Abstract
Proper levels of Hedgehog (HH) signaling are essential during embryonic development and adult tissue homeostasis. A central mechanism to control HH pathway activity is through the regulation of secreted HH ligands at the plasma membrane. Recent studies have revealed a collective requirement for the cell surface co-receptors GAS1, CDON and BOC in HH signal transduction. Despite their requirement in HH pathway function, the mechanisms by which these proteins act to promote HH signaling remain poorly understood. Here we focus on the function of the two structurally related co-receptors, CDON and BOC. We utilized an in vivo gain-of-function approach in the developing chicken spinal cord to dissect the structural requirements for CDON and BOC function in HH signal transduction. Notably, we find that although CDON and BOC display functional redundancy during HH-dependent ventral neural patterning, these molecules utilize distinct molecular mechanisms to execute their HH-promoting effects. Specifically, we define distinct membrane attachment requirements for CDON and BOC function in HH signal transduction. Further, we identify novel and separate extracellular motifs in CDON and BOC that are required to promote HH signaling. Together, these data suggest that HH co-receptors employ distinct mechanisms to mediate HH pathway activity.
Keywords: CDON, BOC, neural patterning, Hedgehog, co-receptor
INTRODUCTION
Hedgehog (HH) signaling is required throughout embryonic development for the proper patterning and growth of a multitude of tissues and organs (McMahon et al, 2003). The actions of secreted HH ligands are tightly regulated at the cell surface through complex interactions with myriad cell surface-associated and extracellular matrix proteins (reviewed in (Briscoe & Therond, 2013). Core cell surface components of the HH signaling pathway include the canonical twelve-pass transmembrane receptor Patched (PTCH1) that antagonizes the function of the GPCR-like protein Smoothened (SMO), which mediates downstream signal transduction upon HH pathway activation (Chen & Struhl, 1996; Marigo et al, 1996; Stone et al, 1996; van den Heuvel & Ingham, 1996). In addition to these core components that are conserved across species, we have recently defined an essential role for two vertebrate-specific cell surface antagonists of the pathway, HH-interacting protein 1 (HHIP1) and PTCH2, that act redundantly with PTCH1 to limit HH pathway function (Holtz et al, 2013). These molecules oppose the activity of three HH pathway co-receptors: CAM-related/down-regulated by oncogenes (CDON), brother of CDON (BOC), and growth arrest-specific 1 (GAS1) that together are essential to transduce HH signaling during embryonic development (Allen et al, 2011). Despite their requirement in vertebrate HH signal transduction, the mechanisms by which these cell surface components control HH signaling remain largely unexplored.
CDON and BOC are conserved from Drosophila (ihog and boi) to mammals (Kang et al, 2002; Lum et al, 2003). These proteins consist of a series of extracellular immunoglobulin (IG) and fibronectin type III (FN) domains, a single-pass transmembrane (TM) domain, and a large, divergent cytoplasmic (CD) domain (Kang et al, 1997; Kang et al, 2002). Recent studies in Drosophila have identified essential roles for ihog and boi in HH signal transduction in the wing imaginal disc (Camp et al, 2010; Zheng et al, 2010). However, in mammals, these co-receptors function redundantly with a third, vertebrate-specific cell surface protein, GAS1, to mediate HH-dependent ventral neural patterning (Allen et al, 2011). These co-receptors function not only during spinal cord development, but also in other HH-dependent processes, including cerebellar development, digit specification, and craniofacial development (Allen et al, 2011; Allen et al, 2007; Cole & Krauss, 2003; Izzi et al, 2011; Zhang et al, 2011). CDON mutations have been identified in human holoprosencephaly (Bae et al, 2011), while craniofacial defects in Cdon mutant mice can be modified by environmental factors (Hong & Krauss, 2013) as well as Boc deletion (Zhang et al, 2011). In contrast to the redundancy observed during craniofacial development, BOC, but not CDON, mediates SHH-mediated axon guidance (Fabre et al, 2010; Okada et al, 2006).
Currently, the prevailing paradigm is that CDON and BOC promote HH signaling through calcium-dependent interactions with HH ligands via a membrane-proximal FN domain, FNIII(3) (McLellan et al, 2008), and interactions with the canonical receptor PTCH1 mediated by two distal FN repeats (Bae et al, 2011; Izzi et al, 2011). However, to date, a comprehensive assessment of the structural determinants in CDON and BOC that are required to mediate HH pathway function, have not been explored.
Here we dissect the domains of CDON and BOC that are required to promote HH signaling through detailed structure-function analyses in the developing spinal cord. We define multiple motifs in these proteins that are required to promote HH signaling. Surprisingly, we find that CDON and BOC require different modes of membrane attachment and utilize distinct extracellular domains to mediate HH pathway function. Together, these data indicate that CDON and BOC employ separate mechanisms to promote HH pathway function.
RESULTS
Distinct membrane attachment requirements for CDON- and BOC-mediated promotion of HH-dependent neural patterning
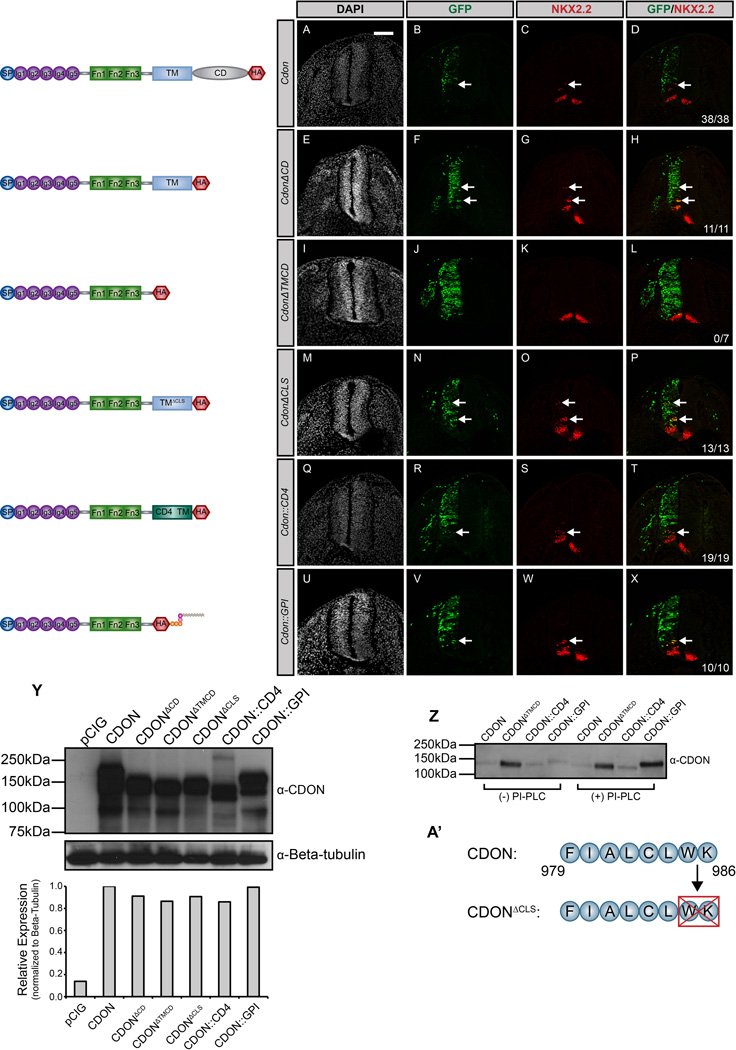
To dissect the structural requirements of CDON and BOC in HH pathway function, we utilized a gain-of-function approach in the developing chicken spinal cord, one of the best-studied sites of HH signaling. In agreement with previous studies (Allen et al, 2011; Tenzen et al, 2006), electroporation of a full-length Cdon construct promotes cell autonomous ectopic expression of the HH-dependent interneuron progenitor (pV3) marker NKX2.2 (Fig. 1A–D). Similarly, electroporation of a truncated Cdon construct lacking the cytoplasmic domain (CdonΔCD) also promotes ectopic NKX2.2 expression (Fig. 1E–H). In contrast, electroporation of a secreted form of CDON (CdonΔTMCD) does not alter HH-dependent neural patterning (Fig. 1I–L). Together with previous work (Tenzen et al, 2006), these data suggest that membrane anchoring of CDON is required to mediate HH signal transduction.
Figure 1. General membrane tethering is sufficient for CDON-mediated HH signaling.
(A–X) Antibody detection of NKX2.2 (red) in forelimb level transverse sections of Hamburger-Hamilton stage 21–22 chicken neural tubes electroporated with Cdon (A–D), CdonΔCD (E–H), CdonΔTMCD (I–L), CdonΔCLS (M–P), Cdon::CD4 (Q–T), and Cdon::GPI (U–X). GFP+ cells (green) designate electroporated cells and DAPI (grayscale) marks nuclei. Merged images are shown on the right, including quantitation of the number of embryos that display ectopic NKX2.2 expression (denoted by arrows). Scale bar, 10µm. (Y) Western blot analysis of COS-7 cell lysates following transfection with the specified constructs and probed with anti-CDON antibody and anti-Beta-tubulin as a loading control. ImageJ quantitation of relative expression levels is expressed as a histogram below the western blots. (Z) Western blot analysis of cell supernatants collected from COS-7 cells transfected with Cdon, CdonΔTMCD, Cdon::CD4, and Cdon::GPI. Cells were untreated (left lanes) or PI-PLC treated (right lanes) prior to supernatant collection. (A’) Schematic illustrating membrane-proximal amino acids that were deleted to generate the CdonΔCLS construct.
Previous studies have described the ciliary localization of membrane-associated HH pathway components, including SMO, PTCH1 and PTCH2 (Corbit et al, 2005; Holtz et al, 2013; Rohatgi et al, 2007; Santos & Reiter, 2014). While we do not detect localization of CDON in primary cilia (data not shown), CDON does possess a potential cilia localization sequence at the carboxy-terminus of the transmembrane domain (Fig. 1A’)(Corbit et al, 2005). Notably deletion of this sequence in CDON (CdonΔCLS) does not compromise the promotion of HH-dependent neural patterning (Fig. 1M–P).
Given the requirement for CDON membrane attachment in the promotion of HH signaling, we next sought to assess whether the mode of membrane attachment affects CDON function. Replacement of the transmembrane domain of CDON with the well-characterized transmembrane domain of the cell surface protein CD4 (Cdon::CD4) (Maddon et al, 1985) does not alter ectopic HH-dependent NKX2.2 specification (Fig. 1Q–T). Remarkably, expression of a GPI-anchored CDON protein (Cdon::GPI) also promotes ectopic HH pathway activation (Fig. 1U–X). Significantly, we observed identical results with immunodetection of other HH pathway targets, including the motor neuron progenitor (pMN) marker OLIG2 (Fig. S1) as well as the class-II transcription factor NKX6.1 (Fig. S2).
We utilized PI-PLC treatment (Low, 1987) to confirm that CDON::GPI is anchored to the cell surface through phosphatidylinositol (Fig. 1Z). Secreted CDON (CdonΔTMCD) is constitutively detected in cell supernatants, while CDON::GPI accumulates only in PI-PLC-treated supernatants. In contrast, PI-PLC treatment does not promote the accumulation of either full-length CDON or CDON::CD4 (Fig. 1Z).
We also employed SDS-PAGE and western blot analysis to determine the relative expression of each Cdon construct (Fig. 1Y). Further, we engineered a Hemagglutinin (HA) epitope tag into each construct to permit protein detection in situ in the chicken neural tube (Fig. S3A–L). Collectively, these data suggest that while CDON requires membrane attachment to promote HH signaling, there are no specific requirements in the type of membrane association needed to mediate CDON-dependent HH pathway activation.
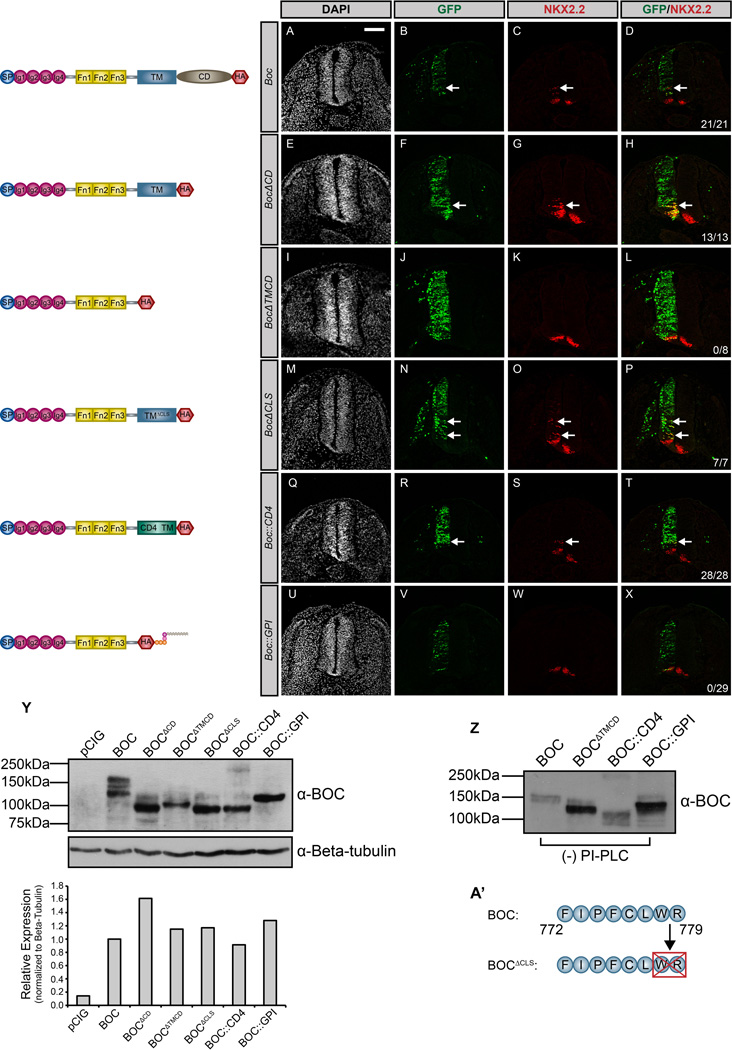
To assess whether membrane anchoring is generally required for other HH co-receptors, we performed an analogous set of experiments for BOC (Fig. 2). Similar to CDON, full-length BOC promotes cell-autonomous ectopic neural progenitor specification (Fig. 2A–D). Additionally, the cytoplasmic domain of BOC (BocΔCD) is dispensable for the promotion of HH signaling (Fig. 2E–H), while a secreted version of BOC (BocΔTMCD) is unable to mediate ectopic HH-dependent progenitor specification (Fig. 2I–L). Further, deletion of a putative cilia localization sequence at the carboxy-terminus of the BOC transmembrane domain (BocΔCLS) does not perturb BOC function (Fig. 2M–P, A’). These data suggest that BOC and CDON both require membrane tethering to promote HH pathway function.
Figure 2. BOC requires integral membrane anchoring to mediate HH signal transduction.
(A–X) Forelimb level transverse sections of Hamburger-Hamilton stage 21–22 chicken neural tubes electroporated with Boc (A–D), BocΔCD (E–H), BocΔTMCD (I–L), BocΔCLS (M–P), Boc::CD4 (Q–T), and Boc::GPI (U–X) stained with anti-NKX2.2 antibody (red). GFP+ cells (green) denote electroporated cells and nuclei are stained with DAPI (grayscale). Merged images are shown at the right, including quantitation of the number of embryos that display ectopic NKX2.2 expression (denoted by arrows). Scale bar, 10µm. (Y) Western Blot Analysis of COS-7 cell lysates following transfection with the specified constructs and probed with anti-BOC antibody and anti-Beta-tubulin as a loading control. ImageJ quantitation of relative expression levels is expressed as a histogram below the western blots. (Z) Western blot analysis of cell supernatants collected from COS-7 cells transfected with Boc, BocΔTMCD, Boc::CD4, and Boc::GPI. (A’) Schematic illustrating membrane-proximal amino acids that were deleted to generate the BocΔCLS construct.
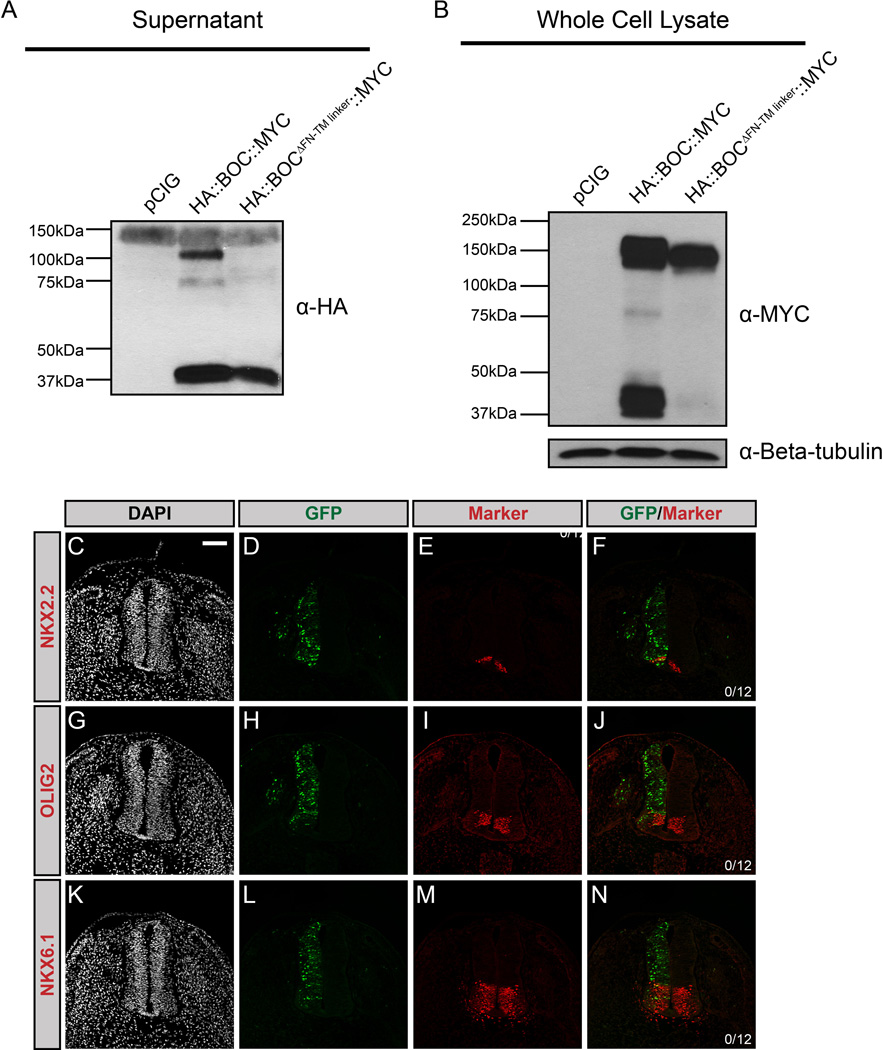
To further assess the requirement for BOC membrane tethering, we generated Boc::CD4 and Boc::GPI constructs. Electroporation of Boc::CD4 promotes ectopic NKX2.2+ cell specification in the ventral neural tube (Fig. 2Q–T). However, Boc::GPI does not induce HH pathway activation (Fig. 2U–X). This failure to promote signaling was not due to reduced protein expression, as western blot analysis indicated equal expression of full-length BOC and BOC::GPI (Fig. 2Y). Further, we used antibody detection of epitope (HA)-tagged BOC in the chicken neural tube to confirm BOC::GPI expression (Fig. S4A–L). However, western blot analysis revealed that secreted BOC protein is detected in supernatants collected from cells expressing full-length BOC, as well as BOC::CD4 and BOC::GPI, even in the absence of PI-PLC treatment (Fig. 2Z). These data contrast with CDON (cf. Fig. 1Z), and suggest that BOC undergoes proteolytic cleavage, resulting in its release from the cell surface. Notably, sequence analysis of BOC and CDON identified a putative matrix metallopeptidase 9 recognition site present in a juxtamembrane region of BOC, but not CDON (Song et al, 2012). To test whether this represents a functional cleavage site, we generated a Boc construct in which the juxtamembrane sequence containing the putative protease cleavage site was deleted (BocΔFN-TM linker). We also included an N-terminal HA epitope tag and a C-terminal MYC tag to validate this result. Strikingly, we detected two populations of secreted BOC (~100kDa and ~37kDa) in supernatants from cells expressing full-length BOC (HA::Boc::MYC; Fig. 3A). In contrast, deletion of the juxtamembrane region (HA::BocΔFN-TM linker::MYC) abrogates the detection of the ~100kDa band in cell supernatants (Fig. 3A). Similarly, we detected a truncated (~37kDa) C-terminal BOC fragment in whole cell lysates from cells expressing full-length BOC (Fig. 3B), that is absent in lysates from cells expressing a version of BOC lacking the putative cleavage site (Fig. 3B). These data suggest an extracellular cleavage event that results in release of a large BOC fragment from the cell surface and the generation of a truncated, membrane tethered cytoplasmic fragment. Importantly, electroporation of this construct (HA::BocΔFN-TM linker::MYC) completely abrogates BOC-mediated promotion of HH signaling in the developing chicken neural tube, including the specification of targets that require low levels of HH signaling (Fig. 3C–N). These data indicate that, similar to CDON, membrane tethering of BOC is a key determinant of its function. However, unlike CDON, BOC contains a putative extracellular cleavage site that may regulate its activity during HH-dependent neural patterning.
Figure 3. BOC undergoes extracellular proteolytic cleavage mediated by a juxtamembrane domain.
(A) Western blot analysis of supernatants from COS-7 cells transfected with pCIG, HA::Boc::MYC, or HA::BocΔFN-TM linker::MYC, probed with anti-HA antibody. Note the presence of a 100kDa band in the HA::BOC::MYC lane, but not the HA::BOCΔFN-TM linker::MYC lane, while a 37kDa band is detected in both lanes. (B) Western blot analysis of whole cell lysates probed with anti-MYC antibody and anti-Beta-tubulin as a loading control. (C–N) Forelimb level transverse sections of Hamburger-Hamilton stage 21–22 chicken neural tubes electroporated with BocΔFN-TM linker, and stained with DAPI (grayscale; C, G, K), and antibodies against NKX2.2 (red; E, F), OLIG2 (red; I, J), or NKX6.1 (red; M, N). GFP+ cells denote electroporated cells (green; D, H, L). Merged images are shown at the right, including quantitation of the number of embryos that display ectopic NKX2.2, OLIG2 or NKX6.1 expression. Scale bar (C), 10µm.
The FNIII domains of CDON are necessary and sufficient to promote the HH signaling
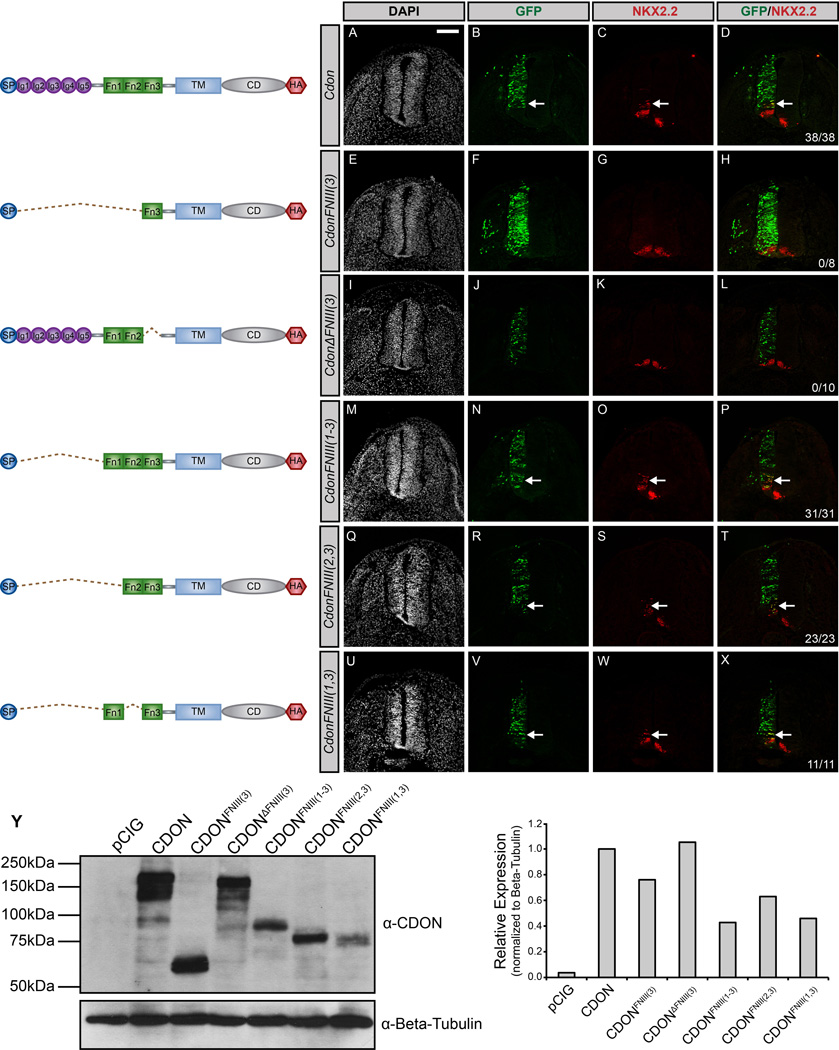
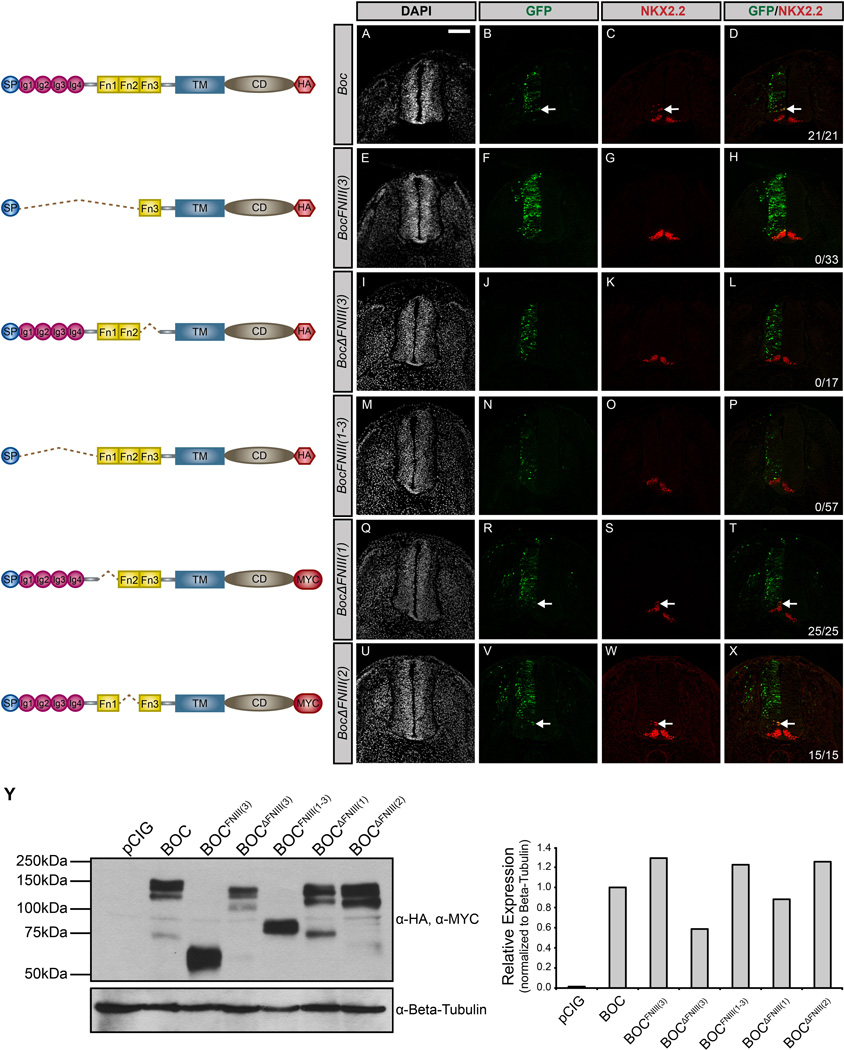
The different membrane attachment requirements for CDON and BOC function suggest that these proteins do not convey HH signals identically. Instead these data suggest a model where distinct elements in the extracellular domains of CDON and BOC contribute to the promotion of HH pathway activity. To test this notion, we again utilized gain-of-function experiments in the chicken neural tube (Fig. 4). The extracellular domain of CDON consists of five amino-terminal IG domains and three membrane-proximal FNIII repeats (Kang et al, 1997). In support of previous studies (Tenzen et al, 2006), and in contrast to full-length CDON (Fig. 4A–D), deletion of the HH ligand-binding FNIII(3) domain of CDON [CdonΔFNIII(3)] abrogates ectopic HH-dependent neural progenitor specification (Fig. 4I–L). Further, the FNIII(3) domain of CDON is not sufficient to promote HH pathway activation in the chicken neural tube (Fig. 4E–H). These data suggest that additional extracellular domains are required to mediate CDON function. Recent studies indicate that the FNIII(1) and FNIII(2) domains of CDON and BOC mediate interactions with the canonical HH receptor PTCH1, as well as with all three co-receptors (GAS1, CDON and BOC) (Bae et al, 2011; Izzi et al, 2011). To test whether the FNIII domains of CDON are sufficient to stimulate HH signaling, we generated a construct lacking all five IG domains [CdonFNIII(1–3)]. Our data indicate that CdonFNIII(1–3) is indeed sufficient to promote HH-dependent neural patterning (Fig. 4M–P). To examine potential functional redundancy between the FNIII(1) and FNIII(2) repeats, we generated two additional constructs: CdonFNIII(2,3) and CdonFNIII(1,3). Following electroporation in the chicken neural tube, either construct promotes ectopic NKX2.2+ cell specification (Fig. 4Q–X), suggesting that, together with the ligand-binding FNIII(3) domain, either the FNIII(1) or the FNIII(2) domain of CDON is sufficient to promote HH signaling.
Figure 4. The FNIII domains of CDON are sufficient to promote HH signaling.
(A–X) Forelimb level transverse sections of Hamburger-Hamilton stage 21–22 chicken neural tubes electroporated with Cdon (A–D), CdonFNIII(3) (E–H), CdonΔFNIII(3) (I–L), CdonFNIII(1–3) (M–P), CdonFNIII(2,3) (Q–T), and CdonFNIII(1,3) (U–X) stained with anti-NKX2.2 antibody (red). GFP expression (green) indicates electroporated cells and DAPI (grayscale) identifies nuclei. Merged images are shown on the right, including quantitation of the number of embryos that display ectopic NKX2.2 expression (denoted by arrows). Scale bar, 10µm. (Y) Western blot analysis of COS-7 cell lysates following transfection with the specified constructs and probed with anti-CDON antibody and anti-Beta-tubulin as a loading control (left panel). ImageJ quantitation of relative expression levels is expressed as a histogram (right panel).
Differential extracellular requirements for BOC-mediated promotion of HH signaling
We employed the same approach to evaluate the extracellular requirements for BOC-mediated promotion of HH signaling. Again, full-length BOC induces cell autonomous expression of ectopic NKX2.2 (Fig. 5A–D); similar to CDON, the HH ligand-binding domain of BOC, FNIII(3), is necessary, but not sufficient to promote HH signaling (Fig. 5E–L). Strikingly, and in contrast to CDON, expression of a construct containing all three BOC FNIII domains, BocFNIII(1–3), does not activate HH signaling (Fig. 5M–P). Importantly, BOCFNIII(1–3) protein is expressed equivalently to full-length BOC protein in vitro as assessed by western blot analysis (Fig. 5Y), while immunofluorescent staining confirms expression of BOCFNIII(1–3) in the chicken neural tube (S4Q–R). These data suggest that BOC requires additional extracellular determinants to promote HH signaling. Notably, individual deletion of either the FNIII(1) or FNIII(2) domains of BOC does not alter BOC function (Fig. 5Q–X), suggesting that unlike the FNIII(3) domain, these domains are not required to promote HH signal transduction. We also generated a construct deleting both FNIII(1) and FNIII(2); however, this construct, BocΔFNIII(1,2), is not efficiently expressed in the chicken neural tube (data not shown), precluding further analysis of the combined requirement for these domains. Taken together, these results support the view that, compared to CDON, BOC requires additional extracellular determinants to successfully activate the HH pathway.
Figure 5. The FNIII domains of BOC are not sufficient to promote HH signaling.
(A–X) Forelimb level transverse sections of Hamburger-Hamilton stage 21–22 chicken neural tubes electroporated with Boc (A–D), BocFNIII(3) (E–H), BocΔFNIII(3) (I–L), BocFNIII(1–3) (M–P), BocΔFNIII(1) (Q–T), and BocΔFNIII(2) (U–X) stained with anti-NKX2.2 antibody (red). Green (GFP+) cells denote electroporated cells and nuclei are identified by DAPI (grayscale). Merged images are shown on the right, including quantitation of the number of embryos that display ectopic NKX2.2 expression (denoted by arrows). Scale bar, 10µm. (Y) Western Blot Analysis of COS-7 cell lysates following transfection with the specified constructs and probed with anti-HA and anti-MYC antibodies and anti-Beta-tubulin as a loading control (left panel). ImageJ quantitation of relative expression levels is expressed as a histogram (right panel).
Chimeric constructs reveal functional differences between the FNIII domains of CDON and BOC
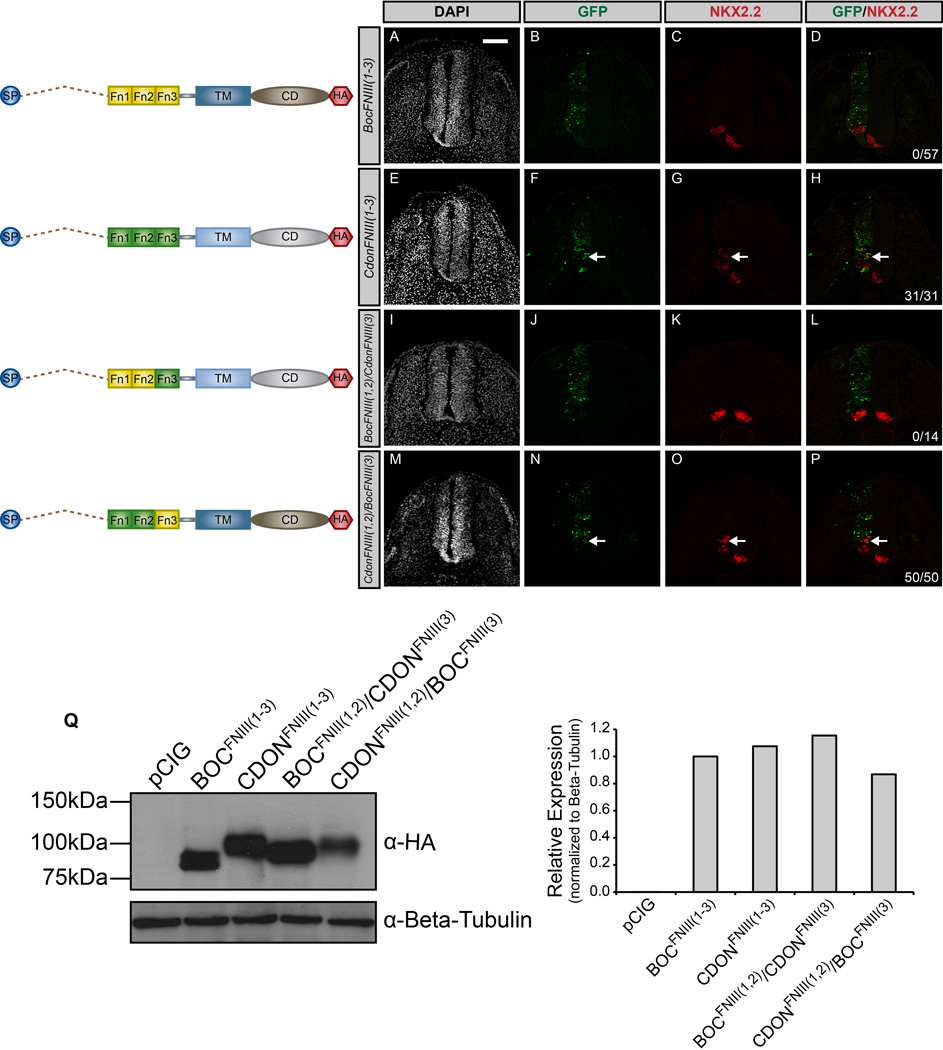
To further dissect the discrepancy in CDON and BOC function, we generated chimera constructs by interchanging the first two FNIII repeats of CDON and BOC. The first construct, BocFNIII(1,2)/CdonFNIII(3), was produced by inserting FNIII(1) and FNIII(2) of BOC into the CdonFNIII(3) construct. That is, the signal peptide, FNIII(3) domain, transmembrane domain, and cytoplasmic domain were all from CDON, while the first two FNIII repeats were from BOC. We also generated a similar chimeric construct, CdonFNIII(1,2)/BocFNIII(3), in which the first two FN domains of BOC were replaced with those from CDON (Fig. 6).
Figure 6. Chimeric constructs reveal functional differences between the FNIII domains of CDON and BOC.
(A–P) Forelimb level transverse sections of Hamburger-Hamilton stage 21–22 chicken neural tubes electroporated with BocFNIII(1–3) (A–D), CdonFNIII(1–3) (E–H), BocFNIII(1,2)/CdonFNIII(3) (I–L), and CdonFNIII(1,2)/BocFNIII(3) (M–P) stained with anti-NKX2.2 antibody (red). Green (GFP+) cells depict electroporated cells and nuclei are identified by DAPI (grayscale). Merged images are shown on the right, including quantitation of the number of embryos that display ectopic NKX2.2 expression (denoted by arrows). Scale bar, 10µm. (Q) Western Blot Analysis of COS-7 cell lysates following transfection with the specified constructs and probed with anti-HA antibody and anti-Beta-tubulin as a loading control (left panel). ImageJ quantitation of relative expression levels is expressed as a histogram (right panel).
Unlike the CdonFNIII(1–3) construct that promotes HH signaling (Fig. 6E–H), expression of the BocFNIII(1,2)/CdonFNIII(3) chimeric construct in the chicken neural tube fails to promote HH signaling (Fig. 6I–L). Conversely, the reciprocal chimeric construct, CdonFNIII(1,2)/BocFNIII(3), does promote ectopic HH pathway activity (Fig. 6M–P), in direct opposition to the BocFNIII(1–3) construct that does not induce HH signaling (Fig. 6A–D). Notably, these constructs are expressed at similar levels in vitro as assessed by western blot analysis (Fig. 6Q), and expression of the chimeric constructs in the chicken neural tube was confirmed by immunofluorescent staining (Fig S5M–P). These data suggest that the FNIII(1) and FNIII(2) domains of CDON can promote HH signal transduction while the homologous domains of BOC cannot, even in the heterologous context of CDON.
To explore whether this difference in CDON and BOC function are due to differences in association with the canonical receptor PTCH1, we performed immunoprecipitation experiments with the various CDON and BOC FNIII chimeras (Fig. 7). We initially confirmed that PTCH1 co-precipitates with CDON and BOC (Fig. 7A), similar to previous studies (Bae et al, 2011; Izzi et al, 2011). Surprisingly, we found that PTCH1 co-precipitates with both CDONFNIII(1–3) and BOCFNIII(1–3) (Fig. 7B), even though BOCFNIII(1–3) does not promote HH signaling (cf. Fig. 6A–H). Counterintuitively, we also observed that PTCH1 co-precipitates with BOCFNIII(1,2)/CDONFNIII(3) (which does not promote HH signaling), but does not co-precipitate with CDONFNIII(1,2)/BOCFNIII(3) (which does promote HH signaling; Fig. 7B). Together these data suggest that 1) differential association with PTCH1 does not explain the difference in CDON and BOC function, and that 2) association with PTCH1 is not required for the promotion of HH-dependent neural patterning.
Figure 7. CDON and BOC association with PTCH1 does not correlate with the promotion of HH signaling.
(A) Immunoprecipitation of CDON::HA and BOC::HA from COS-7 cells co-expressing PTCH1::MYC. Immunoprecipitates (IP) and whole cell lysates (WCL) were subjected to SDS-PAGE and western blot analysis (IB) using antibodies directed against MYC (α-MYC) and HA (α-HA). (B) Immunoprecipitation of HA-tagged CDON and BOC FNIII constructs from COS-7 cells co-expressing PTCH1::MYC. Note that PTCH1::MYC co-precipitates with both CDONFNIII(1–3) and BOCFNIII(1–3).
Identification of a novel motif in BOC required to promote HH signal transduction
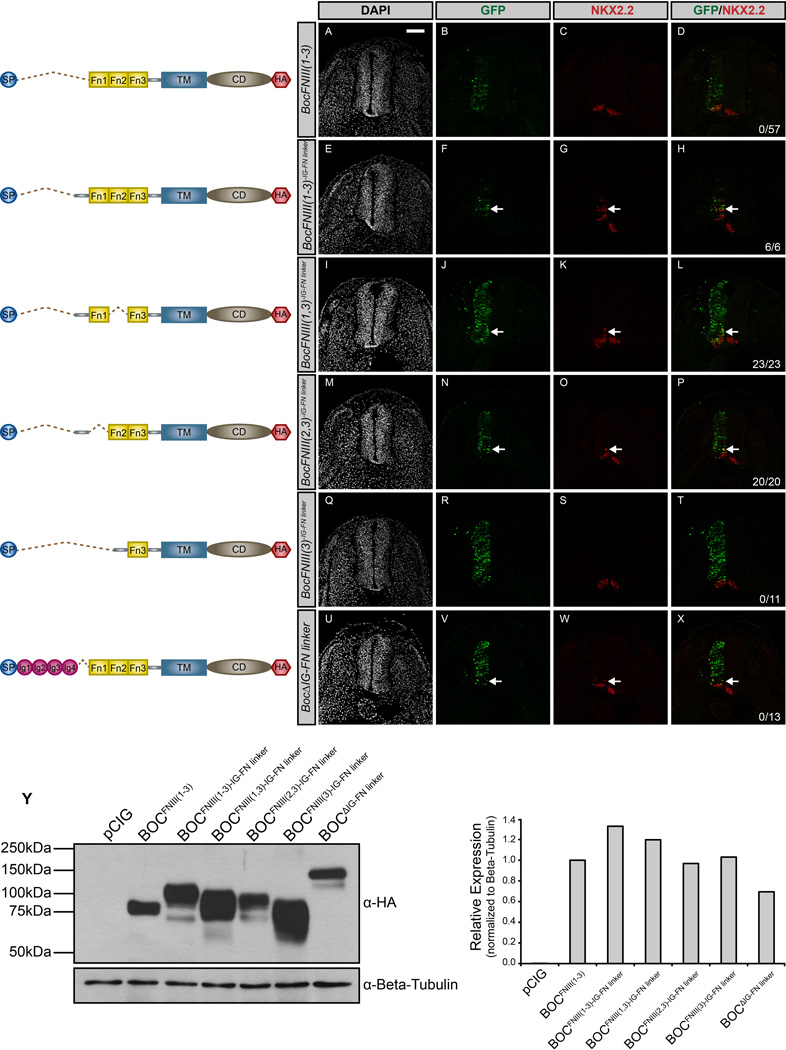
The discovery that the FNIII domains of BOC are not sufficient to promote HH pathway activity suggests an additional extracellular requirement for BOC function. Through a series of additional structure-function experiments (data not shown), we identified a 78-amino acid region in BOC bridging the conserved IG and FNIII domains (IG-FN linker) as a candidate regulator of HH activity. To test whether this IG-FN linker is required for BOC-mediated promotion of HH signaling, we generated a construct containing the linker region along with all three FNIII domains of BOC, referred to as BocFNIII(1–3)IG-FN linker (Fig. 8).
Figure 8. Identification of a novel motif in BOC required to promote HH signal transduction.
(A–X) Forelimb level transverse sections of Hamburger-Hamilton stage 21–22 chicken neural tubes electroporated with BocFNIII(1–3) (A–D), BocFNIII(1–3)IG-FN linker (E–H), BocFNIII(1,3)IG-FN linker (I–L), BocFNIII(2,3)IG-FN linker (M–P), BocFNIII(3)IG-FN linker (Q–T), and BocΔIG-FN linker (U–X) stained with anti-NKX2.2 antibody (red). GFP+ cells denote electroporated cells (green) and DAPI (grayscale) marks nuclei. Merged images are shown on the right, including quantitation of the number of embryos that display ectopic NKX2.2 expression (denoted by arrows). Scale bar, 10µm. (Y) Western Blot Analysis of COS-7 cell lysates following transfection with the specified constructs and probed with anti-HA antibody and anti-Beta-tubulin as a loading control (left panel). ImageJ quantitation of relative expression levels is expressed as a histogram (right panel).
Significantly, we find that BocFNIII(1–3)IG-FN linker induces HH pathway activity equivalently to full-length BOC, and in contrast to BocFNIII(1–3) lacking this region (Fig. 8A–H). Further, we performed analogous experiments to CDON, generating constructs lacking either the FNIII(1) or FNIII(2) repeats, but with an intact linker region. Both constructs, BocFNIII(1,3)IG-FN linker, and BocFNIII(2,3)IG-FN linker, promote HH signaling (Fig. 8I–P), similar to previous constructs lacking only the FNIII(1) or FNIII(2) domains (cf. Fig. 5Q–X). To assess whether the linker region is sufficient to foster HH-dependent neural patterning, we generated a membrane-anchored construct containing the linker region and the ligand-binding FNIII(3) domain, BocFNIII(3)IG-FN linker. Expression of this construct in the chicken neural tube does not rescue SHH signal transduction (Fig. 8Q–T). Conversely, to determine the necessity of the linker region, we deleted this region from full-length BOC (BocΔIG-FN linker). This construct still promotes ectopic NKX2.2+ progenitor cell specification (Fig. 8U–X), suggesting that the linker region is not necessary to promote HH signaling in the context of full-length BOC protein. We confirmed the expression of these constructs via immunostaining and western blot analysis (Fig. S5A–J and Fig. 8Y). Taken together, these data indicate that the IG-FN linker region in BOC can promote HH signaling in cooperation with either FNIII(1) or FNIII(2), but is not required to activate signaling in the context of the full-length BOC protein.
DISCUSSION
CDON and BOC require distinct modes of membrane attachment to promote HH signaling
In this study we employed a gain-of-function approach in the developing chicken neural tube to define the structural requirements for CDON and BOC in the promotion of HH signaling (summarized in Fig. 9). Previous reports suggested that CDON and BOC require cell surface attachment to mediate HH signal transduction (Tenzen et al, 2006). Our data confirm and extend these results to demonstrate that CDON and BOC can promote HH signaling when anchored via an integral (CD4) transmembrane domain. Surprisingly, we find that CDON, but not BOC, can induce ectopic HH signaling when anchored via a peripheral (GPI) membrane anchor. These data are the first to suggest that, despite their functional redundancy and structural similarities, these proteins utilize distinct motifs to mediate HH signal transduction.
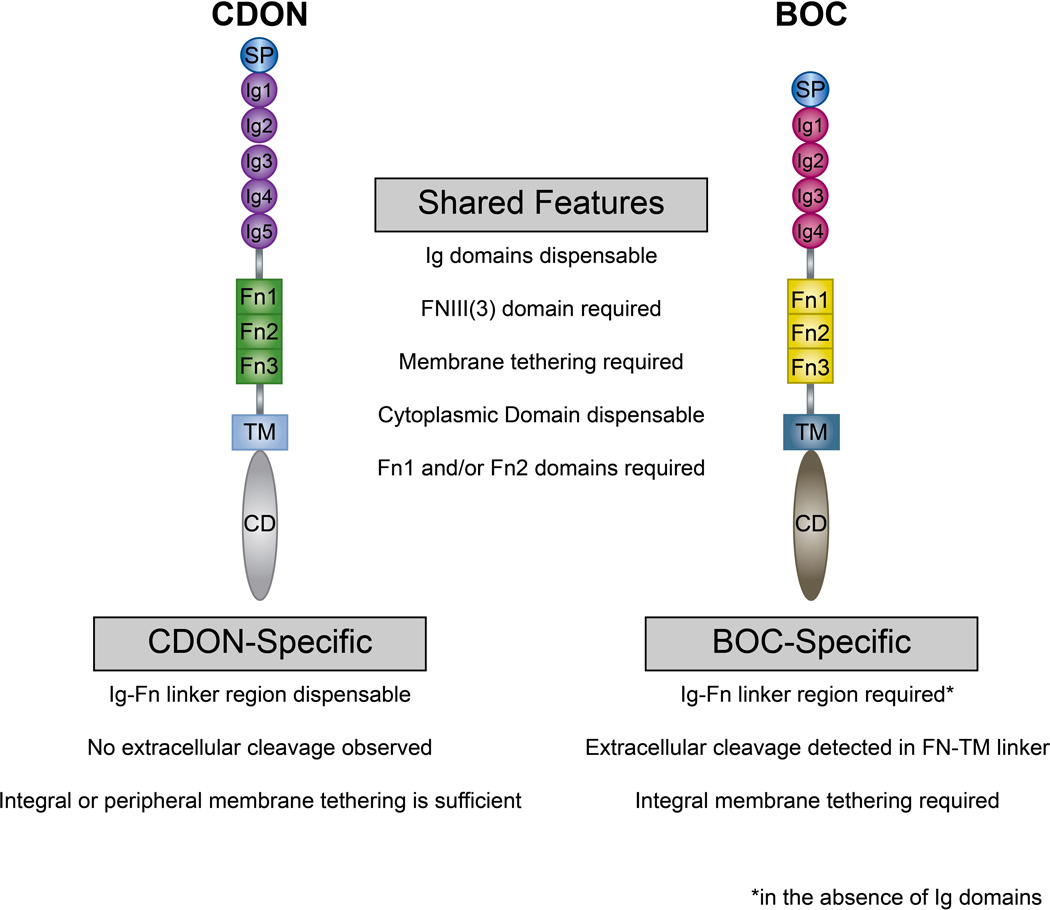
Figure 9. Summary of the structural requirements for CDON and BOC in HH signal transduction.
Schematic summarizing the structural domain requirements for CDON- and BOC-mediated promotion of HH signaling. Shared features are listed in the center, while CDON- and BOC-specific features are listed below each protein. While the IG domains are dispensable, the FNIII domains are essential for both CDON- and BOC-mediated HH signal transduction. In the absence of the Ig domains, BOC requires an additional linker region N-terminal to the FNIII domains, whereas CDON does not require this domain to induce HH signaling. Further, BOC requires integral membrane tethering, whereas CDON effectively drives HH signaling when tethered via either integral or peripheral membrane anchors. BOC also possesses an extracellular proteolytic cleavage site that may regulate its promotion of HH signaling. Notably, the cytoplasmic domains of both CDON and BOC are dispensable for the promotion of HH signaling during neural tube patterning.
The inability of BOC to promote HH pathway activity when tethered via a peripheral membrane anchor reveals an important aspect of BOC function- namely that the subcellular localization of BOC may be especially critical to its activity as a HH co-receptor. Given that GPI-anchored proteins are frequently targeted to the apical surface of polarized epithelial cells (Chan et al, 2011; Megason & McMahon, 2002), these results suggest that during HH-dependent neural patterning BOC requires targeting to a particular membrane domain to promote HH signaling. In support of this idea, recent studies in the limb indicate that CDON and BOC localize to specific domains within filopodial extensions (Kim et al, 2011). Further, in Drosophila, the CDON and BOC homolog Ihog controls the cell surface localization of Ptc (Zheng et al, 2010). While the ability of CDON and BOC to regulate the subcellular distribution of PTCH1 has not been examined, it is possible that these proteins regulate HH signaling through modulation of PTCH1 distribution at the cell surface. It is tempting to speculate that CDON and BOC may regulate the ciliary localization of PTCH1 (Rohatgi et al, 2007), although our data indicate that deletion of a putative ciliary localization sequence (Corbit et al, 2005) in these proteins does not affect the function of either CDON or BOC.
One caveat associated with these experiments is that endogenous CDON and BOC could complicate our interpretation of the results. At the time of electroporation (Hamburger-Hamilton stage 11–13) and subsequently at the time of neural patterning analysis (stage 21–22), Cdon expression is restricted to the floor plate and dorsal neural tube, while Boc expression extends further ventrally, but is still largely excluded from the cells analyzed in these experiments (Allen et al, 2011; Tenzen et al, 2006). However, we cannot formally exclude the possibility that endogenous CDON and BOC impact the activity of the Cdon and Boc expression constructs used in this study.
CDON and BOC employ distinct extracellular determinants to promote HH signaling
In addition to distinct membrane tethering requirements for CDON and BOC, our data also identify functional differences in the extracellular domains of these proteins in HH signal transduction. We find that the FNIII domains of CDON are necessary and sufficient to promote HH signaling. This corroborates previous studies demonstrating physical interactions between the FNIII(3) domain of CDON with HH ligands (Kang et al, 1997; Tenzen et al, 2006) and between the FNIII(1) and FNIII(2) domains with PTCH1, and with other co-receptors (Bae et al, 2011; Izzi et al, 2011). Surprisingly, the FNIII domains of BOC are not sufficient to induce HH signaling; instead BOC requires an additional motif- a 78 amino acid region that links the IG and FNIII domains of BOC and works in concert with either the FNIII(1) or FNIII(2) domains to promote HH signaling. BOC also requires an additional juxtamembrane sequence, which contains a putative matrix metallopeptidase 9 recognition site, to drive ectopic HH signal transduction. While this domain mediates the release of the BOC extraellular domain from the cell surface, it remains unknown which protease mediates this release. Further, it is unclear whether this cleavage is actually required to promote BOC-mediated HH signal transduction. Given that the expression of a secreted BOC protein is not sufficient to induce HH signaling, one prediction is that proteolytic cleavage of BOC is not required to promote HH pathway activity. However, further studies that more precisely delete this cleavage site will be needed to test this hypothesis.
It is interesting to note that the FNIII(1) and FNIII(2) domains of both BOC and CDON appear to function redundantly in HH signaling, but they are not interchangeable. That is, either the FNIII(1) or FNIII(2) domains of CDON can cooperate with the FNIII(3) domain to promote HH signaling, as can the analogous domains of BOC; however the FN domains of BOC further require an additional linker motif to drive HH pathway function. Surprisingly, the ability the FNIII domains to promote HH signaling appear to be independent of interactions with PTCH1. In particular, BOCFNIII(1–3), which does interact with PTCH1, does not promote HH signaling; conversely CDONFNIII(1,2)/BOCFNIII(3), which does not associate with PTCH1, does induce HH pathway activity. Future studies are required to more precisely define the requirements within the FNIII domains of CDON and within the linker domain of BOC in order dissect their distinct functions in HH pathway activity.
While CDON and BOC appear to function redundantly with GAS1 during HH-dependent neural patterning (Allen et al, 2011) and craniofacial development (Pineda-Alvarez et al, 2012; Seppala et al, 2007; Zhang et al, 2011), our data indicate that these proteins employ distinct mechanisms to activate HH signaling. This suggests that CDON and BOC may exert distinct effects on HH signaling in different contexts. In support of this notion, genetic studies indicate that Boc, but not Cdon, controls HH-dependent digit specification (Allen et al, 2011; Allen et al, 2007). Similarly, BOC, but not CDON is required for SHH-dependent axon guidance in mice (Fabre et al, 2010; Okada et al, 2006). Conversely, CDON, but not BOC, is proposed to act as a dependence receptor that activates apoptosis in the absence of HH ligand (Kang et al, 2002). Our data provide mechanistic evidence in support of distinct roles for these proteins in different HH-dependent developmental processes and correlate with initial studies of these molecules that suggested distinct functions of the extracellular domains of CDON and BOC in myogenesis (Kang et al, 2002).
Modeling cell surface interactions of CDON and BOC during HH signal transduction
CDON and BOC are essential cell surface modulators of the HH pathway (Kang et al, 1997; Schneider et al, 2012; Tenzen et al, 2006). However, there are a host of cell surface-associated proteins that control HH signaling, ranging from the core pathway components SMO and PTCH1 to the co-receptor GAS1 (Allen et al, 2011), and LRP2, which promotes HH signaling during forebrain development (Christ et al, 2012). Cell surface antagonists of the pathway include PTCH1, PTCH2, and HHIP1 (Holtz et al, 2013), while glypicans act to both promote and inhibit HH signaling in different contexts (Capurro et al, 2008; Filmus & Capurro, 2014; Williams et al, 2010).
Given that CDON and BOC interact not only with all HH ligands (Kavran et al, 2010), but also with each other and with multiple cell surface HH pathway components, the nature of these interactions, and the context in which these interactions occur (i.e. from the tissue-level to the sub-cellular distribution of these proteins) is likely to be a critical determinant in their function in HH signal transduction. Additionally, it is likely that these interactions, and therefore the structural requirements for CDON and BOC function in HH signal transduction will change in a tissue- and stage-specific manner. Therefore, it will be essential to precisely map these sites of interaction to better understand the nature of these interactions and how perturbation of these interactions affects HH-dependent embryonic development. Notably, impairment of these interactions has been implicated in developmental diseases such as holoprosencephaly (Bae et al, 2011). Disruption of these interactions may also mediate abnormal HH signaling in the context of cancer; HH-responsive Boc expression has been demonstrated in medulloblastoma (Lee et al, 2010) while a potential tumor suppressor role for CDON has also been proposed (Kang et al, 2002). Thus, the development of tools and reagents that allow for the precise targeting of specific domains of CDON and BOC may provide a means to selectively alter HH signaling in different tissues and in different HH-driven pathologies.
MATERIALS AND METHODS
Cdon and Boc constructs
The generation of various Cdon and Boc constructs was performed using standard molecular biology methods (Sambrook et al, 1989). Cdon and Boc deletion constructs were created via a modified PCR-based mutagenesis protocol (Geiser et al, 2001) using primers designed to flank the region desired for deletion. All constructs were cloned into the pCIG vector, which contains the CMV enhancer, chicken beta-actin promoter, and an internal ribosome entry site (IRES) followed by nuclearly localized enhanced green fluorescent protein (3XNLS-EGFP) (Megason & McMahon, 2002). Detailed information for each construct, including the number of embryos analyzed for each construct is presented in Fig. S6.
Chicken In Ovo Electroporations
Chicken in ovo neural tube electroporations were performed as previously described (Tenzen et al, 2006). DNA constructs were injected into Hamburger-Hamilton (HH) stage 12–13 chicken embryos at a concentration of 1µg/µl in sterile PBS with 1% Fast Green (VWR). Embryos were visualized by staining with 1% Neutral Red (Sigma Aldrich) diluted in PBS. 48hr post electroporation, embryos were dissected and fixed in 4% paraformaldehyde (EMS) for 1hr on ice, and equilibrated overnight in 30% sucrose prior to embedding in OCT. Fertilized eggs were obtained from the Michigan State University Poultry Farm.
Neural Patterning Analysis
Immunofluorescent analyses were performed as previously described (Jeong & McMahon, 2005). Sucrose-equilibrated embryos were frozen in OCT compound (Tissue-tek) and cryo-sectioned at a thickness of 12–14µm at the forelimb level for further neural patterning analysis. The primary antibodies used include: 1:20 mouse anti-NKX2.2 (DSHB; 74.5A5), 1:20 mouse anti-NKX6.1 (DSHB; F55A10), and 1:1000 rabbit anti-OLIG2 (Millipore; AB9610). Antibodies were diluted in blocking buffer containing 3% bovine serum albumin (Sigma Aldrich), 1% heat inactivated sheep serum, 0.1% Triton X-100, and 0.02% sodium azide. Alexa 555-conjugated secondary anti-mouse and anti-rabbit antibodies (Invitrogen) were used at 1:500 dilutions. Nuclei were visualized with DAPI (Invitrogen) diluted 1:30,000 in blocking buffer. Sections were incubated in primary antibody overnight at 4°C and in secondary antibody for 1hr at room temperature. All images were collected on a Leica SP5X confocal microscope. For each construct a minimum of 6 different embryos (three sections per embryo) were analyzed. A summary of the number of embryos analyzed per construct is available in Fig. S6.
Immunoprecipitation and Western Blot Analysis
COS-7 cells were transfected with the specified DNA constructs using Lipofectamine 2000 (Life Technologies). Supernatants were collected through aspiration and lysates were collected in RIPA buffer with proteinase inhibitor 48hr post transfection. Lysates were sonicated and protein concentration was assessed by BCA assay (Pierce). 50µg of cell lysate and 30µl of supernatant were subjected to SDS-PAGE on a 7.5% polyacrylamide gel. The transferred membranes were blotted with the following primary antibodies: mouse anti-HA (Covance, 1:1000) or rabbit anti-HA (Bethyl, 1:10,000), rabbit anti-MYC (Bethyl, 1:1000), goat anti-CDON (R&D systems, AF2429, 1:1000), goat anti-BOC (R&D systems, AF2385, 1:1000), and mouse anti-Beta-Tubulin (a gift from Dr. Kristen Verhey, 1:1000). All antibodies were diluted in blocking buffer containing TBST, 0.02% sodium azide and 3% bovine serum albumin (Sigma Aldrich). Following primary antibody incubation, the blots were subjected to peroxidase-conjugated secondary antibody incubation, with either HRP-conjugated anti-mouse, anti-rabbit, or anti-goat antibodies (Jackson ImmunoResearch) all diluted 1:10,000 in blocking buffer. The blots were incubated for 1hr at room temperature for both primary and secondary antibodies. After secondary antibody incubation, the blots were subjected to a 5-minute-incubation in HRP substrate (Millipore) and developed onto x-ray film. Immunoprecipitation experiments were performed as previously described (Holtz et al, 2013; Okada et al, 2006). Quantitation of western blots was performed using ImageJ (Schneider et al, 2012). Band intensity was measured, background corrected, and normalized to Beta-Tubulin intensity (also background corrected) for each individual lane. Relative expression levels were compared to full-length CDON (Fig. 1 and Fig. 4), BOC (Fig. 2 and Fig. 5), or BOCFNIII(1–3) (Fig. 6 and Fig. 8), all of which were set to 1.
Phosphatidylinositol-specific Phospholipase C (PI-PLC) Treatment
COS-7 cells were transfected in 6-well plates with the specified DNA constructs using Lipofectamine 2000 (Life Technologies). 48hr post transfection cells were treated with 2.5µl of PI-PLC (Life Technologies) diluted in 250µl of serum-free cell media for 30min at 37°C. Following PI-PLC treatment, the supernatants and cell lysates were collected for subsequent western blot analysis.
Supplementary Material
Highlights.
The means of membrane attachment differentially affects CDON and BOC function
BOC contains a novel extracellular proteolytic cleavage site
The FNIII repeats of BOC and CDON are not functionally equivalent
PTCH1 binding to BOC and CDON does not correlate with HH pathway activity
BOC requires an additional linker region to promote HH signal transduction
ACKNOWLEDGEMENTS
We are grateful to Dr. R. Krauss (Icahn School of Medicine at Mount Sinai) for the full-length CDON construct. We also thank members of the Allen lab for insightful comments and helpful suggestions. The NKX2.2 and NKX6.1 antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA. Confocal microscopy was performed in the Microscopy and Image Analysis Laboratory at the University of Michigan. J.Y.S. is supported by a research team grant from the University of Michigan Center for Organogenesis. A.M.H. is supported by an NIH predoctoral fellowship (F31 NS081806). This work was supported by an American Heart Association scientist development grant (11SDG6380000) and by NIH grants (R21 CA167122 and R01 DC014428) to B.L.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.Y.S. and B.L.A. conceived and designed the experiments. J.Y.S. executed the experiments and collected the data. A.M.H. and J.Y.S performed the immunoprecipitation experiments. J.M.P. assisted with confocal imaging analysis. J.Y.S. and B.L.A. analyzed and interpreted the data. J.Y.S. and B.L.A. wrote and edited the manuscript.
REFERENCES
- Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, BOC in SHH pathway function. Developmental cell. 2011;20:775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BL, Tenzen T, McMahon AP. The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes & development. 2007;21:1244–1257. doi: 10.1101/gad.1543607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae GU, Domene S, Roessler E, Schachter K, Kang JS, Muenke M, Krauss RS. Mutations in CDON, encoding a hedgehog receptor, result in holoprosencephaly and defective interactions with other hedgehog receptors. American journal of human genetics. 2011;89:231–240. doi: 10.1016/j.ajhg.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews Molecular cell biology. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- Camp D, Currie K, Labbe A, van Meyel DJ, Charron F. Ihog and Boi are essential for Hedgehog signaling in Drosophila. Neural development. 2010;5:28. doi: 10.1186/1749-8104-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Developmental cell. 2008;14:700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Chan HY, V S, Xing X, Kraus P, Yap SP, Ng P, Lim SL, Lufkin T. Comparison of IRES and F2A-based locus-specific multicistronic expression in stable mouse lines. PloS one. 2011;6:e28885. doi: 10.1371/journal.pone.0028885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- Christ A, Christa A, Kur E, Lioubinski O, Bachmann S, Willnow TE, Hammes A. LRP2 is an auxiliary SHH receptor required to condition the forebrain ventral midline for inductive signals. Developmental cell. 2012;22:268–278. doi: 10.1016/j.devcel.2011.11.023. [DOI] [PubMed] [Google Scholar]
- Cole F, Krauss RS. Microform holoprosencephaly in mice that lack the Ig superfamily member Cdon. Current biology : CB. 2003;13:411–415. doi: 10.1016/s0960-9822(03)00088-5. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Fabre PJ, Shimogori T, Charron F. Segregation of ipsilateral retinal ganglion cell axons at the optic chiasm requires the Shh receptor Boc. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:266–275. doi: 10.1523/JNEUROSCI.3778-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filmus J, Capurro M. The role of glypicans in Hedgehog signaling. Matrix biology : journal of the International Society for Matrix Biology. 2014 doi: 10.1016/j.matbio.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Geiser M, Cebe R, Drewello D, Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. BioTechniques. 2001;31:88–90. 92. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- Holtz AM, Peterson KA, Nishi Y, Morin S, Song JY, Charron F, McMahon AP, Allen BL. Essential role for ligand-dependent feedback antagonism of vertebrate hedgehog signaling by PTCH1, PTCH2 and HHIP1 during neural patterning. Development. 2013;140:3423–3434. doi: 10.1242/dev.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Krauss RS. Rescue of holoprosencephaly in fetal alcohol-exposed Cdon mutant mice by reduced gene dosage of Ptch1. PloS one. 2013;8:e79269. doi: 10.1371/journal.pone.0079269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi L, Levesque M, Morin S, Laniel D, Wilkes BC, Mille F, Krauss RS, McMahon AP, Allen BL, Charron F. Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Developmental cell. 2011;20:788–801. doi: 10.1016/j.devcel.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, McMahon AP. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development. 2005;132:143–154. doi: 10.1242/dev.01566. [DOI] [PubMed] [Google Scholar]
- Kang JS, Gao M, Feinleib JL, Cotter PD, Guadagno SN, Krauss RS. CDO: an oncogene-, serum-, and anchorage-regulated member of the Ig/fibronectin type III repeat family. The Journal of cell biology. 1997;138:203–213. doi: 10.1083/jcb.138.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Mulieri PJ, Hu Y, Taliana L, Krauss RS. BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. The EMBO journal. 2002;21:114–124. doi: 10.1093/emboj/21.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Ward MD, Oladosu OO, Mulepati S, Leahy DJ. All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. The Journal of biological chemistry. 2010;285:24584–24590. doi: 10.1074/jbc.M110.131680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Ji H, Ouyang Z, Zhou B, Ma W, Vokes SA, McMahon AP, Wong WH, Scott MP. Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9736–9741. doi: 10.1073/pnas.1004602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low MG. Biochemistry of the glycosyl-phosphatidylinositol membrane protein anchors. The Biochemical journal. 1987;244:1–13. doi: 10.1042/bj2440001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- Maddon PJ, Littman DR, Godfrey M, Maddon DE, Chess L, Axel R. The isolation and nucleotide sequence of a cDNA encoding the T cell surface protein T4: a new member of the immunoglobulin gene family. Cell. 1985;42:93–104. doi: 10.1016/s0092-8674(85)80105-7. [DOI] [PubMed] [Google Scholar]
- Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidence that patched is the Hedgehog receptor. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, Leahy DJ. The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature. 2008;455:979–983. doi: 10.1038/nature07358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Current topics in developmental biology. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Okada A, Charron F, Morin S, Shin DS, Wong K, Fabre PJ, Tessier-Lavigne M, McConnell SK. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature. 2006;444:369–373. doi: 10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- Pineda-Alvarez DE, Roessler E, Hu P, Srivastava K, Solomon BD, Siple CE, Fan CM, Muenke M. Missense substitutions in the GAS1 protein present in holoprosencephaly patients reduce the affinity for its ligand, SHH. Human genetics. 2012;131:301–310. doi: 10.1007/s00439-011-1078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Sambrook J. Molecular cloning : a laboratory manual. 2nd edn. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Santos N, Reiter JF. A central region of Gli2 regulates its localization to the primary cilium and transcriptional activity. Journal of cell science. 2014 doi: 10.1242/jcs.139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppala M, Depew MJ, Martinelli DC, Fan CM, Sharpe PT, Cobourne MT. Gas1 is a modifier for holoprosencephaly and genetically interacts with sonic hedgehog. The Journal of clinical investigation. 2007;117:1575–1584. doi: 10.1172/JCI32032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Tan H, Perry AJ, Akutsu T, Webb GI, Whisstock JC, Pike RN. PROSPER: an integrated feature-based tool for predicting protease substrate cleavage sites. PloS one. 2012;7:e50300. doi: 10.1371/journal.pone.0050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Tenzen T, Allen BL, Cole F, Kang JS, Krauss RS, McMahon AP. The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Developmental cell. 2006;10:647–656. doi: 10.1016/j.devcel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Ingham PW. smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- Williams EH, Pappano WN, Saunders AM, Kim MS, Leahy DJ, Beachy PA. Dally-like core protein and its mammalian homologues mediate stimulatory and inhibitory effects on Hedgehog signal response. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5869–5874. doi: 10.1073/pnas.1001777107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Hong M, Bae GU, Kang JS, Krauss RS. Boc modifies the holoprosencephaly spectrum of Cdo mutant mice. Disease models & mechanisms. 2011;4:368–380. doi: 10.1242/dmm.005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Mann RK, Sever N, Beachy PA. Genetic and biochemical definition of the Hedgehog receptor. Genes & development. 2010;24:57–71. doi: 10.1101/gad.1870310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.