Abstract
Background:
The periplasmic overexpression of recombinant human interferon beta (rhIFN-β)-1b using a synthetic gene in Escherichia coli BL21 (DE3) was optimized in shake flasks using Response Surface Methodology (RSM) based on the Box-Behnken Design (BBD).
Objectives:
This study aimed to predict and develop the optimal fermentation conditions for periplasmic expression of rhIFN-β-1b in shake flasks whilst keeping the acetate excretion as the lowest amount and exploit the best results condition for rhIFN-β in a bench top bioreactor.
Materials and Methods:
The process variables studied were the concentration of glucose as carbon source, cell density prior the induction (OD 600 nm) and induction temperature. Ultimately, a three-factor three-level BBD was employed during the optimization process. The rhIFN-β production and the acetate excretion served as the evaluated responses.
Results:
The proposed optimum fermentation condition consisted of 7.81 g L-1 glucose, OD 600 nm prior induction 1.66 and induction temperature of 30.27°C. The model prediction of 0.267 g L-1 of rhIFN-β and 0.961 g L-1 of acetate at the optimum conditions was verified experimentally as 0.255 g L-1 and 0.981 g L-1 of acetate. This agreement between the predicted and observed values confirmed the precision of the applied method to predict the optimum conditions.
Conclusions:
It can be concluded that the RSM is an effective method for the optimization of recombinant protein expression using synthetic genes in E. coli.
Keywords: Periplasmic, Acetic acid, Escherichia coli, Interferon-beta
1. Background
Human Interferon β (IFN-β) is a cytokine that mediate the early innate immune response to viral infections. The recombinant human interferon beta (rhIFN-β) produced in E. coli has 165 amino acid residues, which has a molecular mass of 18 kDa. It is not glycosylated although it is physiologically active. Some studies are shown that IFN-β has antiviral, anticancer and immunomodulatory properties (1-3). Different clinical trials have been done on rhIFN-β and now it is approved for the treatment of multiple sclerosis (4, 5), chronic viral hepatitis (6), rheumatoid arthritis (7, 8), and as a potential adjuvant in prophylactic vaccines against infectious diseases (9, 10). A large quantity of the required rhIFN-β is produced in Escherichia coli and Chinese Hamster Ovary (CHO) cell lines.
The Gram-negative bacterium of E. coli has developed into a highly successful system for the production of a variety of heterologous proteins thanks to its rapid growth to high cell densities on inexpensive substrates and simple process scale up. Moreover,, its genetic and physiology is well-studied, and various cloning vectors and host strains have been developed to use E. coli as an expression host for foreign proteins (11-13). Generally, the presence of an expressing plasmid in the host cell causes a metabolic burden, which may reduce the specific growth rate and biomass content and plasmid instability (14). On the other hand, the specific growth rate has an upper limit which is determined by the onset of glucose overflow metabolism (15) and acetate formation (16) which is detrimental to recombinant protein production (17). Therefore, obtaining an optimum condition for overexpression of recombinant proteins is very important.
The fermentation medium defines the chemical and nutritional environment of the host cell during the production of foreign proteins. The components of fermentation medium directly affect the productivity and the process economics (18). The type of carbon source and its amount in culture medium is essential for high level production of the recombinant protein. It involves in the microorganism biosynthetic pathways and provides the required energy for the host cell to perform its physiological activity. Thermal or chemical inducers are developed for simple and cost-effective promoter induction. The sugar, Isopropyl-β-D-thiogalactopyranoside (IPTG) is a powerful and widely used chemical inducer for recombinant protein expression. The optical density at induction time is also a critical parameter in protein overexpression procedure. Because the productivity (i.e. the amount of product formed per unit volume per unit time) is related to the biomass level of the host cells (19). Moreover, environmental factors, such as fermentation temperature have a major effect on the cell metabolism and consequently the total protein production (20).
Varying single factors at a time to reach an apparent optimum point for optimization of production conditions during overproduction of recombinant proteins are labor intensive, unable to identify interactions between the different factors involved, and fail in identifying the true optimal conditions for protein overexpression. In contrast, using a Design of Experiments (DOE) methodology helps to identify the possible interactions between multiple factors which lead to a more reliable prediction of the true optimum conditions. Several studies have been done to adopt a statistical DOE methodology in order to optimize the rhIFN-β expression in its foreign hosts. In an investigation, a Response Surface Methodology (RSM) based on a Box-Behnken design (BBD) (21) was used during beta- interferon production from E. coli BL21-SI to reach to maximum production of 61 mg L-1 (22). In another study a three-level factorial RSM method was applied to optimize the induction condition of rhIFN-β in an E. coli BL21 (DE3) host to obtain 0.151 g L-1 h of beta interferon (23).
2. Objectives
In this study, we applied DOE using a RSM method based upon the BBD for optimization of the fermentation conditions in order to improve the final yield of the rhIFN-β-1b during expression in shake flasks culture by the E. coli BL21 (DE3) strain and eliminate the acetate secretion as an undesirable by-product. Three factors; the glucose concentration as carbon source (5-15 g L-1), intensity of the cell density prior induction (OD 600 nm = 0.7 - 3.5) and induction temperature (24 - 32°C), alone and in combination, were investigated using the RSM. The amount of the periplasmic rhIFN-β production after SDS-PAGE Sodium dodecyl sulphate-Polyacrylamide gel electrophoresis (SDS-PAGE) analysis was measured by gel densitometry with an accuracy of greater than 95%. The amount of acetate production was determined using an appropriate kit. The best result was repeated in batch culture experiment using a bench top bioreactor.
3. Materials and Methods
3.1. Bacterial Strain and Vector System
Escherichia coli BL21 F- ompT hsdSB (rB-mB-) gal dcm (DE3) (Invitrogen, CA, USA) harboring a recombinant plasmid (24) was used as host for production of the requested protein. A synthetic construct (6.8 kbp) encoding rhINFIFN-β was inserted into the XhoI and MscI cloning sites (Biomatik, ON, Canada). The plasmid contains the strong inducible T7 promoter under the control of lac-operator sequence and an ampicillin resistance cassette. It contains also, the ColE1 origin of replication and the strong transcription terminator of phage λ. The heat shock method was used for transformation of the prokaryotic host cells and the transforming clones were selected using Luria-Bertani (LB) agar plates containing 100 µg L-1 of ampicillin. Aliquots containing transformed bacterium were stored in 20% (v/v) glycerol at −70°C for long-term usage.
3.2. Media and Culture Condition
Recombinant cells were precultured as 50 mL cultures in 500 mL Erlenmeyer flasks on a NB-205 V incubator (N-BIOTEK, Bucheon-si, South Korea) at 160 rpm with orbital shaking at 37°C and incubated overnight. For antibiotic selection, ampicillin was added at a concentration of 100 µg L-1. The first preculture in TB medium was inoculated to a second preculture for 4 hours at 37°C at 160 rpm, which was then inoculated to an OD 600 nm of 0.7 to the main cultures of shake flask. The culture medium was supplemented with equal amounts of glucose and yeast extract as carbon and nitrogen sources, respectively, to keep the carbon-nitrogen ratio equal in all experiments. Each experiment was performed under different conditions of glucose concentration, cell density prior induction and temperature as described in experimental design (Table 1).
Table 1. Independent Variables and Their Coded and Actual Values Used for Optimization.
| Independent Variable | Symbol | Code Levels | ||
|---|---|---|---|---|
| -1 | 0 | 1 | ||
| Glucose concentration, g L -1 | X 1 | 5 | 10 | 15 |
| Optical density, OD 600 nm | X 2 | 0.7 | 2.1 | 3.5 |
| Induction temperature, °C | X 3 | 24 | 28 | 32 |
3.3. Bioreactor Experiment
A single colony of E. coli BL21 (DE3) containing the recombinant plasmid was inoculated in Terrific Broth (TB) culture medium containing 100 µg mL-1 of ampicillin overnight at 37°C and shaken at 160 rpm. The inoculum with OD 600 nm = 1.0 was aseptically transferred to the bioreactor. Batch cultivation was carried out in a 5-L bench-top Minifors stirred bioreactor (Infors HT, Basel, Switzerland) with a working volume of 2L, including two six-blade Rushton impellers with a speed range of 50 - 500 rpm.
Growth was initiated by sparging air into the bioreactor at 1.0 vvm, and the dissolved oxygen concentration was maintained above 30% of air saturation throughout the experiment by varying the stirrer speed. The pH was controlled at 7.0 by automatically feeding 1.0 M NaOH and 1.0 M HCl. The temperature was maintained at the specified value. Glucose as the sole carbon source was added at the indicated amount. Ampicillin, when required, was added to the medium at the concentration of 100 µg mL-1. The foam was controlled by the addition of a silicone-based anti-foaming reagent. Samples were taken at different times and analyzed for OD 600 nm, acetate excretion and rhINFIFN-β production.
3.4. Induction and Expression of Recombinant Human Interferon Beta
IPTG (Fermentas, Vilnius, Lithuania) was added to a final concentration of 0.2 mM as soon as the OD 600 nm value reached about the requested value. Growth was continued for additional 5 hours at 37°C. The cell pellet was collected by centrifugation at 5000 g for 10 minutes at 4°C and frozen.
3.5. Experimental Design and Optimization by Response Surface Methodology
Box-Behnken design was used to evaluate the main, interaction and quadratic effects of three independent variables on rhIFN-β overexpression and acetate secretion. Box-Behnken Design was specifically selected since it requires fewer runs than a Central Composite Design (CCD) in the cases of three or four variables. The experimental design and statistical analysis were performed using the statistical package Design-Expert, version 7.0.0 (Stat-Ease, Inc. Minneapolis, MN, USA). After selection of independent variables involved in the overexpression of rhIFN-β, the preliminary range of the independent parameters were determined, and the proper ranges of the requested variables were defined. A three-level three-factor BBD was chosen to evaluate the combined effect of three independent variables the glucose concentration (factor A), OD 600 nm (factor B), and induction temperature (factor C) coded as X1, X2 and X3, respectively. The minimum and maximum values for glucose concentration were set at 5 and 15 g L-1, OD 600 nm between 0.7 and 3.5, and induction temperature between 24°C and 32°C (Table 1). The response values were the amount of produced rhIFN-β in mg L-1 of culture medium and excreted acetate in g L-1. The complete design consisted of 15 combinations, including three replicates of the center point (Table 2). The responses function (Y) was partitioned into linear, quadratic and interactive components:
Table 2. Box-Behnken Design With the Values of the Independent Variables in Natural and Coded Units, the Glucose Concentration (Factor A), OD 600 nm (Factor B), and Induction Temperature (Factor C) and the Experimental Results for the Production of Recombinant Human Interferon Beta Using Densitometry Method (mg L-1) and Acetate Excretion (g L-1) Using the Acetate Kit.
| Experiment | Factor A, g L-1 | Factor B, OD600 | Factor C, °C | Acetate Accumulation, g L-1 | Production of IFN, g L-1 | Maximum Growth Cell, g L-1 |
|---|---|---|---|---|---|---|
| X 1 | X 2 | X 3 | ||||
| 1 | 10 (0) | 2.1 (0) | 28 (0) | 1.23 | 0.32 | 1.52 |
| 2 | 10 (0) | 0.7 (-1) | 32 (1) | 1.22 | 0.22 | 1.05 |
| 3 | 15 (1) | 2.1 (0) | 32 (1) | 2.43 | 0.28 | 1.21 |
| 4 | 10 (0) | 2.1 (0) | 28 (0) | 1.22 | 0.31 | 1.56 |
| 5 | 10 (0) | 3.5 (1) | 24 (-1) | 1.34 | 0.28 | 1.19 |
| 6 | 10 (0) | 0.7 (-1) | 24 (-1) | 1.62 | 0.34 | 1.71 |
| 7 | 5 (-1) | 2.1 (0) | 24 (-1) | 0.73 | 0.28 | 1.27 |
| 8 | 5 (-1) | 2.1 (0) | 32 (1) | 1.05 | 0.20 | 0.96 |
| 9 | 15 (1) | 2.1 (0) | 24 (-1) | 2.24 | 0.36 | 1.87 |
| 10 | 10 (0) | 2.1 (0) | 28 (0) | 1.23 | 0.31 | 1.57 |
| 11 | 5 (-1) | 3.5 (1) | 28 (0) | 0.89 | 0.19 | 0.91 |
| 12 | 10 (0) | 3.5 (1) | 32 (1) | 1.52 | 0.24 | 1.18 |
| 13 | 15 (1) | 0.7 (-1) | 28 (0) | 2.43 | 0.26 | 1.49 |
| 14 | 5 (-1) | 0.7 (-1) | 28 (0) | 0.68 | 0.25 | 1.25 |
| 15 | 15 (1) | 3.5 (1) | 28 (0) | 2.61 | 0.29 | 1.42 |
| (1) |
Where β0 is defined as the constant, Bi the linear coefficient, Bii the quadratic coefficient and Bij the cross-product coefficient. Xi and Xj are levels of the independent variables, while ε is the residual error. The analysis of variance (ANOVA) tables were generated and the effect and regression coefficients of individual linear, quadratic and interaction terms were determined. The significances of all terms in the polynomial were judged statistically by computing the F-value at a probability (P) of 0.001, 0.01, or 0.05. The regression coefficients were then used to make statistical calculations to generate contour maps from the regression models. The significance of each coefficient (linear or quadratic) was determined with the Student’s t-test, at 0.05 probability level. The optimal values were obtained solving the regression equation by the Newton-Raphson method and analyzing the response surface contour.
3.6. Analytical Procedures
The cell growth profile was followed by optical density measurements at 600 nm in a CT-1500 spectrophotometer (E-Chrome Tech, Taipei, Taiwan). The culture was diluted to the linear range before measuring the absorbance. Acetate was analyzed enzymatically using the appropriate kit (Megazyme, Wicklow, Ireland), according to procedures suggested by the supplier. The total protein concentration was determined by Bradford method using Bovine Serum Albumin (BSA) (Merck Millipore, Darmstadt, Germany) as the standard. The rhINFIFN-β protein was extracted from the periplasm by osmotic shock treatment. Proteins then were separated in a 17.5% sodium dodecyl sulphate polyacrylamide gel electrophoresis using a vertical electrophoresis unit (omniPAGE mini, Cleaver Scientific, UK). A commercial rhIFN-β (ZiferonTM, Zist Daru Danesh, Tehran, Iran) was used as the standard. Proteins were stained with Coomassie Brilliant Blue R-250 (Merck Millipore, Darmstadt, Germany). The prepared gels were analyzed using a Bio-Rad Gel Doc 2000 densitometric gel scanner to measure the concentration of the rhIFN-β.
4. Results
4.1. Data Analysis and Evaluation of the Models
Response surface methodology is a collection of useful mathematical and statistical procedures, based on the fit of a polynomial equation to the experimental data (25) which reduces the number of experimental trials in comparison to one factor at a time methods. Compared to the other experimental design methodologies, such as CCD and Doehlert Matrix (DM), Box-Behnken employs a design that allows testing a large number of effective factors with fewer experimental points (three levels per factor) and high efficiency (26). After preliminary studies, a three-factor three-level RSM of BBD was used to analyze the effects of three parameters, glucose concentration (X1), optical density (X2), and induction temperature (X3) on the rhIFN-β production (Y1) and acetate excretion (Y2).
The experimental design and results were shown in Table 2. The whole design consisted of 15 experimental points carried out in a random order, which included 12 factorial points and 3 central points. Three replicates (runs 4, 11, 13) at the center of the design were used to allow for estimation of a pure error sum of squares. An approximate regression model for predicting the yield (Y) of recombinant protein production based on the experimental results was calculated and expressed by the following second-order polynomial Equation (in terms of actual values):
| (2) |
In Equation 2, Y1 is the amount of rhIFN-β production, X1, X2 and X3 are corresponding coded variables of glucose concentration, optical density and induction temperature, respectively. Moreover, suggested a regression model to predict the amount of acetate excretion in terms of actual value was as follows:
| (3) |
In Equation 3, Y2 is the amount of acetate excretion, and the other parameters are the same as Equation 2.
To determine the statistical significance of the regression model, the F-distribution analysis was performed which measures the ratio of two chi-square distributions. The analysis of variance (ANOVA) for experimental results was shown in Table 3. The ANOVA of regression model implies the model terms are highly significant, as is shown in Fisher’s F-test with a very low probability value (P value < 0.05). The R2 coefficient was determined to check the fitting of the model. The closer values of R2 to 1, means stronger the model and the better prediction of the response. As described in Table 3, the R2 value, adjusted R2 value (R2adj) and predicted R2 value (R2pred) of 0.9885, 0.9679 and 0.8434, respectively which show that the regression model for rhIFN-β overexpression fits to the experimental values (Figure 1). Besides, as it has been shown in Table 3, the R2 value, adjusted R2 value (R2adj) and predicted R2 value (R2pred) for amount of acetate excretion were 0.9779, 0.9381 and 0.6464, respectively.
Table 3. Analysis of Variance for the Effect of the Independent Variables on the Dependent Variables and the Regression Coefficients, of the Fitted Quadratic Equations Obtained Obtained From Experimental Results; Regression Coefficients, R2, and P or Probability Values for Four Dependent Variables for rhIFN-β Production a.
| Source | DF | rhIFN-β Production, mg L-1 | Acetate Excretion, g L-1 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sum of Squares | Mean Square | F Value | P Value | Sum of Squares | Mean Square | F Value | P Value | ||
| Model | 9 | 0.034 | 3.754E - 003 | 47.92 | 0.0003 | 5.57 | 0.62 | 24.57 | 0.0013 |
| X1 | 1 | 9.112E - 003 | 9.112E - 003 | 116.33 | 0.0001 | 5.06 | 5.06 | 200.90 | ˂ 0.0001 |
| X 2 | 1 | 6.125E - 004 | 6.125E - 004 | 7.82 | 0.0382 | 0.021 | 0.021 | 0.83 | 0.4028 |
| X 3 | 1 | 0.013 | 0.013 | 163.4 | ˂ 0.0001 | 0.011 | 0.011 | 0.42 | 0.5466 |
| X 1 X 2 | 1 | 2.025E - 003 | 2.025E - 003 | 25.85 | 0.0038 | 2.250E - 004 | 2.250E - 004 | 8.940E - 003 | 0.9283 |
| X 1 X 3 | 1 | 0.000 | 0.000 | 0.000 | 1.0000 | 4.225E - 003 | 4.225E - 003 | 0.17 | 0.6990 |
| X 2 X 3 | 1 | 1.600E - 003 | 1.600E - 003 | 20.43 | 0.0063 | 0.084 | 0.084 | 3.34 | 0.1271 |
| X 1 2 | 1 | 2.878E - 003 | 2.878E - 003 | 36.73 | 0.0018 | 0.35 | 0.35 | 13.80 | 0.0138 |
| X 2 2 | 1 | 5.308E - 003 | 5.308E - 003 | 67.77 | 0.0004 | 0.052 | 0.052 | 2.08 | 0.2085 |
| X 3 2 | 1 | 1.083E - 004 | 1.083E - 004 | 1.38 | 0.2925 | 0.023 | 0.023 | 0.92 | 0.3817 |
| Residual | 5 | 3.917E - 004 | 7.833E - 005 | 0.13 | 0.025 | ||||
| Lack of fit | 3 | 3.250E - 004 | 1.083E - 004 | 3.25 | 0.2441 | 0.13 | 0.042 | 1257.75 | 0.008 |
| Pure error | 2 | 6.667E - 005 | 3.333E - 005 | 6.667E - 005 | 3.333E - 005 | ||||
| Cor. total | 14 | 0.034 | 5.69 | ||||||
| SD | 8.851E - 003 | 0.16 | |||||||
| Mean | 0.28 | 1.50 | |||||||
| CV, % | 3.21 | 10.60 | |||||||
| PRESS | 5.350E - 003 | 2.01 | |||||||
| R 2 | 0.9885 | 0.9779 | |||||||
| R 2 adj | 0.9679 | 0.9381 | |||||||
| R 2 pred | 0.8434 | 0.6464 | |||||||
| Adeq. precision | 23.698 | 13.452 | |||||||
a Abbreviations: DF, degree of freedom; PRESS, predicted residual sums of squares; CV, Coefficient of variation.
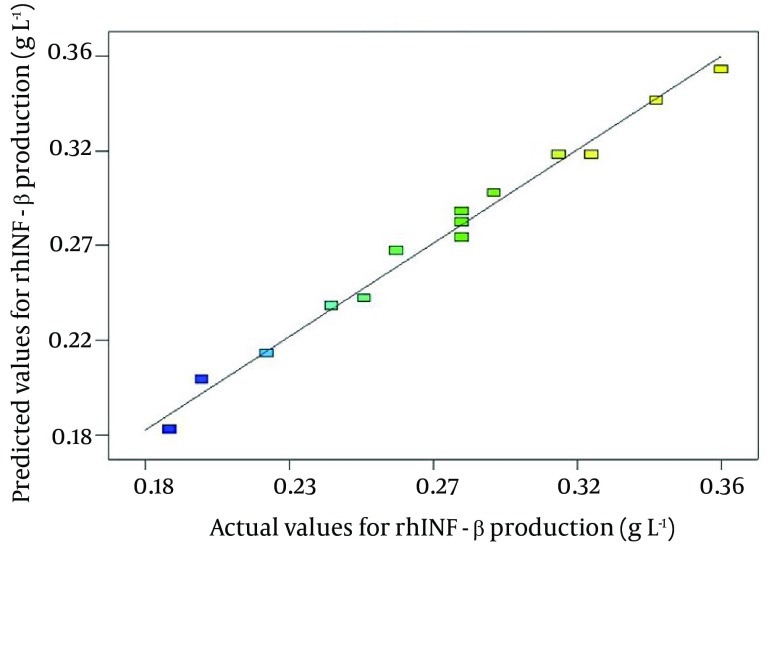
Figure 1. Predictability of the Obtained Model for Recombinant Human Interferon-Beta Overexpression Was Studied Using the Scatter Plot of the Predicted Versus Measured Experimental Densitometric Values.
The fit to the line of parity was shown with R2 = 0.9885, which indicated that the regression model was able to predict output data with high reliability.
4.2. Graphical Interpretation of the Response Surface Model
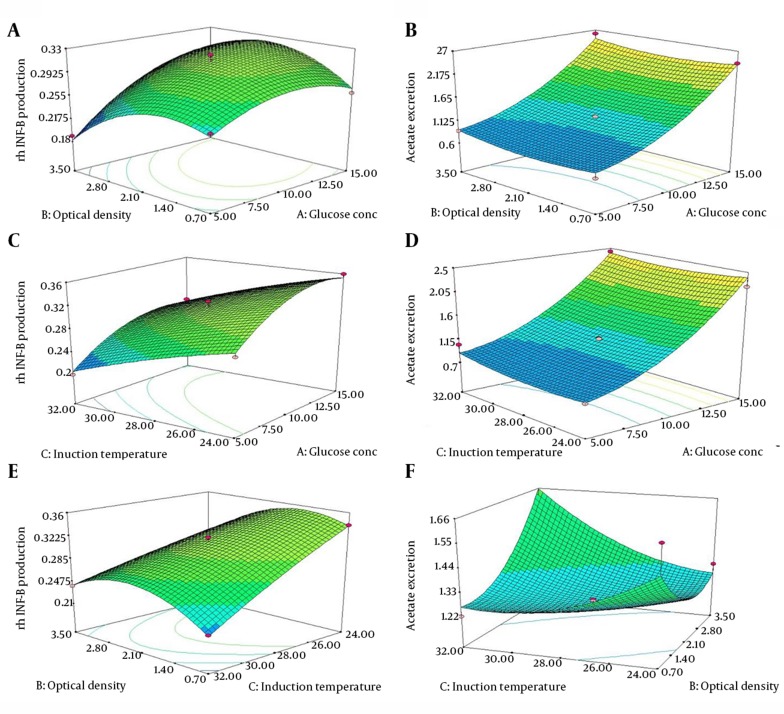
The response surface curves are plotted to explore the effects of changing the factor levels on the response and to determine the optimum level of each variable to reach a maximum amount of the requested recombinant protein whilst keeping the minimum amounts of acetate excretion. Three-Dimensional (3D) surface plots and contour plots were constructed as shown in Figure 2 A-F. The 3D surface plots show the effects and interaction of two independent variables on the response while the other independent variable was remained unchanged.
Figure 2. Response Surface Plots (3D) Showing the Influence of Three Studied Variables on the rhINFIFN-β Production Reflected by the Densitometric Measurement and Acetate Using Kit, When Optimizing the Following Pair of Parameters, While the Other Parameter Was Kept Constant at a Central Point (zero level): (A and B) Interaction Between Glucose Concentration and Optical Density; (C and D) Interaction Between Glucose Concentration and Induction Temperature; and (E and F) Interaction Between Optical Density and Induction Temperature.
As shown in Figure 2 A and 2C, increasing the amounts of glucose concentration favored the rhIFN-β overexpression to an extent. More concentrations of glucose disfavored the recombinant protein production. This observation may be explained by the fact that the presence of higher concentration of glucose causes the bacterial Crabtree effect. As a consequence of Crabtree effect, more than 15% of the glucose will be excreted as acetate (16). Figures 2 B and 2D prove this observation. The acetate excretion level was increased in the presence of higher amounts of glucose. Besides, more values of optical density to some extents led to an increase in the recombinant protein production (Figure 2 A). Meanwhile, increasing the induction temperature in presence of the higher glucose amounts slightly decreased the levels of rhIFN-β overexpression (Figure 2 C).
As demonstrated by the data depicted in Figure 2 E, increasing the level of induction temperature, the rhIFN-β overexpression was found to increase rapidly at the beginning but with a slower rate toward the end. As the optical density increased, the rhIFN-β overexpression increased obviously, especially at the higher induction temperature. It was demonstrated that an extended increase in optical density disfavors the production of rhIFN-β (Figure 2 E). This might be due to the acidification of culture medium after acetate excretion due to higher growth rates of the recombinant cells and subsequent inhibition of recombinant protein production. As can be seen in Figure 2 F, higher acetate levels were observed after induction at higher optical densities. The higher values of temperature at the induction point were slightly decreased the rhINFIFN-β overexpression. Considering all the responses, it is evident that the glucose concentration, optical density and the induction temperature had a significant effect on the rhINFIFN-β overexpression. The results agree well with presented data in Table 3.
4.3. Model Validation and Experimental Confirmation
The second-order polynomial model obtained described in Equations 2 and 3 were utilized for response optimization using Design Expert software (version 7.0.0, Stat-Ease, Inc. Minneapolis, MN, USA). The maximum rhIFN-β production as 0.267 g L-1 and minimum amount of acetate excretion as 0.961 g L-1 were determined at the following optimum conditions: glucose concentration 7.81 g L-1, induction at optical density at 600 nm of 1.66 and induction temperature of 30.27°C. To confirm the validity of model for predicting the maximum rhIFN-β overexpression and minimum amount of acetate excretion, an additional experiment using this optimum operation conditions was performed in a 5-L bench top bioreactor. The average value of recombinant protein production and observed acetate secretion obtained experimentally were 0.255 g L-1 and 0.981 g L-1, which were very similar to the predicted yield (0.267 g L-1 and 0.961 g L-1 respectively), thus confirming the validity of Equations 2 and 3 within the specified range of process parameters and the existence of an optimal production point for rhIFN-β production in the prokaryotic host while keeping the acetate excretion to the minimum level.
4.4. Cell Growth, Acetate and Recombinant Protein Production
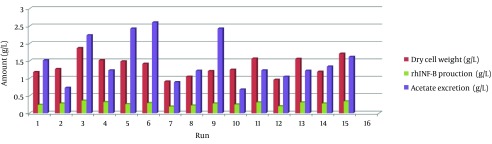
The growth profile of the prokaryotic host cell was determined in each experiment using spectroscopy method. The formation of by-product acetic acid was determined enzymatically and the productivity was detected using densitometry method and reported as grams per liter. The maximum values for the cell growth, acetate and the rhIFN-β production observed in each experiment are shown in Figures 3 and 4. A complete agreement between the maximum cell growth and the acetate production level was observed.
Figure 3. Growth Profile Measured as Optical Density at 600 nm, Acetate Production and Recombinant Protein Production of (a) Wild Type and (b) Recombinant Escherichia. coli BL21 (DE3) Cells for 6 Hours in Shake Flask Experiment.
Cells were induced by 0.2 mM IPTG at OD600 nm 0.7.
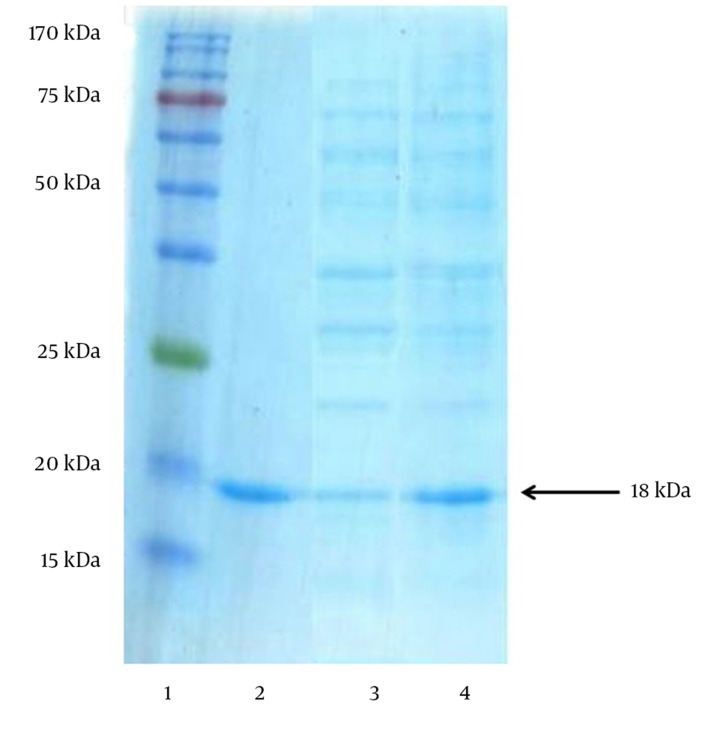
Figure 4. Visualization of the Increased Periplasmic rhIFN-β-1b Overexpression Yield in Escherichia. coli After the Optimization Study Using SDS–PAGE (17.5%) Analysis.
Lane 1: Chromatine prestained protein ladder (SinaClon, Tehran, Iran), Lane 2: rhIFN-β as positive control (ZiferonTM, Zist Daru Danesh, Tehran, Iran), Lane 3: Cell lysates from nonoptimized conditions (consisted of 5 g L-1 glucose, OD600 nm prior induction 3.5 and induction temperature of 37°C) and Lane 4: Optimized conditions (consisted of 7.81 g L-1 glucose, OD600 nm prior induction 1.66 and induction temperature of 30.27°C). The additional band with molecular weight of 18 kDa in periplasmic fraction after induction corresponds to rhINFIFN-β. The arrow indicates the position of rhIFN-β-1b.
5. Discussion
Box and Behnken design as a RSM was originally developed in the 60’s (27). The BBD is a very efficient model, which requires a small number of runs; therefore, is an important alternative avoiding time-consuming and laborious experiments (28). At present, BBD is extensively used to predict and improve the optimization issues involving several variables simultaneously in different fields of study. Box and Behnken design is a spherical, rotatable, or nearly rotatable second-order RSM design that consists of a central point and with the middle points of the edges of the cube circumscribed on the sphere. It could be considered as three interlocking 22 factorial designs with points lying on the surface of a sphere surrounding the center of the design (29). Its missing corners may be useful when the combined factor extremes should be avoided. This property prevents potential loss of data in those cases.
The number of experimental points (N) is defined using following Equation:
| (4) |
Where k is the number of variables and C0 is the number of center points (30). An equation is used to describe how the test variables affect the response and determine the interrelationship among the variables. In order to find the appropriate fermentation conditions to enhance the recombinant protein production, several parameters should be considered. Among them the induction point, the initial point at which the recombinant protein accumulates in the periplasmic space of the prokaryotic host is a critical parameter. Many researches have proposed the mid-log phase as the optimum point for induction (31, 32). Although some other has mentioned the stationary phase as the best point (33). Indeed the optimum induction point seems to be strain dependent.
In some strains the induction at early log-phase will utilize the whole cellular machinery to recombinant protein production instead of expressing the essential proteins for the cell proliferation (34). Hence, the late-log or stationary phase induction is preferred to provide the high-cell densities of the recombinant host. In other cases, induction at the lower specific growth rates makes the carbon/energy level as the limiting factor for recombinant protein overexpression (35). Consequently, the induction points between the early to middle-log phase are the favored to enhance the protein production. Similarly, the obtained results in this study propose the middle log-phase as the best induction point for rhIFN-β overexpression. On the other hand, the induction temperature which defines the activity of cellular enzymes and solubility of the produced protein also has a significant role to achieve the maximum production yield of the protein production. Although the maximal activity of the lac promoter occurs at higher temperatures (36), but in the case of periplasmic proteins the transport machinery will be the limiting factor (37). Therefore, the lower temperatures are favorable in which facilitate the rate of protein transfer through the cellular membranes.
As it has been shown, more accumulation of acetate in the presence of higher amounts of glucose was expected. Because during a fermentation experiment, the excess amounts of glucose will be diverted towards the formation of acetate due to overflow metabolism instead of biomass and energy generation through the respiratory chain and proton motive force (38). The optimal fermentation condition achieved from the current study may be applied to the scale-up of rhIFN-β production using E. coli BL21 (DE3) in a high-cell density cultivation experiment using a fed-batch cultivation strategy. Besides, using such a system will be helpful to save the time and increase the cost-effectiveness of the fermentation condition; because, the less amounts of the undesirable by-product acetate will be accumulated and the carbon flux will be diverted to biomass and recombinant protein production.
In conclusion, the RSM experiment predicted that a set level of 7.81 g L-1 glucose, induction at OD 600 nm = 1.66, and induction temperature of 30.27°C would provide the overall optimum region for overexpression of the requested recombinant protein with the maximum productivity. The obtained amount of IFN-β production and acetate excretion under optimized conditions showed 95.50% and 97.96% productivity, showing a good coincidence with the predicted value, and thus indicating the adequacy of the fitted model. This study showed that the BBD and RSM could be successfully used in optimizing fermentation condition operating variables for overexpression of rhIFN-β.
Acknowledgments
The authors would like to thank research deputy of Shahid Beheshti University of Medical Sciences in Tehran, Iran, for the financial support of this research.
Footnotes
Authors’ Contributions:Valiollah Babaeipour, Hossein Vahidi and Hamid Rajabi Memari designed the study. Hamid Rajabi Memari and Mohammad Hossein Morowvat performed plasmid design and molecular works. Mohammad Hossein Morowvat carried out the analysis and wrote the manuscript draft. Valiollah Babaeipour and Hossein Vahidi directed the research and coordinated the study. All authors read and approved the final manuscript.
Funding/Support:This study was a part of PhD thesis of Mohammad Hossein Morowvat, proposed and approved in faculty of pharmacy, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
References
- 1.Elco CP, Sen GC. Interferon Receptors. In: William JL, Lane MD, editors. Encyclopedia of Biological Chemistry. Waltham: Academic Press; 2013. pp. 619–23. [Google Scholar]
- 2.Larocque L, Bliu A, Xu R, Diress A, Wang J, Lin R, et al. Bioactivity determination of native and variant forms of therapeutic interferons. J Biomed Biotechnol. 2011;2011:174615. doi: 10.1155/2011/174615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.George PM, Badiger R, Alazawi W, Foster GR, Mitchell JA. Pharmacology and therapeutic potential of interferons. Pharmacol Ther. 2012;135(1):44–53. doi: 10.1016/j.pharmthera.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Sottini A, Capra R, Serana F, Chiarini M, Caimi L, Imberti L. Interferon-beta therapy monitoring in multiple sclerosis patients. Endocr Metab Immune Disord Drug Targets. 2009;9(1):14–28. doi: 10.2174/187153009787582379. [DOI] [PubMed] [Google Scholar]
- 5.Lim SY, Constantinescu CS. Current and future disease-modifying therapies in multiple sclerosis. Int J Clin Pract. 2010;64(5):637–50. doi: 10.1111/j.1742-1241.2009.02261.x. [DOI] [PubMed] [Google Scholar]
- 6.Ahn SH, Lee HW, Kim YS, Kim JK, Han KH, Chon CY, et al. Recombinant interferon-Beta-1alpha plus ribavirin for the treatment of chronic HCV infection: a prospective, randomized, comparative pilot study. Gut Liver. 2009;3(1):20–5. doi: 10.5009/gnl.2009.3.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smeets TJ, Dayer JM, Kraan MC, Versendaal J, Chicheportiche R, Breedveld FC, et al. The effects of interferon-beta treatment of synovial inflammation and expression of metalloproteinases in patients with rheumatoid arthritis. Arthritis Rheum. 2000;43(2):270–4. doi: 10.1002/1529-0131(200002)43:2<270::AID-ANR5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 8.van Holten J, Plater-Zyberk C, Tak PP. Interferon-beta for treatment of rheumatoid arthritis? Arthritis Res. 2002;4(6):346–52. doi: 10.1186/ar598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toporovski R, Morrow MP, Weiner DB. Interferons as potential adjuvants in prophylactic vaccines. Expert Opin Biol Ther. 2010;10(10):1489–500. doi: 10.1517/14712598.2010.521495. [DOI] [PubMed] [Google Scholar]
- 10.Bracci L, La Sorsa V, Belardelli F, Proietti E. Type I interferons as vaccine adjuvants against infectious diseases and cancer. Expert Rev Vaccines. 2008;7(3):373–81. doi: 10.1586/14760584.7.3.373. [DOI] [PubMed] [Google Scholar]
- 11.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72(2):211–22. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 12.Tripathi N, Sathyaseelan K, Jana AM, Rao PVL. High Yield Production of Heterologous Proteins with Escherichia coli. Defence Sci J. 2009;59(2):137–46. doi: 10.14429/dsj.59.1501. [DOI] [Google Scholar]
- 13.Swartz JR. Advances in Escherichia coli production of therapeutic proteins. Curr Opin Biotechnol. 2001;12(2):195–201. doi: 10.1016/s0958-1669(00)00199-3. [DOI] [PubMed] [Google Scholar]
- 14.DeLisa MP, Valdes JJ, Bentley WE. Quorum signaling via AI-2 communicates the "Metabolic Burden" associated with heterologous protein production in Escherichia coli. Biotechnol Bioeng. 2001;75(4):439–50. doi: 10.1002/bit.10034. [DOI] [PubMed] [Google Scholar]
- 15.Valgepea K, Adamberg K, Nahku R, Lahtvee PJ, Arike L, Vilu R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst Biol. 2010;4:166. doi: 10.1186/1752-0509-4-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69(1):12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eiteman MA, Altman E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006;24(11):530–6. doi: 10.1016/j.tibtech.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Greasham R. Chemically defined media for commercial fermentations. Appl Microbiol Biotech. 1999;51(4):407–21. doi: 10.1007/s002530051411. [DOI] [Google Scholar]
- 19.Shojaosadati SA, Varedi Kolaeil SM, Babaeipour V, Farnoud AM. Recent advances in high cell density cultivation for production of recombinant protein. Iranian J Biotech. 2008;6(2):63–84. [Google Scholar]
- 20.Caspeta L, Flores N, Perez NO, Bolivar F, Ramirez OT. The effect of heating rate on Escherichia coli metabolism, physiological stress, transcriptional response, and production of temperature-induced recombinant protein: a scale-down study. Biotechnol Bioeng. 2009;102(2):468–82. doi: 10.1002/bit.22084. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira SL, Bruns RE, Ferreira HS, Matos GD, David JM, Brandao GC, et al. Box-Behnken design: an alternative for the optimization of analytical methods. Anal Chim Acta. 2007;597(2):179–86. doi: 10.1016/j.aca.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Maldonado LM, Hernandez VE, Rivero EM, Barba de la Rosa AP, Flores JL, Acevedo LG, et al. Optimization of culture conditions for a synthetic gene expression in Escherichia coli using response surface methodology: the case of human interferon beta. Biomol Eng. 2007;24(2):217–22. doi: 10.1016/j.bioeng.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Tabandeh F, Khodabandeh M, Yakhchali B, Habib-Ghomi H, Shariati P. Response surface methodology for optimizing the induction conditions of recombinant interferon beta during high cell density culture. Chem Eng Sci. 2008;63(9):2477–83. doi: 10.1016/j.ces.2008.02.003. [DOI] [Google Scholar]
- 24.Morowvat MH, Babaeipour V, Rajabi-Memari H, Vahidi H, Maghsoudi N. Overexpression of Recombinant Human Beta Interferon (rhINF-beta) in Periplasmic Space of Escherichia coli. Iran J Pharm Res. 2014;13(Suppl):151–60. [PMC free article] [PubMed] [Google Scholar]
- 25.Bezerra MA, Santelli RE, Oliveira EP, Villar LS, Escaleira LA. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta. 2008;76(5):965–77. doi: 10.1016/j.talanta.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 26.Ferreira SL, Bruns RE, da Silva EG, Dos Santos WN, Quintella CM, David JM, et al. Statistical designs and response surface techniques for the optimization of chromatographic systems. J Chromatogr A. 2007;1158(1-2):2–14. doi: 10.1016/j.chroma.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 27.Box GEP, Behnken DW. Some New Three Level Designs for the Study of Quantitative Variables. Technometrics. 1960;2(4):455–75. doi: 10.1080/00401706.1960.10489912. [DOI] [Google Scholar]
- 28.Kaiser S, Verza SG, Moraes RC, Pittol V, Peñaloza EMC, Pavei C, et al. Extraction optimization of polyphenols, oxindole alkaloids and quinovic acid glycosides from cat's claw bark by Box–Behnken design. Ind Crop Prod. 2013;48:153–61. doi: 10.1016/j.indcrop.2013.04.026. [DOI] [Google Scholar]
- 29.Kumar A, Prasad B, Mishra I M. Process Parametric Study for Ethene Carboxylic Acid Removal onto Powder Activated Carbon using Box-Behnken Design. Chem Eng Tech. 2007;30(7):932–7. doi: 10.1002/ceat.200700084. [DOI] [Google Scholar]
- 30.Zolgharnein J, Shahmoradi A, Ghasemi JB. Comparative study of Box-Behnken, central composite, and Doehlert matrix for multivariate optimization of Pb (II) adsorption ontoRobiniatree leaves. Chemometrics J. 2013;27(1-2):12–20. doi: 10.1002/cem.2487. [DOI] [Google Scholar]
- 31.Vásquez-Bahena JM, Vega-Estrada J, Santiago-Hernández JA, Ortega-López J, Flores-Cotera LB, Montes-Horcasitas MC, et al. Expression and improved production of the soluble extracellular invertase from Zymomonas mobilis in Escherichia coli. Enzyme Microb Tech. 2006;40(1):61–6. doi: 10.1016/j.enzmictec.2005.11.051. [DOI] [Google Scholar]
- 32.Peng L, Xu Z, Fang X, Wang F, Cen P. High-level expression of soluble human β-defensin-2 in Escherichia coli. Process Biochem. 2004;39(12):2199–205. doi: 10.1016/j.procbio.2003.11.011. [DOI] [Google Scholar]
- 33.Yildir C, Önsan ZI, Kirdar B. Optimization of starting time and period of induction and inducer concentration in the production of the restriction enzyme EcoRI from recombinant Escherichia coli 294. Turk Chem J. 1998;22(3):221–6. [Google Scholar]
- 34.Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodriguez-Carmona E, Baumann K, et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Azaman SNA, Ramanan RN, Tan JS, Rahim RA, Abdullah MP, Ariff AB. Screening for the optimal induction parameters for periplasmic producing interferon-α2b in Escherichia coli. Afr J Biotechnol. 2010;9(38):6345–54. [Google Scholar]
- 36.Donovan RS, Robinson CW, Glick BR. Review: optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter. J Ind Microbiol. 1996;16(3):145–54. doi: 10.1007/BF01569997. [DOI] [PubMed] [Google Scholar]
- 37.Mergulhao FJ, Monteiro GA. Analysis of factors affecting the periplasmic production of recombinant proteins in Escherichia coli. J Microbiol Biotechnol. 2007;17(8):1236–41. [PubMed] [Google Scholar]
- 38.Shiloach J, Rinas U. 2009. pp. 377–400.Glucose and Acetate Metabolism in E. coli – System Level Analysis and Biotechnological Applications in Protein Production Processes. [DOI] [Google Scholar]