCherry [20] and DYNLT1-GFP. At 18 hr post transfection, cells were washed 3 times in cold PBS, scraped into 1.5 mls of 0.5% Zwittergent 3–14 in PBS, agitated for 20 minutes on ice, and incubated with protein-A beads followed by centrifugation to pre-clear the lysate. Protein-A agarose beads were incubated with rabbit antibodies against R. rickettsii or CT850 for 4 hours at room temperature. The beads were rinsed 3 times with PBS and mixed with the pre-cleared cell lysate. After gentle agitation overnight at 4°C, the beads were washed 3 times with 1% Zwittergent 3–14 in PBS and proteins were eluted into 1x SDS-PAGE sample buffer. The eluted proteins were separated by SDS-PAGE on a 12% acrylamide gel for immunoblotting as previously described [17].
3. RESULTS
3.1. Identification of DYNLT1 as a binding partner of CT850
CT850 ectopically expressed in HeLa cell associates with centrosomes independently of other chlamydial components of the microdomains [17]. A yeast two-hybrid analysis was therefore employed to identify eukaryotic host cell interacting partners for CT850 (Fig. 1). The C-terminus (amino acids 121–405) of CT850 was cloned into pGBKT7 to create pGBKT7-CT805 and transformed into yeast strain AH109. Expression of the CT850 construct was confirmed by immunoblot analysis using an anti-myc antibody. AH109 containing pGBKT7-CT850 was used as bait to screen yeast strain Y187 containing a random normalized HeLa cDNA library. Two independent clones identified in a high stringency screen expressed fragments of the dynein light chain, DYNLT1. To verify the interaction between CT850 and DYNLT1, DYNLT1 was cloned into pGADT7 and used in a targeted screen against pGBKT7-CT850. Full-length DYNLT1 also interacted with CT850 in the yeast 2-hybrid system and thus validated this interaction.
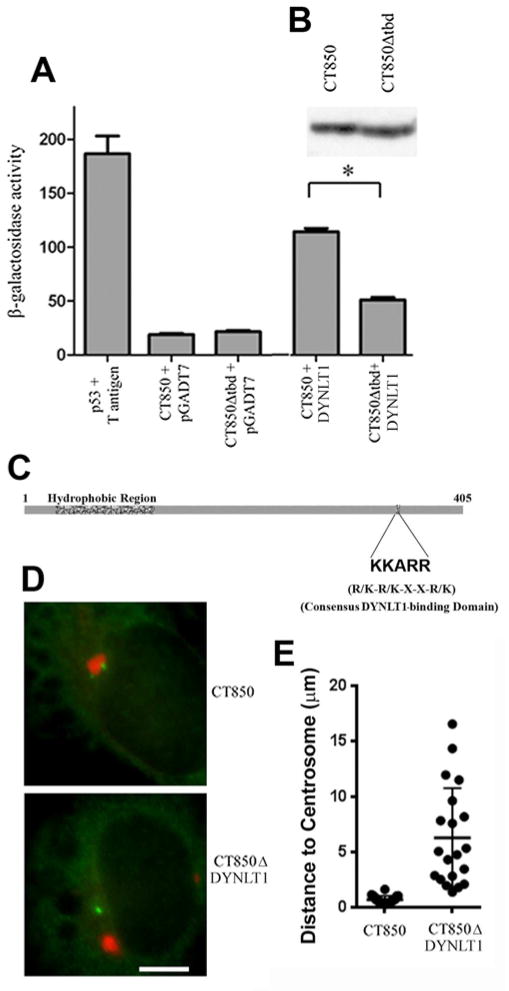
Figure 1.
Interaction of CT850 with DYNLT1. A. Yeast 2-hybrid identification of DYNLT1 as a binding partner of CT850. Quantitative β-galactosidase assays were performed as described in the text. Yeast strains expressing p53 and T-antigen serve as a positive control. Yeast strains expressing CT850 or a derivative lacking a putative DYNLT1 consensus binding domain (CT850Δtbd) mated to empty vector plasmid pGADT7 represent negative controls. Yeast strains expressing CT850 or CT850Δtbd mated to yeast strain Y190 containing pGADT7/ DYNLT1 were used to show the role of the consensus DYNLT1 binding domain. * indicates statistical significance at a level of p < 0.05 using a one way analysis of variance and a Newman-Keuls posttest.. B. Immunoblot of CT850 and CT850Δtbd expression in yeast to confirm equivalent levels of expression for use in the quantitative β-galactosidase assays. C. Schematic of C. trachomatis CT850 to illustrate the location of the characteristic inclusion membrane protein hydrophobic domain and the consensus DYNLT1 binding domain. D. Ectopic expression of CT850- and CT850Δtbd-mCherry fusions (red) with co-staining for centrosomes (green) to demonstrate the requirement for the DYNLT1 binding domain to promote recruitment to the MTOC and association with centrosomes. E. Quantitative assessment of the association of ectopically expressed CT850- and CT850Δtbd-mCherry fusions with centrosomes. Data from a single representative experiment (out of 3) are presented as a scatter plot with the mean and SEM indicated.
3.2. CT850 contains a predicted DYNLT1 binding motif
CT850 contains an amino acid sequence motif KKARR that is consistent with a consensus DYNLT1 binding domain (R/K-R/K-X-X-R/K) in several different proteins (Fig. 1C) [21,22,23]. To explore the role of the predicted DYNLT1 binding domain in CT850, we constructed site-directed mutants in which this five amino acid sequence was deleted to create pGBKT7CT850Δtbd. Expression of the new construct was verified in S. cerevisiae AH109 (Figure 1A,B). Targeted interaction between DYNLT1 and CT850 and CT850Δtbd was performed in parallel using the yeast two-hybrid system. The interaction between CT850Δtbd and DYNLT1 was significantly decreased (p < 0.05) compared to the original CT850 and DYNLT1 (Fig 1A).
The requirement for the putative DYNLT1 binding domain of CT850 was confirmed in HeLa cells. CT850-mCherry expressed in HeLa cells forms aggregates [20] that show a high rate of association with centrosomes (>90%) as compared to less than 5% observed with any of the three other C. trachomatis Incs found in inclusion membrane microdomains [17]. We therefore expressed CT850-mCherry and CT850Δtbd-mCherry in HeLa cells (Fig. 1D) and evaluated association of the respective aggregates with centrosomes in the transfected cells by measuring distance to the nearest centrosome (Fig. 1E). CT850-mCherry associated tightly with centrosomes which were typically observed less than 1 μm from the aggregates whereas CT850Δtbd-mCherry was randomly distributed. Collectively, the data suggest that the DYNLT1 binding domain is important in the interaction of CT850 with DYNLT1.
3.3. CT850 and DYNLT1 interact in vivo
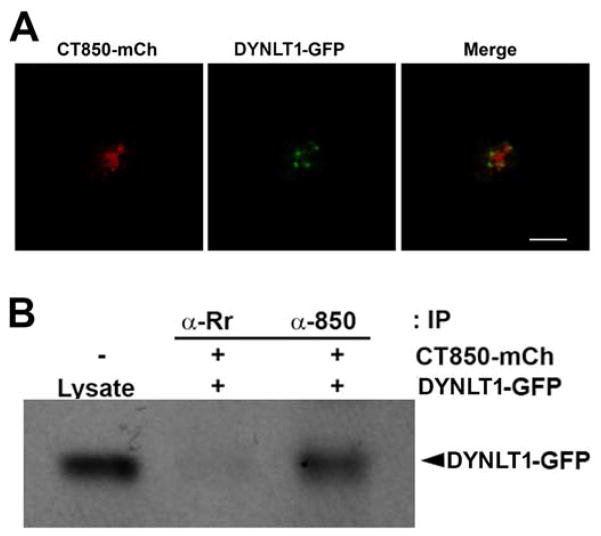
Co-transfection of CT850-mCherry and DYNLT1-GFP demonstrates recruitment of DYNLT1 to CT850 in the absence of other chlamydial microdomain constituents (Fig. 2A). To confirm an interaction of CT850 and DYNLT1 in vivo, we co-expressed CT850-mCherry and DYNLT1-GFP in HeLa cells and performed co-immunoprecipitation experiments (Fig. 2B). Lysates were immunoprecipitated with a rabbit polyclonal anti-CT850 or a control antibody. The immunoprecipitates were separated by SDS-PAGE and Western blots were then probed for DYNLT1. DYNLT1 was co-precipitated with CT850 but not by an irrelevant antiserum. CT850 thus interacts with DYNLT1 in the cytosol of eukaryotic cells.
Figure 2.
Association of CT850 with DYNLT1 in vivo. A. DYNLT1-GFP associates with CT850-mCherry co-expressed ectopically in HeLa cells. B. Immunoblot of DYNLT1 co-immunoprecipitated from lysates of HeLa cells co-expressing DYNLT1-GFP and CT850-mCherry. A rabbit polyclonal antisera to R. rickettsii (α-Rr) served as an irrelevant negative control antibody.
3.4. DYNLT1 is enriched at C. trachomatis inclusion microdomains
To examine the association of DYNLT1 with the chlamydial inclusion, exogenous GFP-tagged and endogenous DYNLT1 were localized in C. trachomatis-infected cells. DYNLT1-GFP was ectopically expressed in HeLa cells and its association with mature inclusions examined at 18 hr post-infection (Fig. 3). The C. trachomatis inclusion membrane microdomains, which are a focal point for microtubules, dynein, and centrosomes [17], also show recruitment of DYNLT1-GFP.
Figure 3.
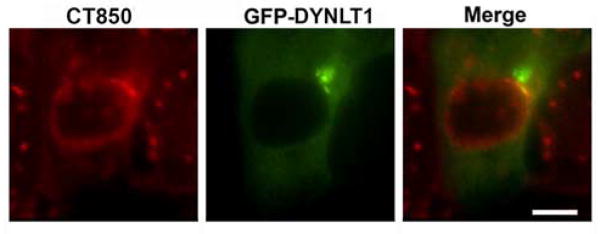
DYNLT1 is associated with C. trachomatis inclusion membrane microdomains. Ectopically expressed DYNLT1-GFP (green) is co-localized at inclusion microdomains identified by immunostaining with antibodies to one of the microdomain components, CT850 (red). Bar = 10 um.
3.5. DYNLT1 knockdown disrupts inclusion positioning at the MTOC
To assess the requirement for DYNLT1, we examined the characteristic dynein dependent association of inclusions with centrosomes at the MTOC in HeLa cells transfected with a non-targeting, negative control siRNA or with siRNAs against DYNLT1. Depletion of DYNLT1 was confirmed by immunoblotting (Fig. 4A). Because the interaction between the inclusion membrane and the dynein motor is retained throughout the chlamydial developmental cycle, the inclusion remains tightly associated with centrosomes [24]. When this interaction is disrupted, the average nearest distance from the inclusion membrane to centrosomes is increased (Fig. 4B,C). Thus the requirement for DYNLT1 in the positioning of the C. trachomatis inclusion at the MTOC was verified.
Figure 4.
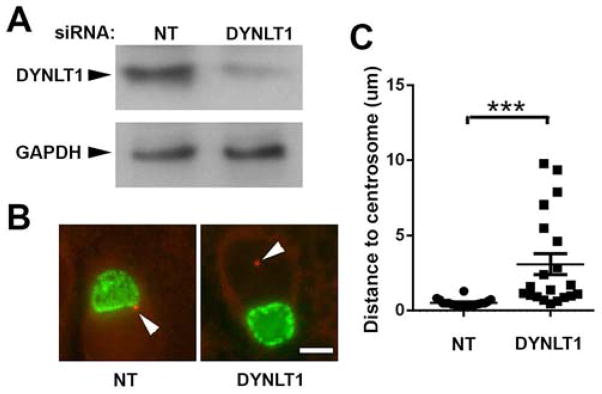
DYNLT1 knockdown interrupts inclusion trafficking and CT850 association with centrosomes. A. Immunoblot showing effects of siRNA knockdown of DYNLT1. A non-targeting siRNA (NT) was included as a negative control. Immunoblots were probed for DYNLT1 or GAPDH as a loading control. B. Localization of inclusions at the MTOC in non-targeting control cultures and in cells depleted of DYNLT1. HeLa cells were treated with siRNA for 48 and then infected with C. trachomatis. Eighteen hours later, the cells were fixed and stained for EBs and γ-tubulin to identify centrosomes. In DYNLT1 depleted cells, the distances from the inclusion membrane to the nearest centrosome were typically greater than in control cells treated with non-targeting siRNA (NT). Bar = 10 μm. C. The average distance to centrosomes was measured in cells transfected with non-targeting (NT) or DYNLT1 siRNA. Data from a typical experiment are shown and the Mean +/− S.E.M.; n = 20. are indicated. *** indicates statistical significance at a level of p < 0.0001 using an unpaired t test with Welch’s correction.
It has previously been shown that disruption of C. trachomatis trafficking to the MTOC had no negative effect on replication (9). Depletion of DYNLT1 similarly had no demonstrable effect on replication with 1.6 x 107 +/− 4.7 x 106 progeny IFU/ml from cells harvested at 44 hr post-infection in cells transfected with non-targeting siRNA vs. 1.7 x 107 +/− 6.7 x 106 progeny IFU/ml in DYNLT1 depleted cells.
4. DISCUSSION
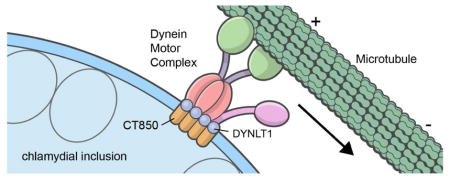
C. trachomatis is transported in a dynein dependent fashion to the MTOC following modification of the inclusion membrane by the insertion of chlamydial proteins [8,9,10]. The absence of a requirement for an intact dynactin cargo-linking complex, coupled with a requirement for chlamydial protein synthesis, led us to hypothesize that an unknown chlamydial protein may subsume the role of dynactin [9]. Recently, microdomains have been identified on the C. trachomatis inclusion membrane that are enriched in activated Src family kinases, cholesterol, and the Incs, IncB, CT101, CT222, and CT850. Of these C. trachomatis Incs, only CT850 associates with centrosomes when expressed in HeLa cells [17]. Here, we demonstrate that CT850 interacts with the dynein light chain DYNLT1. This interaction appears to be promoted by a predicted DYNLT1 binding domain on CT850. CT850 interacts with DYNLT1 in the cytsosol and depletion of DYNLT1 decreases association of the chlamydial inclusion with the MTOC. The interaction between the chlamydial inclusion membrane protein CT850 and the dynein light chain DYNLT1 therefore suggests a mechanism by which the chlamydial inclusion may subvert the dynein motor to traffic along microtubules in the absence of an intact dynactin complex
The circumvention of a requirement for an intact dynactin complex to support minus-end trafficking appears to be a mechanism unique to C. trachomatis. Many viruses exploit dynein and the microtubular network to effect a centripetal migration to a perinuclear location. There are numerous examples of viral proteins that directly interact with dynein polypeptides including DYNLT1 (Tctex1), DYNLL (LC8), DYNLRB (roadblock), DYNC1LI (dynein light intermediate chain), and DYNC1I (dynein intermediate chain) (reviewed in: [25,26,27,28]. In all cases where it has been examined, including adenovirus [29,30,31], herpes simplex virus [32], human immunodeficiency virus [33], African swine fever virus, [34], and vaccinia [35], overexpression of p50 dynamitin is inhibitory to trafficking. Because of the multiple means of interacting with dynein, it seems unlikely that the role of dynactin is exclusively for cargo linkage. Dynactin also plays a role in activation and processivity of the dynein motor and such a role has been proposed for adenovirus dynein mediated transport [29].
The interaction of CT850 with DYNLT1 suggests a mechanism by which chlamydiae might avoid the necessity for the cargo-linking function of dynactin but there are several examples of viral pathogens that also directly interact with dynein components and yet require dynactin. In addition to a role in linking cargo to dynein, dynactin also plays a role in processivity of the motor [36]. Although p50-dynamitin appears not to be essential for chlamydial trafficking, at least one other component of the dynactin complex, p150-Glued, has been localized to the nascent inclusion [9]. The mechanism appears reminiscent of adenovirus transport to the nucleus [29]. The adenovirus capsid protein hexon binds directly to dynein intermediate chain and light intermediate chain subunits thus dynactin does not appear essential for recruitment of capsids to the dynein motor complex. Over-expression of p50 dynamitin is, however, inhibitory to adenovirus trafficking to the nucleus suggesting that the role of dynactin is not cargo linking but as a regulator of dynein activity and processivity [29]. The mechanism employed by C. trachomatis similarly involves a direct linkage of a chlamydial protein to a dynein subunit but differs in that overexpression of p50 dynamitin is not inhibitory to trafficking.
The biological function of C. trachomatis association with dynein and localization to the MTOC is unknown and not apparent in cell culture. Inhibition of trafficking and perinuclear localization by depletion of DYNLT1 or incubation in the presence of nocodazole inhibits transport to the MTOC but has no discernible phenotype or effect on replication [9,37,38]. Other chlamydial species, such as the mouse and Guinea pig pathogens C. muridarum and C. caviae, do not exhibit the characteristic transport to the MTOC as seen with C. trachomatis [37]. Other host mechanisms in addition to dynein may be required for C. trachomatis trafficking and positioning at the MTOC. Src-family tyrosine kinases are recruited to the C. trachomatis inclusion membrane and required for C. trachomatis trafficking along microtubules but are absent from the C. muridarum and C. caviae inclusion membranes. Thus while CT850 appears to be necessary for C. trachomatis association with the MTOC, it is not sufficient as other factors, such as Src kinases, are also required. All C. trachomatis serovars yet examined, including ocular, urogenital and lymphogranuloma venereum strains, display centripetal trafficking to a perinuclear region [9,37]. It appears that CT850 interactions with DYNLT1 play an important role in the trafficking of the inclusion to the MTOC. Whether positioning of the inclusion at the MTOC provides a benefit in specific sites or cell types in vivo remains to be be determined.
Supplementary Material
Chlamydia trachomatis inclusions localize to the Microtubule Organizing Center
The microtubule motor complex, dynein, is required for trafficking to the MTOC
The MTOC is associated with specific microdomains on the inclusion membrane
The microdomain protein CT850 interacts with the dynein light chain 1 subunit
Acknowledgments
This work was supported by the Intramural Research Program of the NIAID/NIH. We thank Janet Sager for technical assistance and Drs. D. Bublitz and A. Omsland for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burton MJ, Mabey DC. The global burden of trachoma: a review. PLoS Negl Trop Dis. 2009;3:e460. doi: 10.1371/journal.pntd.0000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schachter J. Infection and disease epidemiology. In: Stephens RS, editor. Chlamydia; Intracellular biology, pathogenesis, and immunity. ASM Press; Washington, D.C: 1999. pp. 139–169. [Google Scholar]
- 3.Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hackstadt T. Cell Biology. In: Stephens RS, editor. Chlamydia: Intracellular biology, pathogenesis, and immunity. ASM Press; Washington, D. C: 1999. pp. 101–138. [Google Scholar]
- 5.Bannantine JP, Griffiths RS, Viratyosin W, et al. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2000;2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- 6.Shaw EI, Dooley CA, Fischer ER, et al. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol. 2000;37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 7.Fields KA, Hackstadt T. The Chlamydial Inclusion: Escape from the Endocytic Pathway. Annu Rev Cell Dev Biol. 2002;14:14. doi: 10.1146/annurev.cellbio.18.012502.105845. [DOI] [PubMed] [Google Scholar]
- 8.Clausen JD, Christiansen G, Holst HU, et al. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol Microbiol. 1997;25:441–449. doi: 10.1046/j.1365-2958.1997.4591832.x. [DOI] [PubMed] [Google Scholar]
- 9.Grieshaber S, Grieshaber N, Hackstadt T. Chlamydia trachomatis uses host cell dynein to traffic to the microtube organizing center in a p50 dynamitin independent process. J Cell Biol. 2003;116:3793–3802. doi: 10.1242/jcs.00695. [DOI] [PubMed] [Google Scholar]
- 10.Higashi N. Electron microscopic studies on the mode of reproduction of trachoma virus and psittacosis virus in cell cultures. Exp Mol Pathol. 1965;76:24–39. doi: 10.1016/0014-4800(65)90021-3. [DOI] [PubMed] [Google Scholar]
- 11.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 12.Allan VJ. Cytoplasmic dynein. Biochem Soc Trans. 2011;39:1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 13.Hirokawa N, Noda Y, Okada Y. Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr Opin Cell Biol. 1998;10:60–73. doi: 10.1016/s0955-0674(98)80087-2. [DOI] [PubMed] [Google Scholar]
- 14.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 15.Burkhardt JK, Echeverri CJ, Nilsson T, et al. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valetti C, Wetzel DM, Schrader M, et al. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mital J, Miller NJ, Fischer ER, et al. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol. 2010;12:1235–1249. doi: 10.1111/j.1462-5822.2010.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutter EI, Barger AC, Nair V, et al. Chlamydia trachomatis inclusion membrane protein CT228 recruits elements of the myosin phosphatase pathway to regulate release mechanisms. Cell Rep. 2013;3:1921–1931. doi: 10.1016/j.celrep.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mital J, Miller NJ, Dorward DW, et al. Role for chlamydial inclusion membrane proteins in inclusion membrane structure and biogenesis. PLoS ONE. 2013;8:e63426. doi: 10.1371/journal.pone.0063426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mok YK, Lo KW, Zhang M. Structure of Tctex-1 and its interaction with cytoplasmic dynein intermediate chain. J Biol Chem. 2001;276:14067–14074. doi: 10.1074/jbc.M011358200. [DOI] [PubMed] [Google Scholar]
- 22.Sachdev P, Menon S, Kastner DB, et al. G protein beta gamma subunit interaction with the dynein light-chain component Tctex-1 regulates neurite outgrowth. EMBO J. 2007;26:2621–2632. doi: 10.1038/sj.emboj.7601716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugai M, Saito M, Sukegawa I, et al. PTH/PTH-related protein receptor interacts directly with Tctex-1 through its COOH terminus. Biochem Biophys Res Commun. 2003;311:24–31. doi: 10.1016/j.bbrc.2003.09.157. [DOI] [PubMed] [Google Scholar]
- 24.Grieshaber SS, Grieshaber NA, Miller N, et al. Chlamydia trachomatis causes centrosomal defects resulting in chromosomal segregation abnormalities. Traffic. 2006;7:940–949. doi: 10.1111/j.1600-0854.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- 25.Radtke K, Dohner K, Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker’s guide to the cell. Cell Microbiol. 2006;8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 26.Smith GA, Enquist LW. Break ins and break outs: viral interactions with the cytoskeleton of Mammalian cells. Annu Rev Cell Dev Biol. 2002;18:135–161. doi: 10.1146/annurev.cellbio.18.012502.105920. [DOI] [PubMed] [Google Scholar]
- 27.Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–3539. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merino-Gracia J, Garcia-Mayoral MF, Rodriguez-Crespo I. The association of viral proteins with host cell dynein components during virus infection. FEBS J. 2011;278:2997–3011. doi: 10.1111/j.1742-4658.2011.08252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bremner KH, Scherer J, Yi J, et al. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe. 2009;6:523–535. doi: 10.1016/j.chom.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suomalainen M, Nakano MY, Keller S, et al. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leopold PL, Kreitzer G, Miyazawa N, et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 32.Dohner K, Wolfstein A, Prank U, et al. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell. 2002;13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald D, Vodicka MA, Lucero G, et al. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alonso C, Miskin J, Hernaez B, et al. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J Virol. 2001;75:9819–9827. doi: 10.1128/JVI.75.20.9819-9827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ploubidou A, Moreau V, Ashman K, et al. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 2000;19:3932–3944. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 37.Mital J, Hackstadt T. Diverse requirements for SRC-family tyrosine kinases distinguish chlamydial species. MBio. 2011;2:e00031–11. doi: 10.1128/mBio.00031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schramm N, Wyrick PB. Cytoskeletal requirements in Chlamydia trachomatis infection of host cells. Infect Immun. 1995;63:324–332. doi: 10.1128/iai.63.1.324-332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.