Abstract
Background:
Helicobacter pylori, causing the most common chronic bacterial infection, exist in two forms; bacilli and coccoid. The coccoid form is identified as viable but non-culturable bacteria.
Objectives:
The current study aimed to conduct culture, polymerase chain reaction (PCR), and loop-mediated isothermal amplification (LAMP) tests to identify coccoid forms of H. pylori.
Materials and Methods:
The PCR and LAMP tests were optimized using specific primers for glmM gene. The sensitivity and specificity of the tests were determined. The current experimental study was conducted on 10 different strains isolated from clinical cases (H1-H10). The isolates were added to tap water and incubated at three different temperatures for one and two months intervals. After pure-culturing of the bacteria, DNAs were extracted and PCR and LAMP were performed.
Results:
Ten copies of targeted DNA were required for PCR detection whereas only five copies gave a positive reaction by LAMP assay, with 100% specificity. Of the 10 isolates inoculated in water for one and two months at three different temperatures 4, 22, and 37°C, only three cases (5%) were found positive in the first month; 13 (21.6%) and 29 cases (48.3%) were also positive by PCR and LAMP tests in the first and second months.
Conclusions:
Results of the current study confirmed that molecular methods such as PCR and LAMP were much more sensitive, rapid, and specific than culturing to identify non-culturable coccoid forms of H. pylori in water.
Keywords: Helicobacter pylori, Coccoid, Culture, LAMP, Polymerase Chain Reaction
1. Background
Helicobacter pylori is a Gram-negative, microaerophilic, spiral and mobile bacilli, which is colonized in stomach of approximately half of the world wide population and is the cause of gastritis, peptic ulcers, gastric cancer, and B-cell lymphoma (1). Three different forms of H. pylori are identified; viable spiral, viable coccoid, and viable but not culturable forms (2, 3). Although the coccoid form of H. pylori are detected both in human stomach (4) and natural environments (5), H. pylori can transform from a culturable spiral-shaped to a non-culturable coccoid form in undesirable conditions (6). Aging, starvation, expose to air, prolonged incubation, improper temperature, proton pump inhibitors and antibiotic treatment are different unfavorable conditions, which may lead to conversion of spiral to coccoid form (7).
Transmission of this organism is usually through oral-fecal ways; thus, water is one of the spreading pathways of these bacteria (8, 9). Therefore, identification of H. pylori is very essential due to high spreading, global distribution, and a large number of its carriers, also due to changing the form from bacilli to coccoid (10, 11). Some PCR based methods were conducted to identify H. pylori in the environmental samples to overcome this problem (12-14). Faster identification and ability to detect small number of bacterial DNA in the samples are the advantages of DNA based techniques. Loop-mediated isothermal amplification (LAMP) is one of the novel techniques developed worldwide, due to its simple and cost effective application, and it seems to substitute the recent available techniques in the future. Devices such as thermocycler are not required in this technique.
2. Objectives
The current study aimed to identify coccoid forms of H. pylori inoculated to water using culturing, PCR and LAMP methods.
3. Materials and Methods
3.1. Preparing the Helicobacter pylori Strain and Culturing Method
H. pylori (No: C30) was obtained from the Liver and Digestive Disease Research Center of Shahid Beheshti University and cultured in enriched brucella blood agar. The plates were incubated under microaerophilic conditions for five to seven days at 37°C using Anaerocult C (Merck, Germany) apparatus (15).
3.2. DNA Extraction From the Standard Strain
DNA was extracted using DNG-plus kit (Sinaclon, Iran, Cat No: DN8118C), and then PCR and LAMP tests were performed for this strain.
3.3. Optimization of Polymerase Chain Reaction
The primers used for PCR test were selected based on glmM genes (16, 17) (Table 1). PCR mixture was prepared as follows: double distilled water (D.D.W): 14 µL, 10X buffer: 2.5 µL, MgCl2 (50 mM): 0.75 µL, dNTP Mix (10 mM): 0.5 µL, (10 µM) forward primer: 1 µL, (10 µM) reverse primer: 1 µL, Taq DNA Polymerase enzyme (BioFlux) (5 u/µL): 0.3 µL. Target DNA (from standard strain): 5 µL and total volume was 25 µL. Further, thermal profile was optimized as follows: The number of thermal cycles was 35 as follows: denaturation, 30 seconds at 93°C; annealing, 60 seconds at 72°C; extension 1 minute at 72°C and final extension, 25 minutes at 72°C. PCR was conducted under optimized conditions and PCR product was analyzed by electrophoresis and visualized on 2% agarose stained with SYBR green (Sinaclon).
Table 1. Primers Designed Based on glmM Gene Used in PCR and Primers Designed for LAMP.
| Primers | Sequence (5' to 3') |
|---|---|
| PCR | |
| H.P-F | 5’-AAGCTT TTAGGGGTGTTAGGGGTTT-3’ |
| H.P-R | 5’-AAGCTTACTTTCTAACACTAACGC-3’ |
| LAMP | |
| F3 | 5’-ACACAATTTAAGACGTAGACTT-3’ |
| B3 | 5’-GCTATGCGACAACATACGG-3´ |
| FIP | 5’-GAGGTAGGTAGGTAGGTAGGTAGGTAAGCGTTAAACAATTCAATGAGAG-3’ |
| BIP | 5’-ATCTGTGAGATGGAAGAATAAACCCAAAAAACACGAGGCACCG-3’ |
| LF | 5’-GTAGGTAGGTAGGTAGGTAGGTAAC-3’ |
| LB | 5’-CCGGGGGATCCATTTTTACG-3’ |
3.4. Polymerase Chain Reaction Product Cloning as Positive Control
The PCR product was purified by chloroform and ethanol precipitation methods. The purified product was cloned into the compatible sites of the T-Vector pTZ57R by T/A cloning kit (Fermentas, cat: K1214). Recombinant plasmids were confirmed by PCR and used as positive control in the tests.
3.5. Identification of Polymerase Chain Reaction Sensitivity and Specificity
A suspension of fresh H. pylori culture was prepared with the concentration of 0.9 × 109 CFU/mL in OD = 600 nm; DNA was extracted using DNG-plus. Extracted DNA was diluted to one copy using dilution method. To evaluate the specificity human, mouse, Saccharomyces cerevisiae, Escherichia coli, Mycoplasma pneumonia, Herpes Simplex Virus, and Mycobacterium tuberculosis DNAs were extracted and loaded in wells as positive control.
3.6. Optimization of Loop-Mediated Isothermal Amplification
LAMP primers were designed for glmM gene using primer explorer V4 software; (http://primerexplorer.jp./e/) (Table 1). LAMP reaction mixture was prepared as follows: D.D.W: 5.2 µL, Betaine 5 Mol: 4 µL, dNTP (10 mM): 3.5 µL, 10X buffer: 2.5 µL, MgSo4 (100 mM): 1.8 µL, Mix Ӏ: 1 µL, Mix ӀӀ: 1 µL, Bst DNA polymerase enzyme (New England BioLabs; Lot:33/110806): 1 µL, target DNA (extracted DNA from standard strain): 5 µL, and total volume was 25 µL. In Mix Ӏ the concentration of FIP and BIP primers were 40, 10 µL D.D.W in 100 µL total volume respectively, and in Mix ӀӀ the concentration of LF, and LB were 20 and 60 µL D.D.W in 100 µL total volume, respectively.
3.7. Identification of LAMP Test Specification and Sensitivity
After preparing the serial dilution of the sample, SYBR green 0.1% was added to each tube and then observed under Ultra Violet light. In order to conduct LAMP test specificity, the extracted DNAs of human, mouse, S. cerevisiae, E. coli, M. pneumonia, Herpes Simplex Virus, and M. tuberculosis were used.
3.8. Preparing Coccoid Form
To inoculate coccoid forms in the current study, 10 different strains (H1-H10) were separated from stomach tissue biopsy, and added to 30 tubes containing 3 mL D.D.W with 1.5 × 108 CFU/mL bacteria. Then, these samples were divided into three groups each containing 10 samples and stored at three different temperatures 4, 22 and 37°C. The test was conducted after one and two months of incubation; 1400 µL of each sample was transferred into 1.5 mL eppendorf tubes, and then the sample was vortexed once and centrifuged at 10000 rpm for five minutes. Then the supernatant was removed and the remained pellet was re-suspended in 200 µL water. Suspension was divided into two 100 µL equivalents for culturing on the brucella blood agar and DNA extraction by boiling method.
3.9. Culturing
The prepared 100 µL solution was cultured on the brucella blood agar under microaerophilic conditions at 37°C for seven days and then the growth was studied.
3.10. DNA Extraction Using Boiling Method
Another 100 µL of the suspension was heated in boiling water. After that, tubes were centrifuged in 12000 rpm for five minutes. The supernatant was removed from the pellet.
3.11. PCR and LAMP Tests
PCR test was performed under optimized conditions using glmM gene based primers on the extracted DNAs to identify coccoid form. In addition, LAMP test was performed on the extracted DNAs as well, with 1 µL SYBR green 0.1% added to each tube and then observed under UV light.
3.12. Coccoid Form in Environmental Water Samples
Ten microliter of 40 samples obtained from the environmental waters of Tehran with different sources, including: drinking and non-drinking waters, air conditioners and sewages, and stored in sterile tubes and immediately transferred to the laboratory on ice. DNAs were extracted by boiling method; 1400 µL of each water sample was transferred to 1.5 mL eppendorf tubes and centrifuged at 10000 rpm for ten minutes. The resulting solution from the centrifuge was removed and 50 µL of double distilled de-ionized water was added to the existing sediment. The tubes were boiled in water for 15 minutes. Finally, PCR test was performed on 5 µL of the boiled waters. Further, LAMP test was performed on the extracted DNAs using specific primers designed for LAMP and finally 1 µL of 0.1% green SYBR was added to each tube and observed under UV light.
4. Results
4.1. Optimization of PCR and LAMP Tests
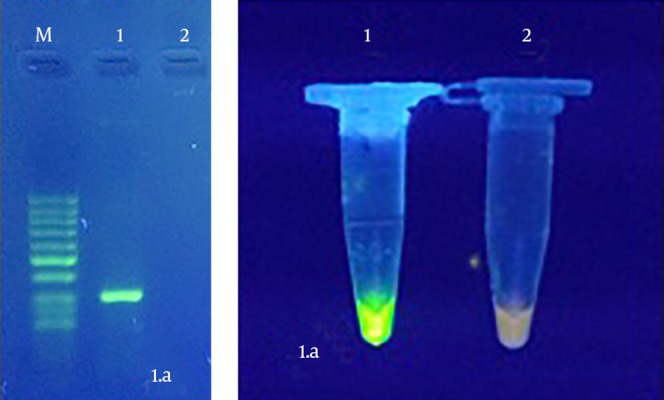
Amplicon of H. pylori (294 base pair) was observed by PCR test on 2% agarose (Figure 1 A). LAMP procedure was optimized at 66°C for 1 hour (Figure 1 B).
Figure 1. A, Optimized PCR Test for glmM Gene of Helicobacter pylori. M: 50 bp DNA Ladder (Thermo Scientific), 1: Amplicon (294 bp) of H. pylori (positive control), 2: Negative Control, B, Optimized LAMP test. 1: Positive Control, 2: Negative Control.
4.2. Specificity and Sensitivity of PCR
PCR performed with different serial dilutions of H. pylori DNAs. The results showed that amplification was performed with only 10 DNA copies. No amplification was observed in less than 10 copies of DNA, which indicated high test sensitivity. Specificity of PCR was measured using human, mouse, S. cerevisiae, E. coli, M. pneumonia, Herpes Simplex Virus, and M. tuberculosis. PCR showed very high specificity and only responded to H. pylori DNA with 100% specificity.
4.3. Specificity and Sensitivity of LAMP
LAMP reaction was conducted by different dilutions of H. pylori DNAs at 66°C for one hour. The sensitivity results of LAMP test showed that amplification was carried out with only five copies of DNA and green color was observed; but in less than five copies it was not observed and the tube remained in light orange, which indicates high sensitivity of the test. LAMP also had 100% specificity.
4.4. PCR, LAMP and Culturing Results at 4°C
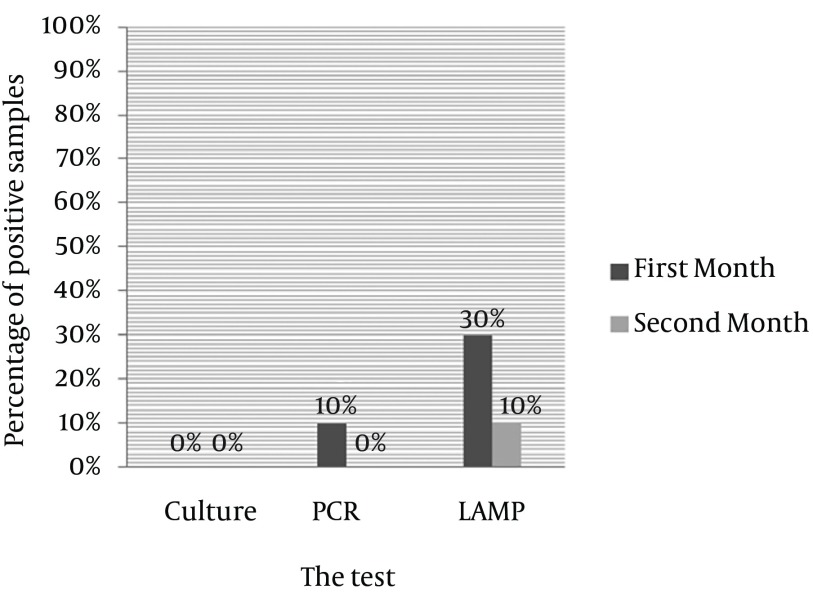
In samples experimentally cultured on brucella blood agar after 30 and 60 days inoculation of H. pylori, no growth was observed after incubation for seven days; but in the first month, 10% of the samples had positive results in PCR while no band was observed in the second month PCR test; LAMP test indicated positive results in 30% of the samples in the first month and 10% in the second month (Figure 2).
Figure 2. Comparing Culturing, PCR, and LAMP Results of Coccoid Samples After 30 and 60 Days sat 4°C.
4.5. PCR, LAMP, and Culturing Results at 22°C
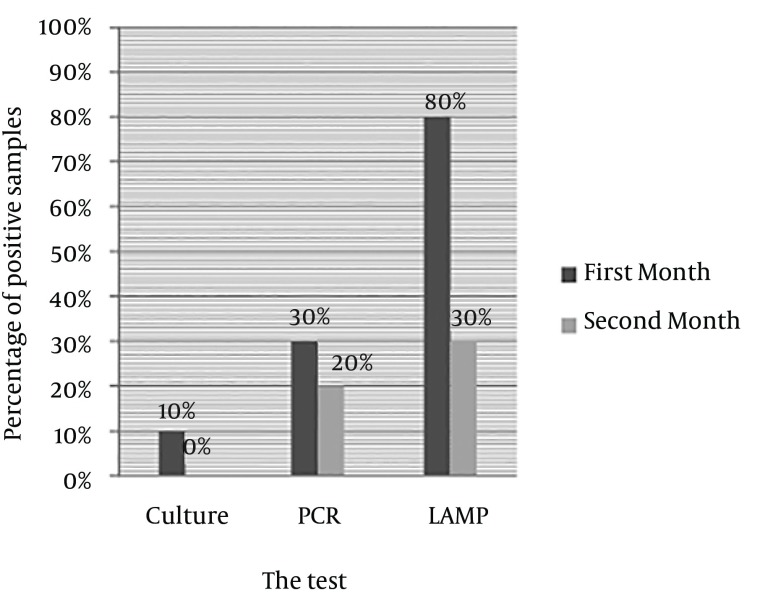
In the samples experimentally cultured on brucella blood agar after 30 and 60 days inoculation of H. pylori, after seven days of incubation, only 10% of the first month samples had growth on the blood agar (Figure 3); while there was no growth in the second month samples; 30% of the first month and 20% of the second month samples had positive PCR results, and 80% of the first month and 30% of the second month samples had positive LAMP results (Figure 4).
Figure 3. Coccoid Form of Helicobacter pylori.
Figure 4. Comparing PCR, and LAMP Results of Coccoid Samples Culturing After 30 and 60 Days at 22°C.
4.6. PCR, LAMP, and Culturing Results at 37°C
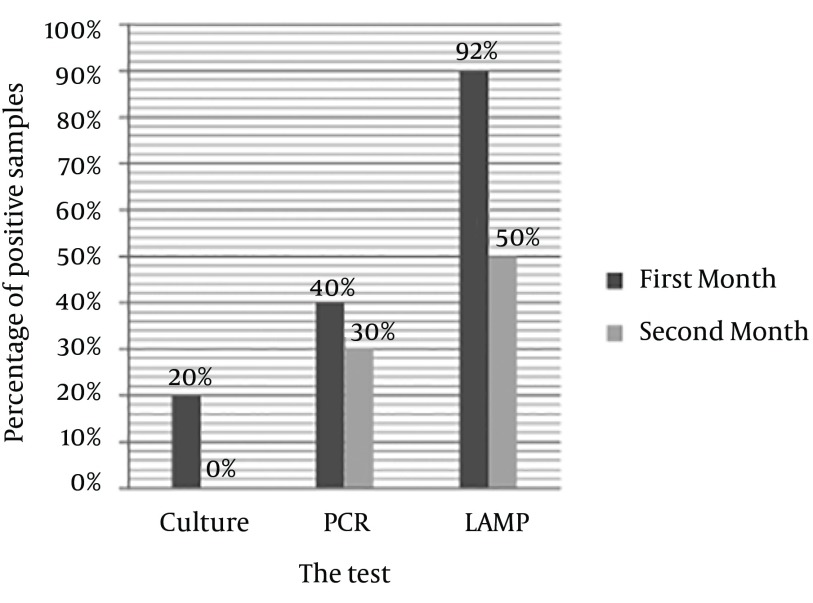
In samples experimentally cultured on brucella blood agar after 30 and 60 days inoculation of H. pylori, after seven days incubation, 20% of the first month samples showed growth; while no samples of the second month had any growth on the culture. Totally, 40% of the first month and 30% of the second month samples had PCR positive results; 90% of the first month and 50% of the second month samples had positive LAMP results (Figure 5).
Figure 5. Comparing PCR, and LAMP Results of Coccoid Samples Culturing After 30 and 60 Days at 37°C.
4.7. Coccoid Forms in Environmental Water
Of the 40 samples, no positive case was observed by PCR and LAMP.
5. Discussions
The primary mode of H. pylori transmission, a human pathogen carried by more than half of the population worldwide, is still unresolved. Some epidemiological data suggest water as a possible transmission route. In the environment, H. pylori transform into coccoid form, which frequently results in the failure to identify these bacteria in the environmental samples by conventional culturing techniques. To overcome limitations associated with culturing, molecular approaches based on DNA amplification by PCR are developed and used to detect H. pylori in clinical and environmental samples (5). Waterborne transmission is not unusual for an enteric pathogen. In vitro experiments show that H. pylori can survive in water for several months. However, H. pylori are not isolated from the environment by culturing techniques. Coccoid forms of H. pylori contain polyphosphate as an energy source, which provides the required energy for certain levels of inter cellular metabolism to save DNA and RNA and construct configurations such as cellular wall and membrane for at least three months (18).
Stability degradation of coccoid forms and revival possibility of this form to bacilli form by animal passage was suggested in 1986 for the first time, addressed in several discussions. If coccoid cells could cause infection in animals, they could also affect human health and play important role in the environmental contamination. Therefore, many researches focus on physiologic status identification and infecting capability of coccoid forms of these bacteria (19, 20). Researchers showed that patients infected with the coccoid forms of H. pylori in their stomach are diagnosed with stomach cancer and stomach ulcers (4). It is also a proven fact that the virulence degradation of inoculated coccoid forms in water still has urease activity and adherence capability to epithelial cells as the spiral forms do.
In addition, flagella, which is considered as one of the components responsible for bacterial infectivity is not visible yet as a cellular configuration in coccoid forms under electron microscope, is able to colonize in stomach mucus, and leads to gastric inflammation (21). The culturing technique is a standard method to identify H. pylori and perform antibiogram for antimicrobial therapy (22). But this method is time consuming, costly, and difficult to perform. Since H. pylori is a fastidious microorganism and transforms to coccoid forms in the environment and water, the culturing technique could not be an appropriate isolation approach in some cases (23). Low sensitivity of the culturing method compared to the molecular techniques is explained by factors such as small number of microorganisms, microorganism death during transferring and culturing, or microorganism transformation to coccoid form (24, 25). Different studies are performed to identify H. pylori from different water samples, including a study in UK conducted by Park et al. on urban water pipe studying prevalence of biofilm formation in different species of Helicobacter including H. pylori. They concluded that H. pylori biofilm could be available in the distributing water worldwide (8). Benson et al. in a study in the US, H. pylori identified from the environmental water samples directly, using PCR and reported the PCR sensitivity as 95% (10). Watson et al. identified H. pylori and biofilm in the UK distributing drinking water using PCR. Results showed that no active bacteria existed in the 151 obtained samples (26).
Bahrami et al. (27) evaluated the prevalence of H. pylori in tap water, dental units’ water, and bottled mineral water in Iran. They collected 200 water samples from Isfahan province, tested the samples for H. pylori by culturing and polymerase chain reaction (PCR) methods using ureC (glmM) gene. Safaei et al. (28) also investigated the existence of the H. pylori antibody and antigen in serum, milk, and faeces samples from 92 lactating Holstein cows in Shahrekord, Iran. The H. pylori antigen and antibodies were detected using enzyme-linked immunosorbent assay (ELISA) test and were confirmed by PCR. Janzon et al. studied the presence of H. pylori DNA in the environmental and drinking water samples of Dakar in Bangladesh; considering the high sensitivity of Real-Time PCR , no trace of H. pylori DNA was found in their study (29).
Compared to the culturing method, PCR is more sensitive and specific but has some limitations such as numerous thermal cycles, expensive thermocycler apparatus, a time consuming and difficult product manifestation and identification method (30). Therefore, a faster and easier assay such as LAMP has seriously been needed. This technique is a one stage amplification reaction, which can produce a large number of copies (109) in less than one hour under isotherm conditions. The most important advantage of this method is that it does not need DNA denaturation (31, 32). Thus, since it takes little time to change temperature, LAMP amplification is an isothermal reaction (33). Another advantage of using LAMP method is based on stem-loop amplification, which leads to accumulation of high amounts of products with different lengths and consequently makes DNA detection much easier (34).
In the only study performed to identify H. pylori in the stomach biopsy samples by LAMP and brushing, by Minami et al. specificity and sensitivity of LAMP test were reported 100% and 102 CFU, respectively (35). In the current study, authors found both PCR and LAMP specificities as 100%, PCR test sensitivity as 10 CFU, and LAMP sensitivity as 5 CFU, which was twice that of the PCR. In the current study, using glmM gene based PCR and LAMP by inoculation of separated H. pylori strains isolated from biopsy tissue samples in water, viable but non-culturable cells in the samples were identified after one and two months, which could not be detected by the culturing method.
In conclusion, evaluation of LAMP, PCR, and culturing techniques showed that molecular techniques used in the current study, especially LAMP, had higher specificity, sensitivity, and accuracy than other techniques and they could be used more easily to identify viable but non-culturable forms of these bacteria.
Acknowledgments
The authors would like to appreciate the Iranian Gene Fanavar (IGF) institute and its personnel, also microbiology laboratory of Baqiyatallah University of Medical Sciences and Hospital, Digestion Center, and Medical Science Research Center of Liver and Digestive Diseases of Shahid Beheshti University of Medical Sciences for their great help and generosity.
References
- 1.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347(15):1175–86. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 2.Benaissa M, Babin P, Quellard N, Pezennec L, Cenatiempo Y, Fauchere JL. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect Immun. 1996;64(6):2331–5. doi: 10.1128/iai.64.6.2331-2335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willen R, Carlen B, Wang X, Papadogiannakis N, Odselius R, Wadstrom T. Morphologic conversion of Helicobacter pylori from spiral to coccoid form. Scanning (SEM) and transmission electron microscopy (TEM) suggest viability. Ups J Med Sci. 2000;105(1):31–40. doi: 10.1517/03009734000000045. [DOI] [PubMed] [Google Scholar]
- 4.Chan WY, Hui PK, Leung KM, Chow J, Kwok F, Ng CS. Coccoid forms of Helicobacter pylori in the human stomach. Am J Clin Pathol. 1994;102(4):503–7. doi: 10.1093/ajcp/102.4.503. [DOI] [PubMed] [Google Scholar]
- 5.Shahamat M, Alavi M, Watts JE, Gonzalez JM, Sowers KR, Maeder DW, et al. Development of two PCR-based techniques for detecting helical and coccoid forms of Helicobacter pylori. J Clin Microbiol. 2004;42(8):3613–9. doi: 10.1128/JCM.42.8.3613-3619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Percival SL, Thomas JG. Transmission of Helicobacter pylori and the role of water and biofilms. J Water Health. 2009;7(3):469–77. doi: 10.2166/wh.2009.070. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson HO, Blom J, Abu-Al-Soud W, Ljungh AA, Andersen LP, Wadstrom T. Effect of cold starvation, acid stress, and nutrients on metabolic activity of Helicobacter pylori. Appl Environ Microbiol. 2002;68(1):11–9. doi: 10.1128/AEM.68.1.11-19.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park SR, Mackay WG, Reid DC. Helicobacter sp. recovered from drinking water biofilm sampled from a water distribution system. Water Res. 2001;35(6):1624–6. doi: 10.1016/s0043-1354(00)00582-0. [DOI] [PubMed] [Google Scholar]
- 9.Massoudian SH, Ghane M, Golijanimoghadam R, Ghorbanimoein F. Absence of Helicobacter pylori in the river waters in the north of Iran. African J Microbiol. 2012;6(8):1790–5. [Google Scholar]
- 10.Benson JA, Fode-Vaughan KA, Collins ML. Detection of Helicobacter pylori in water by direct PCR. Lett Appl Microbiol. 2004;39(3):221–5. doi: 10.1111/j.1472-765X.2004.01555.x. [DOI] [PubMed] [Google Scholar]
- 11.Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10(4):720–41. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1(8336):1273–5. [PubMed] [Google Scholar]
- 13.Bunn JE, MacKay WG, Thomas JE, Reid DC, Weaver LT. Detection of Helicobacter pylori DNA in drinking water biofilms: implications for transmission in early life. Lett Appl Microbiol. 2002;34(6):450–4. doi: 10.1046/j.1472-765x.2002.01122.x. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed KS, Khan AA, Ahmed I, Tiwari SK, Habeeb A, Ahi JD, et al. Impact of household hygiene and water source on the prevalence and transmission of Helicobacter pylori: a South Indian perspective. Singapore Med J. 2007;48(6):543–9. [PubMed] [Google Scholar]
- 15.smaeili D, Mohabati Mobarez A, Hatef Salmanian A, Zavaran HA. Optimization of Helicobacter pylori culture in order to prepare favorable antigens. J Bacteriol Res. 2009;1(9):101–4. [Google Scholar]
- 16.Lu JJ, Perng CL, Shyu RY, Chen CH, Lou Q, Chong SK, et al. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J Clin Microbiol. 1999;37(3):772–4. doi: 10.1128/jcm.37.3.772-774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bickley J, Owen RJ, Fraser AG, Pounder RE. Evaluation of the polymerase chain reaction for detecting the urease C gene of Helicobacter pylori in gastric biopsy samples and dental plaque. J Med Microbiol. 1993;39(5):338–44. doi: 10.1099/00222615-39-5-338. [DOI] [PubMed] [Google Scholar]
- 18.Bode G, Mauch F, Malfertheiner P. The coccoid forms of Helicobacter pylori. Criteria for their viability. Epidemiol Infect. 1993;111(3):483–90. doi: 10.1017/s0950268800057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran AP, Upton ME. Factors affecting production of coccoid forms by Campylobacter jejuni on solid media during incubation. J Appl Bacteriol. 1987;62(6):527–37. doi: 10.1111/j.1365-2672.1987.tb02685.x. [DOI] [PubMed] [Google Scholar]
- 20.Hazeleger W, Arkesteijn C, Toorop-Bouma A, Beumer R. Detection of the coccoid form of Campylobacter jejuni in chicken products with the use of the polymerase chain reaction. Int J Food Microbiol. 1994;24(1-2):273–81. doi: 10.1016/0168-1605(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 21.She FF, Lin JY, Liu JY, Huang C, Su DH. Virulence of water-induced coccoid Helicobacter pylori and its experimental infection in mice. World J Gastroenterol. 2003;9(3):516–20. doi: 10.3748/wjg.v9.i3.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krogfelt KA, Lehours P, Megraud F. Diagnosis of Helicobacter pylori Infection. Helicobacter. 2005;10 Suppl 1:5–13. doi: 10.1111/j.1523-5378.2005.00341.x. [DOI] [PubMed] [Google Scholar]
- 23.Khanolkar-Gaitonde SS, Reubish GJ, Lee CK, Stadtlander CT. Isolation of bacteria other than Helicobacter pylori from stomachs of squirrel monkeys (Saimiri spp.) with gastritis. Dig Dis Sci. 2000;45(2):272–80. doi: 10.1023/a:1005496022881. [DOI] [PubMed] [Google Scholar]
- 24.Kisa O, Albay A, Mas MR, Celasun B, Doganci L. The evaluation of diagnostic methods for the detection of Helicobacter pylori in gastric biopsy specimens. Diagn Microbiol Infect Dis. 2002;43(4):251–5. doi: 10.1016/s0732-8893(02)00409-1. [DOI] [PubMed] [Google Scholar]
- 25.Brooks HJ, Ahmed D, McConnell MA, Barbezat GO. Diagnosis of helicobacter pylori infection by polymerase chain reaction: is it worth it? Diagn Microbiol Infect Dis. 2004;50(1):1–5. doi: 10.1016/j.diagmicrobio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Watson CL, Owen RJ, Said B, Lai S, Lee JV, Surman-Lee S, et al. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J Appl Microbiol. 2004;97(4):690–8. doi: 10.1111/j.1365-2672.2004.02360.x. [DOI] [PubMed] [Google Scholar]
- 27.Bahrami AR, Rahimi E, Ghasemian Safaei H. Detection of Helicobacter pylori in city water, dental units' water, and bottled mineral water in Isfahan, Iran. Sci World J. 2013;2013:280510. doi: 10.1155/2013/280510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Safaei HG, Rahimi E, Zandi A, Rashidipour A. Helicobacter pylori as a zoonotic infection: the detection of H. pylori antigens in the milk and faeces of cows. J Res Med Sci. 2011;16(2):184–7. [PMC free article] [PubMed] [Google Scholar]
- 29.Janzon A, Sjoling A, Lothigius A, Ahmed D, Qadri F, Svennerholm AM. Failure to detect Helicobacter pylori DNA in drinking and environmental water in Dhaka, Bangladesh, using highly sensitive real-time PCR assays. Appl Environ Microbiol. 2009;75(10):3039–44. doi: 10.1128/AEM.02779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Megraud F. Advantages and disadvantages of current diagnostic tests for the detection of Helicobacter pylori. Scand J Gastroenterol Suppl. 1996;215:57–62. doi: 10.3109/00365529609094536. [DOI] [PubMed] [Google Scholar]
- 31.Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15(2):62–9. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parida MM, Santhosh SR, Dash PK, Tripathi NK, Lakshmi V, Mamidi N, et al. Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol. 2007;45(2):351–7. doi: 10.1128/JCM.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida A, Nagashima S, Ansai T, Tachibana M, Kato H, Watari H, et al. Loop-mediated isothermal amplification method for rapid detection of the periodontopathic bacteria Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola. J Clin Microbiol. 2005;43(5):2418–24. doi: 10.1128/JCM.43.5.2418-2424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aonuma H, Yoshimura A, Perera N, Shinzawa N, Bando H, Oshiro S, et al. Loop-mediated isothermal amplification applied to filarial parasites detection in the mosquito vectors: Dirofilaria immitis as a study model. Parasit Vectors. 2009;2(1):15. doi: 10.1186/1756-3305-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minami M, Ohta M, Ohkura T, Ando T, Torii K, Hasegawa T, et al. Use of a combination of brushing technique and the loop-mediated isothermal amplification method as a novel, rapid, and safe system for detection of Helicobacter pylori. J Clin Microbiol. 2006;44(11):4032–7. doi: 10.1128/JCM.00898-06. [DOI] [PMC free article] [PubMed] [Google Scholar]