Abstract
Zebrafish have become an invaluable vertebrate animal model to interrogate small molecule libraries for modulators of complex biological pathways and phenotypes. We have recently described the implementation of a quantitative, high-content imaging assay in multi-well plates to analyze the effects of small molecules on Fibroblast Growth Factor (FGF) signaling in vivo. Here we have evaluated the ability of the assay to identify compounds that hyperactivate FGF signaling from a test cassette of agents with known biological activities. Using a transgenic zebrafish reporter line for FGF activity, we screened 1040 compounds from an annotated library of known bioactive agents including FDA approved drugs. The assay identified two molecules, 8-hydroxyquinoline sulfate and pyrithione zinc that enhanced FGF signaling in specific areas of the brain. Subsequent studies revealed that both compounds specifically expanded FGF target gene expression. Furthermore, treatment of early stage embryos with either compound resulted in dorsalized phenotypes characteristic of hyperactivation of FGF signaling in early development. Documented activities for both agents included activation of extracellular signal-related kinase (ERK), consistent with FGF hyperactivation. To conclude, we demonstrate the power of automated quantitative high-content imaging to identify small molecule modulators of FGF.
Keywords: transgenic zebrafish, small molecule, FGF modulators, high-content analysis
Introduction
In recent years the interrogation of small molecule libraries in whole animal model systems has emerged as a rapid and efficient means to identify novel agents in the future treatment of disease (den Hertog, 2005; Gosai et al., 2010; Peterson et al., 2000; Tomlinson et al., 2009; Tsang, 2010; White et al., 2011; Zon and Peterson, 2005). Among different animal models, zebrafish have attracted most attention owing to their cost effective maintenance, high fecundity, and rapid development. Above all, zebrafish embryos are optically transparent at early stages, which facilitates real-time monitoring of development and early organogenesis. This makes zebrafish a perfect model for automated in vivo high-content analysis (HCA). The most commonly used method to identify compounds that cause a desired biological effect is visual examination of live embryos for phenotypic changes or for changes in gene expression by in situ hybridization (de Groh et al., 2010; Murphey et al., 2006; North et al., 2007; Peterson et al., 2000; Stern et al., 2005; Yeh et al., 2009). However, the relative ease of introducing exogenous reporter genes such as green fluorescent protein (GFP) into zebrafish has enabled both real-time analysis of embryonic development and the observation of molecular events through changes in reporter gene expression. Transgenic embryos that report on Retinoic Acid, Notch, Bone Morphogenetic Protein, Wnt and Nodal signaling are available and serve as biosensors of the activity of these pathways during development (Collery and Link, 2011; Laux et al., 2011; Lorent et al., 2010; Parsons et al., 2009; Perz-Edwards et al., 2001; Swanhart et al., 2010). Our lab has generated a number of transgenic zebrafish lines that serve as reporters for Fibroblast Growth Factor (FGF) signaling through the use of an FGF target gene, Dual Specificity Phosphatase 6 (Dusp6), promoter to drive GFP expression (Molina et al., 2007; Wang et al., 2011). We validated the Tg(dusp6:EGFP) line as a reporter for FGF activity by treating transgenic embryos with known inhibitors of the FGF pathway and showed that GFP expression was suppressed (Molina et al., 2007; Znosko et al., 2010). Moreover, a manual chemical screen of over 5000 compounds identified a novel agent, (E)-2-benzylidene-3-(cyclohexylamino)-2,3-dihydro-1H-inden-1-one (BCI), that enhanced FGF activity (Molina et al., 2009). Recently, we extended these studies by developing automated HCA methodology that permits quantification of reporter gene expression in Tg(dusp6:EGFP) embryos (Vogt et al., 2009; Vogt et al., 2010). The improved method accurately quantified a robust, dose-dependent induction of GFP expression in BCI-treated transgenic embryos. Here we further validated the automated imaging and analysis system by examining its ability to identify FGF hyperactivators from a library of small molecules with known biological activities. We treated Tg(dusp6:EGFP) embryos with 1040 well-characterized compounds from the MicroSource Discovery Systems US Drug Collection and identified two molecules, 8-hydroxyquinoline hemisulfate (Oxyq) and pyrithione zinc (PYZ), which increased GFP expression in specific areas of the brain previously shown to be under the control of active FGF signaling. In follow-up studies, treatment of early stage embryos with either compound resulted in mild dorsalized phenotypes that are known to occur when the FGF pathway is hyperactivated. In addition, expression of dusp6 was mildly expanded in treated embryos, suggesting that both compounds are moderately active enhancers of FGF signaling. The results document the ability of our previously developed HCA method to identify weak activators of FGF signaling from a small molecule library, which would have been difficult if not impossible with manual visual scoring. The results suggest HCA could be a useful tool in the discovery of novel FGF activators from chemically diverse collections of small molecules.
Results
Automated high-content screen for FGF modulators
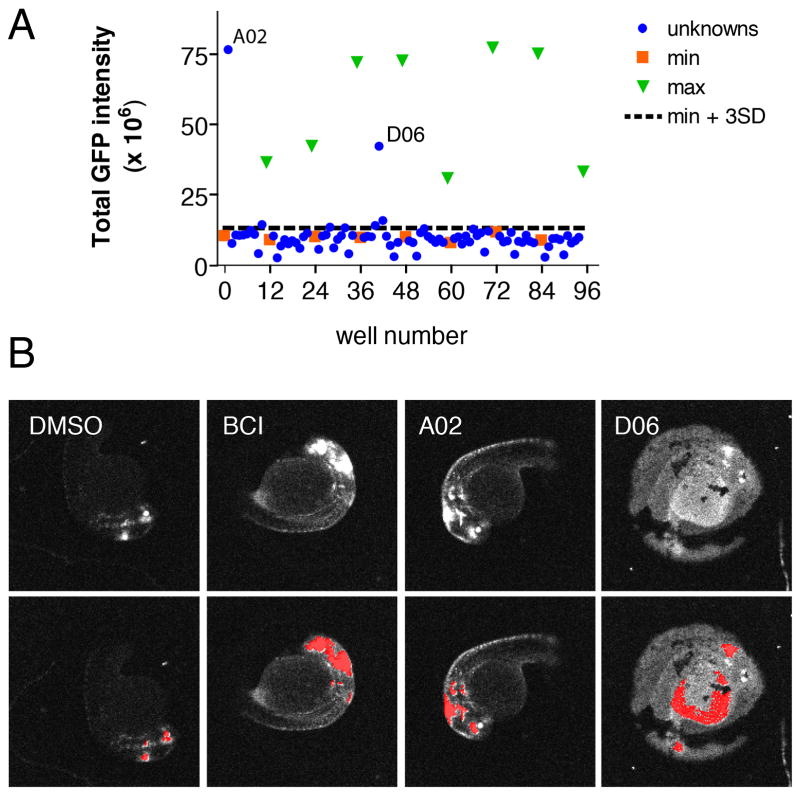
We previously described methodology to automate both image capture and analysis of reporter gene expression in transgenic zebrafish embryos arrayed in multi-well plates (Vogt et al., 2010). Using cognition network technology (CNT, Definiens AG, Germany), we developed a ruleset that quantitatively measured GFP expression in Tg(dusp6:EGFP) embryos. The ruleset delivered graded responses and quantified a dose-dependent increase in GFP expression upon BCI treatment, with a concentration required to elicit a half-maximal response (EC50) of 8.5 μM and saturation above 20 μM (Vogt et al., 2010). We next screened the US drug collection set from MicroSource Discovery Systems, Inc., which contains 1040 diverse chemical structures that have either been approved or have reached clinical trial stages in the USA. Our goal was to validate the quantitative imaging assay by asking whether members of a known well-characterized library of small molecules could hyperactivate FGF signaling. One Tg(dusp6:EGFP) embryo at 24 hours post fertilization (hpf) was arrayed into each well of a 96-well plate containing 30μM of sample compound. For each compound, duplicate assay plates were run in parallel in order to limit biological variability and to nullify the effects of loading artifacts, toxicity, and random off-target fluorescence in the embryos or wells. On each plate, both negative (DMSO) and positive (10 μM BCI) controls were included as to serve as the baseline (minimum) and maximal response, respectively. Embryos were incubated for 5 hours embryos followed by imaging as described in the Materials and Methods Section. GFP fluorescent images were acquired and archived before processing with the CNT ruleset to quantitate GFP intensity. For each well, the total integrated GFP intensity in the head was calculated and GFP intensity from the two replicate plates was averaged. Positives were identified as embryos exhibiting GFP intensity at least three fold greater than the standard deviation (SD) from the mean of all of the samples on the microplate excluding controls (i.e. a z-score > 3), a commonly used hit selection criterion based on plate statistics (Malo et al., 2006). A second criterion was that the compound had to show activity in both duplicate plates in order to be designated as a “hit”. Figure 1A shows a representative plate graph with DMSO control (orange squares), BCI positive controls (green triangles) and sample compounds (blue circles). Visually there was a clear separation of negative and positive controls. The signal to background ratio of BCI to DMSO was 5.7. The coefficients of variation (CV) for the minimum and maximum controls were 12% and 38%, respectively, indicating that the positive controls were the major source of variability. To gauge the degree of separation, we calculated the strictly standardized mean difference (SSMD) between the negative and positive controls, respectively. The SSMD, defined as
Fig 1.
Automated chemical screen in zebrafish identifies Oxyq as a potential activator of FGF signaling. (A) Graph showing mean GFP intensity from two plates together with the mean + 3 SD limits of the negative control wells (B) Morphology of Tg(dusp6:d2eGFP) embryos in the presence of negative control (DMSO), positive control (BCI) and 8-hydroxyquinoline hemisulfate (A02). Well D06 illustrates the appearance of a fluorescent yolk that is not the site of FGF activity and this “hit” was eliminated. Upper panels show original scan images; lower panels show images with CNT analysis applied.
is a statistical parameter that has recently been suggested to be appropriate for quality control of screening assays with high inherent variability, as it measures separation between two populations regardless of their shape of distribution, and has a solid statistical foundation and a clear probabilistic interpretation (Zhang, 2007; 2008). The SSMD provides a numerical value that denotes the size of the mean difference as multiples of the SD of the difference between two populations (Zhang, 2007). As a probability interpretation, SSMDs of 1, 2, and 3 indicate that the probabilities that a value from the first population exceeds a value from the second population are 0.5, 0.95, and 0.975, respectively when the data show a unimodal distribution with finite variance (for a detailed discussion see (Zhang, 2008)). The microplate presented in Figure 1 had an SSMD of 2.2, which would have passed plate QC criteria suggested by Zhang (2008). Consistent with these considerations, the agents in well A02 and D06 had z-scores in both runs that were well above the plate SSMD. Furthermore, as all of the images are archived for future analysis, positive hits can be verified by morphological observation to determine if GFP expression is localized to the known domains of FGF activity in the embryo. In Figure 1A, both wells A02 and D06 were considered potential positives. However, upon analysis of the zebrafish images, D06 was eliminated as the fluorescence was localized in the yolk. In contrast, A02 (8-hydroxyquinoline or isoquinoline, Oxyq) showed the expected increase in GFP expression in the mid-hindbrain boundary (MHB) and in hindbrain rhombomeres, a phenotype similar to BCI treatment (Fig. 1B). Using this approach, we identified a second small molecule activator of FGF (pyrithione zinc, PYZ) that was located on another plate (data not shown).
Oxyq and PYZ are weak activators of FGF signaling
A dose response study was undertaken to determine if these compounds resulted in a dose dependent increase in GFP expression and to derive the concentration required to elicit a half-maximal response (EC50). We exposed transgenic embryos to repurchased, authentical lots of Oxyq (1–50μM range) and PYZ (0.1–5 μM range). For each dose, eight embryos were treated and GFP intensity was measured as described above. We observed statistically significant increases in GFP intensity with both compounds (Fig 2). Numbers in bars shown normalized fold increase over DMSO. The magnitude of response, however, was small compared to BCI and precluded fitting of the curves to a four parameter logistic equation (Fig. 2). These results suggested that both Oxyq and PYZ were weak enhancers of FGF signaling.
Fig 2.
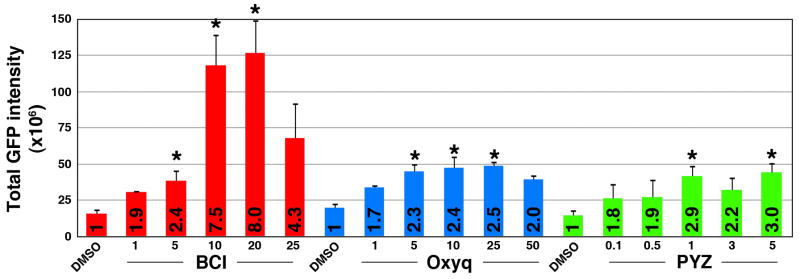
Oxyq and PYZ are mild activators of FGF signaling. A graphical representation of Tg(dusp6:d2eGFP) embryos after drug exposure. The total GFP intensity is expressed as normalized values and treatment response is represented as fold increase over the control (numbers in bars). BCI demonstrated an eight-fold increase whereas Oxyq and PYZ elicited two to three-fold increases in GFP intensity over the control. Treatment bars are represented as mean ± SE. Statistical significance was tested using Student’s t-test (two-tailed assuming unequal variances. *, p<0.05, n=8.
Oxyq is not a Dusp6 inhibitor
Dusp6 is a dual specificity phosphatase that specifically dephosphorylates extracellular signal-related kinase (ERK) and a known feedback inhibitor of the FGF pathway (Keyse, 2008). We previously determined that BCI activates FGF signaling through inhibition of Dusp6 activity (Molina et al., 2009). We therefore reasoned that Oxyq could enhance FGF signaling by inhibiting Dusp6 activity. Cell-based chemical complementation assays were performed to determine if Oxyq could suppress Dusp6 activity. Although BCI could block Dusp6 activity in this assay Oxyq had no effect at concentrations 10 times those that hyperactivated FGF signaling in zebrafish (Fig 3). Thus Oxyq does not appear to inhibit Dusp6 and likely has a mechanism of action different from BCI. At present it is not clear how Oxyq enhances FGF activity; future experiments should focus on this issue.
Fig 3.
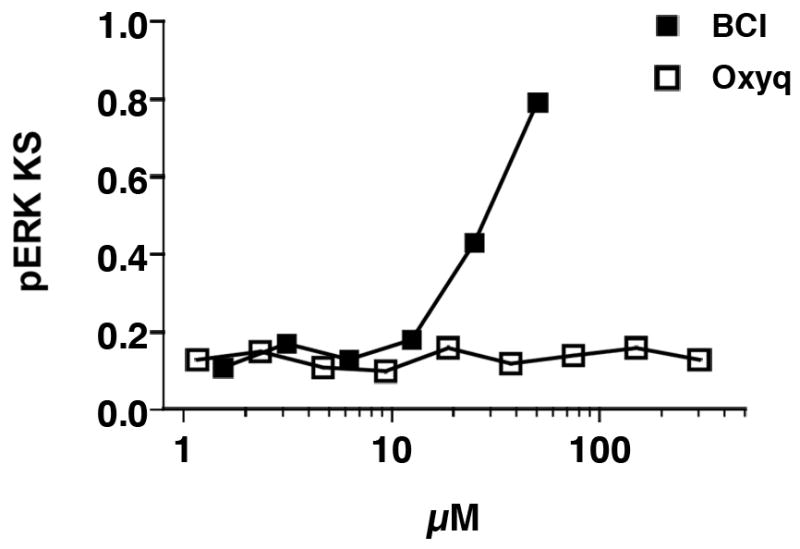
Oxyq is not a DUSP6 inhibitor in a cell based chemical complementation assay quantifying pERK levels in DUSP expressing cells. HeLa cells were transfected with c-Myc tagged DUSP6. Cells with high levels of c-Myc-tagged DUSP6 were identified based on c-Myc staining. pERK levels were measured in the DUSP expressing cells and a cumulative distribution function (cdf) assembled for each well. Cdfs of each well were compared to a reference distribution from 16 vehicle treated cells. Large KS values denote high dissimilarity from control and indicate high pERK levels in the DUSP expressing cells due to phosphatase inhibition. Data are the mean ± SE of four replicates from a single experiment that was repeated once with identical results.
Early embryos treated with Oxyq or PYZ are dorsalized
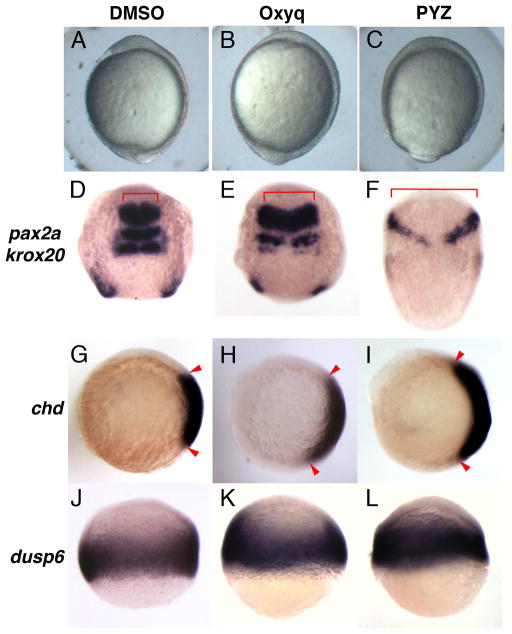
In order to understand the effect of Oxyq and PYZ on embryonic development, we treated with embryos from 1K-cell (3.3hpf) stage until early somitogenesis stage (12hpf). Treated embryos exhibited mildly dorsalized phenotypes (Fig 4 A–C), a phenotype that is similar to knockdown of dusp6 or microinjection of fgf8 mRNA (Furthauer et al., 2004; Tsang et al., 2004). In order to confirm the early effect of dorsalized phenotypes in compound treated embryos, we analyzed the expression of two neural markers, pax2a and krox20, which delineate the mid-hindbrain boundary and hindbrain rhombomeres 3 and 5, respectively (Krauss et al., 1991; Puschel et al., 1992; Woo and Fraser, 1998). Both Oxyq and PYZ resulted in the lateral expansion of pax2a and to a lesser extent, krox20 (also known as egr2b) (Fig. 4D–F). These results recapitulated the phenotypes observed by knockdown of dusp6, by BCI treatment or through injection of fgf8 mRNA (Furthauer et al., 2004; Molina et al., 2009; Tsang et al., 2004). To determine if Oxyq and PYZ treated embryos were dorsalized, we analyzed expression of chordin (chd) at shield stage. Chordin marks the dorsal organizer, and expansion of this gene is indicative of dorsalization. Oxyq and PYZ treated embryos showed a marked expansion of chd expression (Fig 4G–I). Finally, to confirm that FGF signaling was enhanced in Oxyq or PYZ treated embryos, dusp6 expression was measured by in situ hybridization. The FGF target gene, dusp6, was weakly expanded in Oxyq and PYZ treated embryos as compared to DMSO control (Fig. 4J–L). These experiments confirmed that both Oxyq and PYZ are weak enhancers of FGF signaling.
Fig 4.
Oxyq and PYZ treatment induced dorsalization in zebrafish. Morphology of zebrafish embryos treated with Oxyq and PYZ from 1K cells until early somitogenesis stage. The neural markers pax2a and krox20 showed lateral expansion (compare 4D to 4E and F). chd expression was expanded in treated groups over control (compare 4G to 4H and I). dusp6 expression showed mild expansion in OxyQ and PYZ treated groups over control.
Discussion
The generation of transgenic zebrafish reporter lines that respond to signaling pathways with spatially defined fluorescence expression provides a non-invasive tool for the identification of small molecules that modulate signal transduction pathways. HCA is an especially useful approach to measure such locally specific changes in fluorescent reporter gene activation. Besides its ability to detect and quantify fluorescence intensity in defined regions of the embryo, a critical component of HCA is the ability to archive images and data so that it is possible to reexamine each sample at a future date to confirm the quantitative analysis. This is especially useful when analyzing compound-induced changes in a multicellular organism, as it is possible to eliminate artifacts, rule out non-specific effects, and to assess toxicity. Furthermore, the multiparametric nature of HCA makes it uniquely suited to capture the rich and complex biology of a living organism by providing information about target activity, localization, and possibly functional activity, all in the same assay system. The zebrafish is quickly becoming a preferred subject for HCA due to its small size and optical transparency.
Compared with in situ hybridization, a clear advantage of HCA of fluorescent biosensors is the lack of additional labor-intensive post treatment sample processing of each sample as reporter gene expression can be measured directly in the living animal. This more than offsets the need to measure changes within a limited time window. Having shown that it is possible to automate the quantitation of transgene detection in multi-well plates, we now evaluated the ability of the previously developed, HCA assay to identify known or new FGF hyperactivating agents from a test cassette of agents with known biological activities (Vogt et al., 2010).
We selected the US Drug collection from MicroSource Discoveries as this library contains well-characterized compounds such that follow-up studies could be easily carried-out. From the library of 1040 compounds, we identified two positive hits, Oxyq and PYZ. Dose-response studies with repurchased, authentic material confirmed that both compounds were mild activators of FGF signaling as compared to BCI (Fig. 2). 8-hydroxyquinoline hemisulfate (Oxyq), a monoprotic bidentate chelating agent, exhibits antiseptic, disinfectant and pesticide properties (Heeb et al., 2011). Recently, Oxyq derivatives were found to up regulate MAPK signaling when Hela cells were treated in presence of bivalent copper as the chelating agent (Chen et al., 2009). In another report, Oxyq was identified as a molecule that activated ERK and p38 MAPKs, consistent with activation of FGF signaling (Giuliano et al., 2003). Because one known mechanism for ERK activation is the inhibition of MAPK phosphatases (Dusps), we tested Oxyq for inhibition of Dusp6 in mammalian cells but determined that Oxyq does not inhibit Dusp6. This suggests that Oxyq activates the MAPK signaling pathway at points other than phosphatase-mediated feedback regulation. The second positive found in the US Drug collection, PYZ (pyrithione zinc) a co-ordination complex of zinc that was recently described to play a cardioprotective role in ischemia/reperfusion studies using neonatal rat ventricular cardiomyocytes, presumably through ERB2 expression and PI3K/Akt activation (Viswanath et al., 2011). Another group reported that Zn2+ has a role in neuronal cell death through sustained activation of ERK1/2 (Ho et al., 2008). Since both Oxyq and PYZ seem to activate ERK signaling and because our reporter gene is regulated by FGFs, which signal through the ERK pathway, it is satisfying that our assay could successfully capture these molecules in an unbiased screen. Secondary phenotypic characterization studies demonstrated that both Oxyq and PYZ induced a dorsalized phenotype, similar to previous dusp6 knock-down experiments or through overexpression of fgf8 mRNA (Furthauer et al., 2004; Tsang et al., 2004). We have confirmed the dorsalization phenotypes by the lateral expansion of neural markers, pax2a and krox20, supporting the hypothesis that Oxyq and PYZ upregulate FGF signaling. It has been reported that hyperactivation of FGF can induce expression of the BMP4 inhibitor, chd (Koshida et al., 2002). Consistent with this, we showed here that treating embryos prior to gastrulation increased chd expression.
Conclusion
We have developed and validated an automated image capture and analysis routine to identify small molecules that modulate FGF signaling using zebrafish. By applying the system to analyzing an annotated small molecule library of 1040 agents with known biological activities, we identified two molecules, Oxyq and PYZ that mildly enhanced FGF signaling. Such mild activation of FGF signaling would be very difficult to discern with the naked eye. Thus, the work presented here validates the HCA assay as a drug discovery tool and demonstrates the power of automated, image-based analysis in whole animal chemical screens.
Materials and Methods
Zebrafish care and maintenance
All the procedures involving zebrafish were carried out with the prior review and approval by the University of Pittsburgh Institutional Animal Care Use committee. Heterozygous Tg(dusp6:EGFP)pt6 embryos (24hpf) were obtained by single pair homozygous out crossing (Molina et al., 2007). For phenotypic assays, embryos at 1000-cell stage were treated with 8-hydroxyquinoline (H6878 Sigma-Aldrich, St. Louis, MO) or pyrithione zinc (H6377 Sigma-Aldrich, St. Louis, MO) until embryos reached 1-somite stage.
Plate preparation and processing
Each well of a 96-well plate was loaded with a single transgenic embryo in 200 μL E3 (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4). Each microplate contained embryos from a single mating pair. The US drug collection (MicroSource Discovery Systems, Inc., Gaylordsville, CT) was screened at a final concentration of 30 μM in 1% DMSO. Each microplate contained eight wells of vehicle controls (1% DMSO) and eight wells of 10 μM BCI as the positive control (Molina et al., 2007). Each plate was scanned and analyzed in duplicates on the same day to eliminate loading and toxicity artifacts and to further limit variability. Plates were incubated at 28.5 °C for 5 hours. Subsequent dose response studies were carried out with repurchased authentic material to confirm the results from the primary screen and to determine the concentration required to elicit a half-maximal response (EC50).
Automated embryo imaging and analysis
Embryos were anesthetized with MS222 (0.61mM tricaine methanesulfonate, Sigma-Aldrich, St. Louis, MO) at the end of the drug treatment in order to restrict their movement during imaging. Images were acquired on an ImageXpress Ultra high-content reader (Molecular Devices, Sunnyvale, CA) using a 4X objective at excitation/emission wavelengths of 488/525 nm (GFP) (Vogt et al., 2010). Archived fluorescence micrographs were uploaded into Developer (Definiens AG) using the Cellenger module and analyzed for GFP expression in the head using a slightly modified version of our CNT algorithm developed as described previously (Vogt et al., 2010). The algorithm was designed to identify the zebrafish larva irrespective of their orientation in the well and a threshold defined based on overall larval fluorescence. Regions within the zebrafish larva were classified as positive for GFP expression if their fluorescence intensity exceeded this threshold. Total head structures brightness was defined as the integrated GFP intensity of the four brightest head structures.
Cell Culture
HeLa cells were obtained from ATCC (Manassas, VA), and were maintained in Dulbecco’s minimum essential medium containing 10% fetal bovine serum (HyClone, Logan, UT), and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA) in a humidified atmosphere of 5% CO2 at 37 °C.
Chemical complementation assay for DUSP6
Compounds were analyzed for inhibition of DUSP6 in intact cells as described (Molina et al., 2009; Vogt and Lazo, 2007). HeLa cells were transfected in 384 well plates with human c-Myc-DUSP6 using Fugene HD (Roche, Indianapolis, IN). After 48h, cells were treated in quadruplicate wells for 15 min with ten two-fold concentration gradients of BCI or 8-hydroxyquinoline and stimulated for 15 min. with phorbol ester (TPA, 500 ng/ml). Cells were immunostained with a mixture of anti-pERK (1:200 dilution) and anti-c-Myc (1:100 dilution) antibodies. Positive pERK and c-Myc-DUSP6 signals were visualized with AlexaFluor-594 (pERK) and Alexa-488 (c-Myc) conjugated secondary antibodies, respectively. Plates were analyzed by three-channel multiparametric analysis for pERK and c-Myc-DUSP6 intensities in an area defined by nuclear staining using the Target Activation Bioapplication on the ArrayScan II (Cellomics, Pittsburgh, PA). DUSP6 transfected cells were classified as expressors if their average c-Myc staining intensity exceeded a threshold defined as the mean intensity + 2 SD of untransfected cells. pERK levels were quantified in the DUSP6-expressing subpopulation by Kolmogorov-Smirnov (KS) statistics, comparing the cumulative pERK distribution of each test well to a reference distribution from 14 DUSP6-transfected and vehicle-treated wells. High KS values denote large differences in ERK phosphorylation levels compared with vehicle control and indicate suppression of DUSP6 activity. KS values were plotted against compound concentration and IC50 values calculated by fitting curves to a four parameter logistic equation, with the top defined by the maximum KS value obtained in the presence of the highest concentration of the positive control (BCI).
In situ hybridization
In situ protocols were carried out as described previously (Shima et al., 2009). Following probes generated in the previous studies were used in this study: dusp6 (Kawakami et al., 2004; Tsang et al., 2004), chordin (Schulte-Merker et al., 1997). krox20 (Kudoh et al., 2001), pax2a (Krauss et al., 1991).
Acknowledgments
Thanks to Umasankar K. Perunthottathu and Subramaniam Sanker for their help with the preparation of in situ probes. This work was supported by NIH grant NIH1R01HL088016 to MT and NIH1R01HD053287 to MT and AV.
References
- Chen HL, Chang CY, Lee HT, Lin HH, Lu PJ, Yang CN, Shiau CW, Shaw AY. Synthesis and pharmacological exploitation of clioquinol-derived copper-binding apoptosis inducers triggering reactive oxygen species generation and MAPK pathway activation. Bioorg Med Chem. 2009;17(20):7239–7247. doi: 10.1016/j.bmc.2009.08.054. [DOI] [PubMed] [Google Scholar]
- Collery RF, Link BA. Dynamic smad-mediated BMP signaling revealed through transgenic zebrafish. Dev Dyn. 2011;240(3):712–722. doi: 10.1002/dvdy.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groh ED, Swanhart LM, Cosentino CC, Jackson RL, Dai W, Kitchens CA, Day BW, Smithgall TE, Hukriede NA. Inhibition of histone deacetylase expands the renal progenitor cell population. J Am Soc Nephrol. 2010;21(5):794–802. doi: 10.1681/ASN.2009080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hertog J. Chemical genetics: Drug screens in Zebrafish. Biosci Rep. 2005;25(5–6):289–297. doi: 10.1007/s10540-005-2891-8. [DOI] [PubMed] [Google Scholar]
- Furthauer M, Van Celst J, Thisse C, Thisse B. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development. 2004;131(12):2853–2864. doi: 10.1242/dev.01156. [DOI] [PubMed] [Google Scholar]
- Giuliano KA, Haskins JR, Taylor DL. Advances in high content screening for drug discovery. Assay Drug Dev Technol. 2003;1(4):565–577. doi: 10.1089/154065803322302826. [DOI] [PubMed] [Google Scholar]
- Gosai SJ, Kwak JH, Luke CJ, Long OS, King DE, Kovatch KJ, Johnston PA, Shun TY, Lazo JS, Perlmutter DH, Silverman GA, Pak SC. Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin alpha1-antitrypsin Z. PLoS One. 2010;5(11):e15460. doi: 10.1371/journal.pone.0015460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Camara M. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev. 2011;35(2):247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Samarasinghe R, Knoch ME, Lewis M, Aizenman E, DeFranco DB. Selective inhibition of mitogen-activated protein kinase phosphatases by zinc accounts for extracellular signal-regulated kinase 1/2-dependent oxidative neuronal cell death. Mol Pharmacol. 2008;74(4):1141–1151. doi: 10.1124/mol.108.049064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7(1):133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27(2):253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- Koshida S, Shinya M, Nikaido M, Ueno N, Schulte-Merker S, Kuroiwa A, Takeda H. Inhibition of BMP activity by the FGF signal promotes posterior neural development in zebrafish. Developmental biology. 2002;244(1):9–20. doi: 10.1006/dbio.2002.0581. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development. 1991;113(4):1193–1206. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Tsang M, Hukriede NA, Chen X, Dedekian M, Clarke CJ, Kiang A, Schultz S, Epstein JA, Toyama R, Dawid IB. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11(12):1979–1987. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- Laux DW, Febbo JA, Roman BL. Dynamic analysis of BMP-responsive smad activity in live zebrafish embryos. Dev Dyn. 2011;240(3):682–694. doi: 10.1002/dvdy.22558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239(3):855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nature biotechnology. 2006;24(2):167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- Molina G, Vogt A, Bakan A, Dai W, Queiroz de Oliveira P, Znosko W, Smithgall TE, Bahar I, Lazo JS, Day BW, Tsang M. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol. 2009;5(9):680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina GA, Watkins SC, Tsang M. Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Dev Biol. 2007;7:62. doi: 10.1186/1471-213X-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Des. 2006;68(4):213–219. doi: 10.1111/j.1747-0285.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, Daley GQ, Orkin SH, Zon LI. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447(7147):1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, Leach SD. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126(10):898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz-Edwards A, Hardison NL, Linney E. Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev Biol. 2001;229(1):89–101. doi: 10.1006/dbio.2000.9979. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97(24):12965–12969. doi: 10.1073/pnas.97.24.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschel AW, Westerfield M, Dressler GR. Comparative analysis of Pax-2 protein distributions during neurulation in mice and zebrafish. Mechanisms of development. 1992;38(3):197–208. doi: 10.1016/0925-4773(92)90053-m. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Lee KJ, McMahon AP, Hammerschmidt M. The zebrafish organizer requires chordino. Nature. 1997;387(6636):862–863. doi: 10.1038/43092. [DOI] [PubMed] [Google Scholar]
- Shima T, Znosko W, Tsang M. The characterization of a zebrafish mid-hindbrain mutant, mid-hindbrain gone (mgo) Dev Dyn. 2009;238(4):899–907. doi: 10.1002/dvdy.21916. [DOI] [PubMed] [Google Scholar]
- Stern HM, Murphey RD, Shepard JL, Amatruda JF, Straub CT, Pfaff KL, Weber G, Tallarico JA, King RW, Zon LI. Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol. 2005;1(7):366–370. doi: 10.1038/nchembio749. [DOI] [PubMed] [Google Scholar]
- Swanhart LM, Takahashi N, Jackson RL, Gibson GA, Watkins SC, Dawid IB, Hukriede NA. Characterization of an lhx1a transgenic reporter in zebrafish. Int J Dev Biol. 2010;54(4):731–736. doi: 10.1387/ijdb.092969ls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson ML, Rejzek M, Fidock M, Field RA, Wheeler GN. Chemical genomics identifies compounds affecting Xenopus laevis pigment cell development. Mol Biosyst. 2009;5(4):376–384. doi: 10.1039/b818695b. [DOI] [PubMed] [Google Scholar]
- Tsang M. Zebrafish: A tool for chemical screens. Birth Defects Res C Embryo Today. 2010;90(3):185–192. doi: 10.1002/bdrc.20183. [DOI] [PubMed] [Google Scholar]
- Tsang M, Maegawa S, Kiang A, Habas R, Weinberg E, Dawid IB. A role for MKP3 in axial patterning of the zebrafish embryo. Development. 2004;131(12):2769–2779. doi: 10.1242/dev.01157. [DOI] [PubMed] [Google Scholar]
- Viswanath K, Bodiga S, Balogun V, Zhang A, Bodiga VL. Cardioprotective effect of zinc requires ErbB2 and Akt during hypoxia/reoxygenation. Biometals. 2011;24(1):171–180. doi: 10.1007/s10534-010-9371-8. [DOI] [PubMed] [Google Scholar]
- Vogt A, Cholewinski A, Shen X, Nelson SG, Lazo JS, Tsang M, Hukriede NA. Automated image-based phenotypic analysis in zebrafish embryos. Dev Dyn. 2009;238(3):656–663. doi: 10.1002/dvdy.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, Codore H, Day BW, Hukriede N, Tsang M. Development of automated imaging and analysis for zebrafish chemical screens. JoVE. 2010;40 doi: 10.3791/1900. http://www.jove.com/index/details.stp?id=1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, Lazo JS. Implementation of high-content assay for inhibitors of mitogen-activated protein kinase phosphatases. Methods. 2007;42(3):268–277. doi: 10.1016/j.ymeth.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Cadwallader AB, Jang DS, Tsang M, Yost HJ, Amack JD. The Rho kinase Rock2b establishes anteroposterior asymmetry of the ciliated Kupffer’s vesicle in zebrafish. Development. 2011;138(1):45–54. doi: 10.1242/dev.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, Chen F, Long HK, Kramer M, Datta S, Neuberg D, Granter S, Young RA, Morrison S, Wheeler GN, Zon LI. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471(7339):518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K, Fraser SE. Specification of the hindbrain fate in the zebrafish. Developmental biology. 1998;197(2):283–296. doi: 10.1006/dbio.1998.8870. [DOI] [PubMed] [Google Scholar]
- Yeh JR, Munson KM, Elagib KE, Goldfarb AN, Sweetser DA, Peterson RT. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol. 2009;5(4):236–243. doi: 10.1038/nchembio.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 2007;89(4):552–561. doi: 10.1016/j.ygeno.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Zhang XD. Novel analytic criteria and effective plate designs for quality control in genome-scale RNAi screens. Journal of biomolecular screening. 2008;13(5):363–377. doi: 10.1177/1087057108317062. [DOI] [PubMed] [Google Scholar]
- Znosko WA, Yu S, Thomas K, Molina GA, Li C, Tsang W, Dawid IB, Moon AM, Tsang M. Overlapping functions of Pea3 ETS transcription factors in FGF signaling during zebrafish development. Dev Biol. 2010;342(1):11–25. doi: 10.1016/j.ydbio.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4(1):35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]