Abstract
Intensive care monitoring systems are typically developed from population data, but do not take into account the variability among individual patients’ characteristics. This study develops patient-specific alarm algorithms in real time. Classification tree and neural network learning were carried out in batch mode on individual patients’ vital sign numerics in successive intervals of incremental duration to generate binary classifiers of patient state and thus to determine when to issue an alarm. Results suggest that the performance of these classifiers follows the course of a learning curve. After eight hours of patient-specific training during each of ten monitoring sessions, our neural networks reached average sensitivity, specificity, positive predictive value, and accuracy of 0.96, 0.99, 0.79, and 0.99 respectively. The classification trees achieved 0.84, 0.98, 0.72, and 0.98 respectively. Thus, patient-specific modeling in real time is not only feasible but also effective in generating alerts at the bedside.
Keywords: patient-specific adaptivity, real-time batch learning, classification tree, neural network, patient monitoring, critical care
1 Introduction
In the Intensive Care Unit (ICU), an arsenal of medical devices continuously monitor each patient. The most computation-intensive of these devices is the bedside monitor, which takes in patients’ physiological measurements from biosensors and other devices, converts the incoming electrical signals into digitalized waveforms and vital sign numerics, displays these to caregivers, stores and analyzes them to track patients’ physiological state, and sounds alarms whenever its built-in algorithms detect a physiological abnormality. While new biosensors have increased the number and quality of available physiological signals [8], and color touch screens have made bedside monitors more user-friendly, the clinical utility of alarm algorithms that are central to timely detection of adverse conditions have continued to advance at a slower pace than other medical technologies [10, 17, 5, 9, 7].
Until the 1990’s, most alarms were triggered when a specific physiological measurement fell outside pre-set threshold boundaries, without any specific dependence on other signals or, more importantly, the overall state of the patient, which was not represented. Even the most sophisticated algorithms, such as those interpreting electrocardiogram (ECG) variations, examined only one source of data, from ECG leads. Alarm detection in the newer generation of patient monitoring systems is more sophisticated than previously. Although the details of the new algorithms have not been disclosed, from publicly available information we know that they include sensitive artifact detection, noise elimination, pattern recognition of specific disease conditions, such as ventricular fibrillation, and some multiple-signal/multi-channel data analysis. Indeed, we perceive a gradual shift in emphasis of ICU monitoring from issuing alarms toward creating alerts that are part of a larger decision support infrastructure. We will use alerts when we mean this expanded view.
In this paper we address another potential source of improvement: tuning alerting models to specialized patient populations, or, indeed, to the individual patient. After all, in critical care no two patients are the same.1 In fact, many patients behave in highly individual ways that might deviate significantly from the average patient in population-based models. What counts as “normal” for one patient may be highly abnormal if seen in another, and patients’ dynamic responses to changing circumstances also vary greatly from individual to individual [11].
Alarm algorithms today are developed retrospectively, by using previously collected datasets that encompass thousands of patients to build models that detect adverse clinical events, namely medical conditions that could become life-threatening. Once built, these algorithms are applied without further improvement to many patients in the ICU. Yet, a previous study from our laboratory found that some models built from one patient population performed significantly worse on data from two other groups of patients, but simply optimizing the thresholds used in these models to fit data from 9% of these other patients greatly improved the models’ performance [16]. This finding suggests that for patient monitoring to be robust, its algorithms must be able to adapt to a focused patient population or even to the individual patient.
The research reported here explores the most aggressive form of this hypothesis, that we can build effective patient-specific alarming models from a specific individual’s own data. Our preliminary findings were reported in [19, 20]. Obviously, such a “pure” strategy will be quite ineffective before any individual data are collected, so we also study the rate at which this approach can learn to produce accurate detection of clinically relevant events at the bedside, using the system presented in [19]. As an initial investigation of this approach, the present study has numerous limitations, which we address in the discussion.
2 Methods
2.1 Clinical Setting
This research was carried out in collaboration with the pediatric Multidisciplinary ICU (P-MICU) at Boston Children’s Hospital, with the approval by the hospital’s Institutional Review Board. The P-MICU staff allocated a spacious bedspace to the study and assisted with clinical annotations. Before each study session, informed consent was obtained from the patients and their families to ensure that they were willing to participate in the study and felt comfortable with the presence of the computer equipment and a trained observer. The first author served as the trained observer. During study sessions, which took place between 2001 and 2003, between 8 AM and 2 AM, the trained observer sat at the bedspace with a laptop computer connected to the bedside monitor, with a curtain drawn between the patient and the observer whenever necessary. Patient confidentiality and privacy have been protected according to the hospital’s guidelines.
A total of 196 hours of monitoring data were collected and analyzed from 11 different patients ranging in age from infants to adolescents, five of whom were in especially critical condition. Data collection took place during 23 sessions, of which 14 were of at least eight hours’ duration, and four more of which lasted at least four hours. The shortest five sessions lasted between 2 and 3.5 hours.
2.2 Synchronized Data Collection
To support our study, we collected and recorded the following information during each session:
The second-by-second numerics computed from the measured waveform data by the HP Viridia Neonatal Component Monitoring System (CMS) used in the P-MICU. These include the heart rate derived from ECG waveforms, pulse rate from plethysmography, respiration rate, blood pressure (systolic, diastolic and mean) either arterial or measured by non-invasive means, arterial and venous oxygen saturation, and oxygen perfusion.
One-minute running averages of all numerics. These averages are less prone to momentary noise, though they are obviously not as quickly responsive to changing conditions.
Interpretations made by CMS and related information, including (a) clinical alarm status and severity, (b) whether any of the threshold alarms on individual signals have been triggered, (c) sensor or monitor malfunction (INOP) alarm status and severity, (d) monitor status, and (e) alarm suspension (when an alarm has been silenced by the nurse on duty).
-
Clinical events noted and interpreted by the bedside observer, under each of the following circumstances:
- the bedside monitor issues an alarm other than an INOP
- any of our alarm algorithms under investigation issues an alarm
- the patient became irritated and required immediate attention even when no alarm is issued
For each clinical event, the bedside observer records the start and end time of the event, whether the patient is moving, whether a medical procedure (e.g., suctioning) is in process, and the medical staff’s response to the alarm, such as checking the patient, adjusting sensors, or silencing alarms without other intervention. In addition, the observer asks the nurse or physician at the bedside to classify the event into one of three categories:- true positive, clinically relevant (TP-R)
- true positive, clinically irrelevant (TP-I)
- false positive (FP)
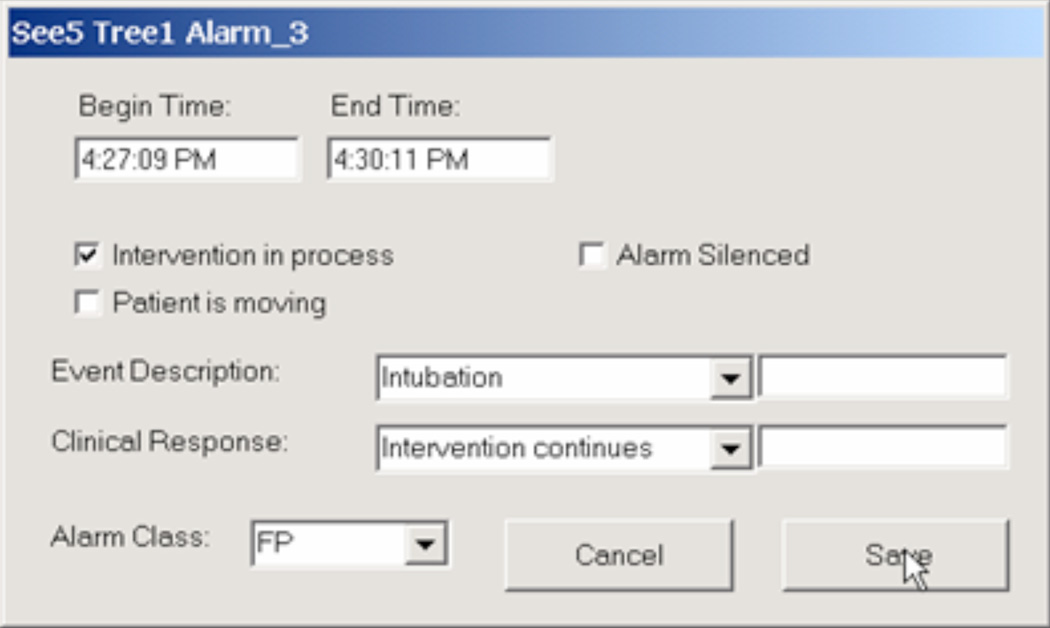
Figure 1 shows the dialog box for annotating one event, in this case resulting from one of the algorithms under investigation issuing an alarm.
Figure 1.
Example of an annotation box that allows the bedside observer to record the nature and duration of an event, as well as an indication of whether the clinical staff consider it a false positive, true positive that is clinically relevant, or true positive that is clinically irrelevant.
2.3 Models Derived from Different Amounts of Past Data
As we described in the Introduction, we investigate the degree to which models learned from a patient’s own data can be effective in interpreting future data points. Rather than doing this continually, we have chosen to construct interpretive models based on all previously collected data at 30 minutes, 1 hour, 2 hours, 4 hours, and 8 hours into each 12-hour recording session.
After each model is built, all subsequent data (i.e., one each second) are interpreted by each of the models and the results are recorded. In addition, we recorded each second the single-signal threshold alarms and the more integrated CMS alarms issued. These records, along with the clinicians’ interpretations of clinical events, are then used both for training of our subsequent models and for evaluation of these models and their comparison against the outputs of the monitoring system.
2.4 Gold Standard Data
The goal of our patient-specific alarm models is to make a binary judgment at each second’s data whether an alarm should or should not be called. To train these models, we assume that the answer should be “alarm” during any clinical event where the clinicians had called the event a true positive, whether or not it was considered clinically relevant. Conversely, the answer should be “stable” at any time that no event occurred or if an event occurred that was annotated as a false positive. Because events could be created not only by an alarm from CMS but also by alarms from our own models or from observations by the clinicians or observer, we are also able to recognize instances of false negatives, where an algorithm should have issued alarm but did not do so. We assume that all data points at times when no clinical event was recorded are true negatives (i.e., when none of the models, CMS, the clinicians or the observer saw an event).
We used one additional method to modify these classifications: if the patient’s condition changed within the 30 minutes following an event, the classification of that event could be revised as appropriate. For example, if an alarm for bradycardia is classified as a false positive, and the patient becomes persistently hypotensive in the next 30 minutes, we would revise the classification to clinically relevant true positive. Thus, the gold standard for our classification tasks consists of human experts’ classification at the time the data become available and either verification or re-classification using subsequently obtained information. In case event classifications changed, we did not re-do training of models that had been built from data about that time period; however, models built at times after the correction would incorporate the revised classification. Such reclassification is rare, having occurred only five times in our 196 hours of data collection.
2.5 Training of Patient-Specific Alarm Algorithms
We chose to investigate two machine learning techniques that had shown potential in generating intelligent alarm algorithms in earlier studies [16, 18], classification trees and artificial neural networks. Classification tree learning is suited for the learning tasks in critical care because it is a useful classification method for problems in which 1) instances are represented by a fixed set of attributes; 2) the classification output is discrete-valued; 3) disjunctive description may be required; 4) training data may contain errors; and 5) the training data may contain missing attribute values [14]. Neural network learning is a general, practical method for learning real-valued, discrete-valued, and vector-valued nonlinear functions. It is especially useful for capturing nonlinear patterns. The training examples may contain errors, and evaluation of the learned target function is fast [12]. These capabilities work well for learning tasks in critical care settings.
For both learning algorithms, the input data consisted of the eight second-by-second numerics and the corresponding minute-by-minute running averages, as described in Section 2.2. The classification label for the training data samples, as well as the output of each learned model, is binary: an “alarm” if the gold standard assignment of the data point was “alarm”, or “stable” otherwise, as described in Section 2.4.
For classification tree learning, we chose See5 [2] to conduct a top-down, greedy search through the space of possible classification trees. See5 is a Windows implementation of C5.0, a new-generation data mining tool that is built upon its predecessors C4.5 and ID3 for generating classification trees and rule-based classifiers more accurately, faster, and with less memory [3]. It includes the ability to handle discrete as well as continuous input values and a variety of pruning methods to try to avoid overfitting. The details of these methods are proprietary, but the software supports a number of user-tunable parameters.
After experimenting with 5 to 15-fold cross-validation and differential misclassification costs (see below), we chose the following settings: 10 trials for boosting, no cross-validation (to speed up training time), no differential misclassification costs, 25% local pruning, global pruning, 2 final minimum cases, no winnowing, no subset selection, and no sampling to obtain more balanced and generalized classification trees. A classification tree that is produced represents a branching sequence of binary decisions that successively subdivide the hyperspace of data points until each terminal region contains only points whose labels are (preponderantly) the same. Figure 2 shows an example.
Figure 2.
An example classification tree. This asymmetric tree encodes a succession of decision criteria, where each condition leads either to a classification (0/stable or 1/alarm) or to a subsequent threshold test. For example, according to this tree, if the patient’s O2Sat exceeds 91, that is considered stable. If it is below 87, that is considered an alarm condition. In-between, we need to examine the heart rate, which, if above 211, means alarm. Otherwise, we continue to apply further threshold tests until we reach a classification. The numbers in parentheses show the total number of training instances that fell into this region of the hyperspace and the number of these that were misclassified. These counts are not generally integers and the number of misclassifications is not zero because of See5’s pruning methods, which try to avoid overfitting. This tree had the smallest training error among a community of classifiers that were built on 14,400 training data points (from 4 monitoring hours) with 10 trials of boosting. Reproduced with permission from [20]. Copyright @ 2007 IEEE.
For neural network learning, we chose a model with a single output node and one layer of hidden nodes in which the number of nodes initially equalled the number of inputs. Each input is connected to each of the hidden nodes, and each hidden node feeds into the output node. We employ back-propagation as the core learning algorithm. Each hidden node is a sigmoid unit that takes in individual inputs x1, …, xn, calculates their weighted sum, and generates an output using the transfer function
for the k-th node. In the equation, wk,i is the weight for input xi, and δk is the neuron offset. The output node, by contrast, is a threshold unit producing a binary result. We employed the software EasyNN-plus because it could run back-propagation as an embedded application in batch-mode [1]. The learning rate was initially set at 1.0 and then optimized over the training cycles. The momentum for the weight-updating rule was first set at 0.8 and then optimized over training cycles. These parameters, as well as the number of nodes in the hidden layer, were adjusted using sequential multi-fold leave one out validation. Thirty percent of the training data were used as test data for the internal optimization of parameters. The training time was capped at 120 seconds. The detailed rationale for the settings for both learning methods is given in [19].
2.6 Implementation
All computations were performed on a Dell laptop computer running Windows 2000 on a 2.2 GHz Pentium 4 CPU with 1GB of RAM. The most demanding computational load arose during the times when new classification models were being built. This occurred five times during each session, and at each time both a classification tree and a neural network model were constructed. At all times, including while building new models, the computer was acquiring data from the bedside monitor, storing these data into its own database, running each previously trained model on the current data, opening annotation windows corresponding to newly detected events from the CMS data, our algorithms, or the observer, and managing the user interface, which could simultaneously display numerous annotation windows if the staff were busy taking care of the patient and had not yet had time to make their interpretations of the events.
To support this highly heterogeneous workload, and to permit interfacing to the communication programs, the database, and the machine learning programs, our program was multi-threaded and relied on the facilities of the operating system to permit the simultaneous execution of all these tasks. Some of the limits imposed on the learning algorithms resulted from the system’s inability to keep up with all the necessary computations if the learning algorithms were allowed to demand more computing power. For example, during execution of the learning algorithms, we did note instances where the CMS interface would miss some incoming data points because the overall system did not respond quickly enough to data appearing in the input buffer.
3 Results
We were able to collect data during 23 sessions from 11 patients over a total of 196 monitoring hours, and to build and test our learning algorithms in real time on these data. We describe the incidence of alarm conditions for the monitored patients and the computer time needed to train our models, and we present the performance of our algorithms in terms of sensitivity, specificity, positive predictive value, and overall accuracy.
3.1 Adverse Events
During the 196 monitoring hours, there were 325 clinical alarms sounded by the bedside monitor and two false negatives observed at the bedside by the trained observer. Of the alarms, 290 were true positives that required clinical interventions, 20 were true positives that did not require clinical intervention, and 15 were false positives. The 312 adverse clinical events generally were both sparse and brief in time, totaling 4.35% of the 196 monitoring hours. The number of such events experienced by each patient in the study varied significantly. Four patients had only one event in more than two hours, three patients experienced an average of three events per hour, and one suffered six events per hour. Although the typical percentage of time each patient spent in an alarm condition was low, these also varied widely, from essentially zero to a high of 42% of the time, for the patient experiencing six alarms per hour.
3.2 Training Time
Table 1 shows the amount of time spent that See5 took to train classification tree models using various numbers of data points and several settings of parameters for one typical 12-hour study session. Training times for other sessions varied by as much as ±50%. We always used the 25% setting for See5’s global pruning, to help avoid overfitting. A 10-fold cross validation is useful for estimating the accuracy of the classification tree, but it also increased the training time approximately three-fold for every doubling in the number of data points. Boosting generally yields a higher predictive accuracy for the classifier, but at the cost of a nonlinear increase in training time.
Table 1.
Training Time for Classification Tree Learning (seconds). Reproduced with permission from [20]. Copyright @ 2007 IEEE.
| Data Points |
Global Pruning 25% | 10-fold Cross-validation Global Pruning 25% |
10 Boosting Trials 10-fold Cross-validation Global Pruning 25% |
|---|---|---|---|
| 1800 | < 0.1 | 0.1 | 0.1 |
| 3600 | 0.1 | 0.2 | 2.5 |
| 7200 | 0.2 | 0.7 | 3.2 |
| 14400 | 0.5 | 2.0 | 11.0 |
| 28800 | 1.1 | 6.0 | 53.5 |
Training neural networks in general took more time than training classification trees. EasyNN-plus does not keep track of training time; thus, precise estimates of training times are not available for neural network learning. A rough estimate based on repeated observations was in the range of a few seconds to several minutes for 1800 to 28800 training data points, corresponding to 30 minutes to 8 monitoring hours. Training time could vary significantly with different training specifications, such as learning rate, momentum, the number of validation cycles, and the target error for each cycle. EasyNN-plus was set to stop training at 120 seconds, despite a toll on modeling accuracy, to prevent its computational demands from disrupting the overall system.
3.3 Performance of Learned Models
We present performance data for our learned models averaged over those ten sessions that lasted at least eleven hours, which accounted for about 120 of the 196 total monitoring hours. We chose this threshold so that there would be adequate test data in each session after computing the final models at the eight-hour time.
Tables 2 and 3 show the performance of the models learned by our methods purely from previous data about the individual patient, using classification trees and neural networks, respectively. For comparison, we also show the performance (in each table) of the CMS algorithm built into the bedside monitor and of a simple threshold algorithm representative of the previous generation of monitors. Both classification tree and neural network derived models have extremely low sensitivities based on only the first half hour of data, and gradually improve as more data become available. With eight hours of data, the two methods attain average sensitivities of 0.84 and 0.96, respectively. Positive predictive value (PPV) starts high for both methods based on very little data, drops dramatically, and then recovers to exceed the PPV of the threshold algorithm after training on eight hours of data. Both specificity and overall accuracy start high, drop slightly, and then increase to close to 1.0 with additional training data. Comparing the average performance measures of the two learning methods shows that for each length of training data the neural network learned models seem better than the classification tree ones.
Table 2.
Performance Comparison for Classification Tree Learning. CMS is the bedside monitor’s alarm algorithm. Threshold stands for a standard threshold-based alarm algorithm. The rightmost five columns are results for the classification models built after the five given times. PPV is positive predictive value. The measures shown are averages from 10 sessions of monitoring, with the ranges of values shown in brackets.
| Metric | CMS | Threshold | Tree built after n hours of data | ||||
|---|---|---|---|---|---|---|---|
|
|
1 | 2 | 4 | 8 | |||
| Sensitivity | 1.00 | 1.00 | 0.00 [.00,.18] | 0.01 [.00,.30] | 0.11 [.04,.36] | 0.43 [.29,.63] | 0.84 [.70,.93] |
| Specificity | 0.99 | 0.88 | 1.00 [.97,1.0] | 0.98 [.69,.99] | 0.98 [.90,.98] | 0.98 [.94,.98] | 0.98 [.96,.99] |
| PPV | 0.82 | 0.70 | 0.63 [0.0,.72] | 0.17 [0.0,.20] | 0.14 [.02,.23] | 0.37 [.15,.57] | 0.72 [.60,.80] |
| Accuracy | 0.99 | 0.96 | 0.96 [.92,.96] | 0.95 [.89,.96] | 0.95 [.93,.98] | 0.97 [.95,.99] | 0.97 [.96,.98] |
Table 3.
Performance Comparison for Neural Network Learning. Labels are as described in the caption for Table 2.
| Metric | CMS | Threshold | ANN built after n hours of data | ||||
|---|---|---|---|---|---|---|---|
|
|
1 | 2 | 4 | 8 | |||
| Sensitivity | 1.00 | 1.00 | 0.05 [0.0,.11] | 0.23 [0.0,.29] | 0.60 [.39,.66] | 0.80 [.61,.90] | 0.96 [.81,.98] |
| Specificity | 0.99 | 0.88 | 0.99 [.97,1.0] | 0.97 [.84,.99] | 0.98 [.94,.99] | 0.99 [.95,.99] | 0.99 [.96,.99] |
| PPV | 0.82 | 0.70 | 0.70 [0.0, .74] | 0.16 [0.0,.24] | 0.37 [.02,.72] | 0.71 [.49,.78] | 0.79 [.63,.80] |
| Accuracy | 0.99 | 0.96 | 0.96 [.92,.96] | 0.95 [.90,.96] | 0.96 [.90,.99] | 0.97 [.92,.99] | 0.99 [.95,1.0] |
4 Discussion
Our goal in this work was to explore the hypothesis that effective classification models for identifying when it is appropriate to alarm during ICU monitoring could be learned from the individual patient’s own history in the ICU and from annotations by the clinical staff of those earlier data. If this approach is valid, we would expect that more sophisticated systems than the ones we have built would combine the best current population-based monitoring algorithms with patient-specific learned models such as we explore here to produce better combined monitors. We have not directly studied this broader expectation, but we believe that our results make a good plausibility argument for it.
4.1 Learning Curve for Patient-Specific Learning
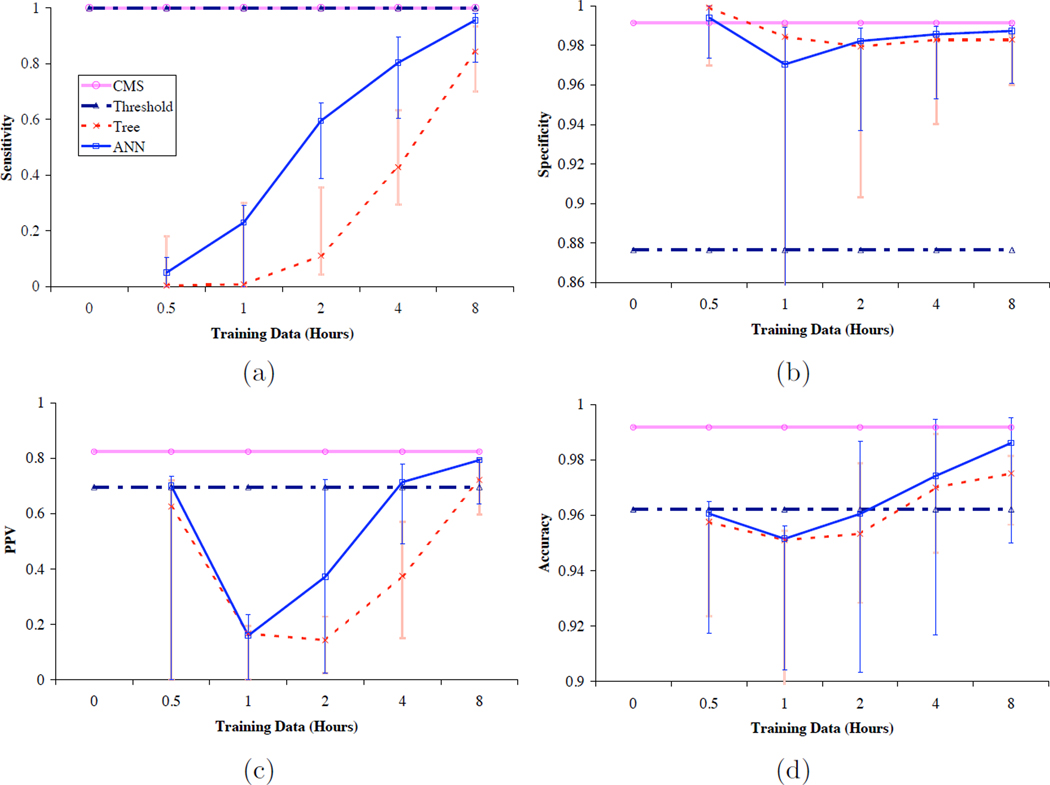
At the onset of this research, we expected that patient-specific learning would exhibit the characteristics of a standard learning curve. Indeed, we see a number of such characteristics in Figure 3, which shows the data in Tables 2 and 3 as plots that demonstrate the changes in average sensitivity, specificity, positive predictive value and accuracy of the patient-specific models as a function of the amount of training data. Although our models as trained on 2 hours or less of patient data generally perform much more poorly than either CMS or a simple threshold model, our models, especially those based on neural networks, exhibit improvements in each of our performance measures that demonstrate significant learning with additional data. In fact, models trained on four and eight hours of data approach (or sometimes surpass, in the case of the threshold algorithm) the performance of systems that have been optimized over large populations and many more data points.
Figure 3.
Plots of averaged (a) sensitivity, (b) specificity, (c) positive predictive value, and (d) accuracy of models trained on increasing amounts of patient-specific data, compared to the performance of the CMS algorithm and a simple threshold algorithm. The horizontal axes are logarithmic in time, and the vertical axes for (b) and (d) are expanded to show differences in a narrow range of values. Vertical bars indicate the ranges of the individual values that comprise the averages. The data come from Tables 2 and 3.
We could not have expected the models based on only a half-hour of data to do very well, because in many monitoring sessions there had been very few alarm events during that brief time. Thus, these models have been unable to learn alarm events, leading to their low sensitivity. To our surprise, however, these models’ specificity is high. One explanation of their high specificity is that they properly (although not very intelligently) call all stable points correctly. Even models built from one, two and four hours of data suffer from this finding. We believe that this same phenomenon probably also accounts for the sharp dip in positive predictive value and the milder dip in accuracy in the models trained on one and two hours of data.
The monotonic increase with time in sensitivity of our models, as shown by Figure 3, suggests that by something like eight hours, the algorithms have encountered most of the alarm conditions that they need to recognize. Although the average performances of the models from both classification tree learning and neural network learning are not as high as that of the current bedside monitors, the models built from eight hours of training data are more specific, accurate, and able to predict correctly than the threshold alarm algorithm does, although their sensitivity remains lower. We had not anticipated this result, though as in most machine learning applications, additional training data tend to lead to better performance. Indeed, we now expect that these performance measures would continue to improve with additional patient-specific training data. The principal limits to such improvement come from the possibility that the models would eventually over-fit the available data and that patients’ physiological state would eventually change so as to make predictions based on past data incorrect.
4.2 Implications for Learning Methods
In our experiments, models built using neural networks do better on almost every measure than those built using classification trees. Perhaps this should not be surprising given the continuous nature of the input data and the greater ability of neural networks to model non-linear interactions among the data. As illustrated by the classification tree in Figure 2, the minute averaged data play a critical role toward the leaves of the tree, probably because they better take into account the context within which each data point is interpreted. Perhaps additional derived features, such as local slopes, or the parameters of linear models fit locally to the data might additionally improve classification, as has been the case in earlier work by Tsien [16].
There are many possible improvements to our methods, which may move us toward more accurate monitoring systems. We have already mentioned the need to combine the best existing models for decision support with patient-specific learning methods. Especially early during a patient’s ICU stay, the general models trained on population data must bear the brunt of recognizing alarm events because the patient-specific learning methods have not yet had a chance to learn much. Later, we should put greater reliance on the learning methods, though there will remain circumstances novel to any individual patient that should be better recognized by a more broadly-trained monitoring algorithm. For any such combined model, it would be helpful for each component to issue not only a classification label for each data point but also some indication of its certainty. For example, our neural network models could use a sigmoid rather than a threshold output unit, so that their results could be combined with other monitoring outputs using some method that relies on continuous risk or probability scores from its inputs.
In retrospect, we believe that a better set of experiments would have learned the shorter-term models from the last rather than the first n hours of data. That would have reflected the most recent history of the patient and thus been a more fair indication of their value. Nevertheless, we suspect that the patient-specific models trained on longer data series would still have performed better.
Our method learned five different sets of models during monitoring session that lasted eight hours or more. Consecutive sets of models were built using twice as much data as the previous set did, and each time we learned new models, we did so by running batch training algorithms over all the previously collected data. Had we run truly incremental (on-line) learning algorithms [6], we would not have had to choose particular training durations because any model would have kept completely up to date to interpret each new data point. However, the feasibility of running incremental learning algorithms for model development in clinical settings such as the ICU still needs to be examined; thus, as a first step in realizing patient-specific learning in real time, this research has focused on the incremental nature of the learning tasks itself and used non-incremental learning algorithms to carry out these tasks. We plan to use truly incremental learning algorithms to develop patient-specific models in the future. Because the first two commandments for implementing clinical information systems are “speed is everything” and “doctors won’t wait for the computer’s pearls” [15], we still face the challenging questions of how to optimize on-line training. Methods that learn more sophisticated models or ones that explore a larger set of parameter settings for learning may be too slow to run on-line. Some delays in using the most recent data may even be desirable if it takes time for clinicians to give their gold standard annotations or for the patient’s future course to modify an annotation.
4.3 Imbalanced Datasets
Because appropriate alarm events are relatively rare, the vast preponderance of data points collected in a study such as this one should be classified as stable. As a result, learning algorithms may be justified to learn to classify all data points as “stable” and to consider the true positives simply as “noise” that could be suppressed by the learners during pruning.
One way to overcome this problem is to use an asymmetric cost function, one that penalizes misclassification of alarm points (false negatives) more heavily than misclassification of stable points (false positives). This seems clinically reasonable, because in a monitoring situation we may be willing to accept more false alarms in order to avoid missing true ones. We did some limited experiments with asymmetric cost functions ranging from 10:1 to 1000:1 for penalizing false negatives, but the resulting models seemed generally inferior to the ones reported above, mainly due to significant decreases in specificity. We do not know what the right cost ratio should be; a careful cost-benefit analysis to determine this ratio has not been performed, to our knowledge.
We also experimented using resampling methods to overcome the problem introduced by the imbalanced datasets, but also without positive results. For example, sub-sampling the “stable” points to equalize the number of alarm and stable training points increased the number of false positives called by our models without notable improvement in other measures. Perhaps by discarding many of the stable points, the models learn fewer of what are considered clinically normal conditions in critical care.
We also tried replicating the data points labeled as alarm, but the resulting training dataset became much bigger in size. While each model did not take much time to classify each new data point, the training time increased significantly with more training examples. Other, more sophisticated resampling methods such as bagging [4] might do a better job at addressing this problem.
4.4 Time
Perhaps the major impediment to further development and deployment of the new methods introduced here lies in the need for correctly annotated data from a very busy, tense, and pressured environment. Clinical staff are unlikely to have the time to annotate all clinical events listed in Section 2.2 or the resources to hire trained observers to perform that task, as done by the first author in our experiments. Therefore, automated methods to annotate clinical events are essential to patient-specific learning in real time. The sources of data we have for developing such annotations are the responses of clinicians to alarms and an ability to judge the appropriateness of an alarm based on what happens in the (near) future course of the patient. These, perhaps combined with data from additional instruments in the ICU, may suffice to provide a basis for learning improved patient-specific models. One capacity of human observers that no automated methods could completely emulate, however, is the timely identification of events where an alarm should have been considered but was not.
Computer time was also an impediment in our experiments, as we have mentioned. A single-processor laptop machine was barely able to keep up with our computational demands. Of course this type of problem is normally overcome by technical advances according to Moore’s Law [13]. For example, the computers being manufactured in 2008 typically have multiple processors, faster memory buses, and both computers and monitors have more robust and faster serial communication ports. Despite these improvements, it appears that added sophistication in the nature of the learning algorithms, the complexity of the models being learned, the amount of training data, and optimization by investigating a space of tunable learning parameters might demand enough additional computing time to overtake even faster computers. For example, we artificially limited the slower neural network training program to 120 seconds of training time; yet Table 1 suggests that training times may increase non-linearly, especially when boosting and cross-validation are used, even in the faster classification tree learner. Furthermore, because variations in parameter settings can lead to significantly different models being constructed, the learning time becomes less predictable.
5 Conclusion
Our expanded system of real-time data collection and algorithm development demonstrated that patient-specific learning in real time is a feasible approach to developing alarm algorithms for monitoring purposes in the ICU. Performance measures of the trained classification trees and neural networks were consistent with the course of a generalized learning process. The ones that are trained with eight hours of monitored numerics data outperformed the standard threshold alarm algorithm, which represented the alarm algorithms in previous generations of patient monitoring systems, and came close in performance to the alarm algorithm in the new-generation monitors. These algorithms are also useful for integrating multiple physiological signals in detecting adverse clinical events and generate informative alerts at the bedside. Our methodology could be used in constructing comprehensive models that, in tracking the state of a patient, generalize over both disease processes and patient populations.
Acknowledgments
The authors would like to thank Adrienne Randolph, Christine Tsien Silvers, Isaac Kohane, David Martin, the P-MICU staff at Boston Children’s Hospital, the patients who participated in the study and their families. This manuscript is based on the first author’s Master’s thesis [19] and is a heavily revised expansion of a conference paper published earlier [20]. The work presented here was supported in part by DARPA contract F30602-99-0509, a MEMP (Medical Engineering/Medical Physics) Fellowship from the Harvard-MIT Division of Health Sciences and Technology, Biomedical Informatics training grant T15-LM07092 from the National Library of Medicine, and research grant R01-EB001659 from the National Institute of Biomedical Imaging and BioEngineering.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Roger G. Mark, MIT, personal communication, 2000.
References
- 1.EasyNN-plus help manual. Neural Planner Software; 2003. Technical report. [Google Scholar]
- 2.See5 help manual. RuleQuest Research; 2003. Technical report. [Google Scholar]
- 3.Data mining tools see5 and c5.0. [accessed 3/18/2008]; http://rulequest.com/see5-info.html.
- 4.Breiman L. Bagging predictors. Machine Learning. 1996;24(2):123–140. [Google Scholar]
- 5.Chambrin MC, Ravaux P, Calvelo-Aros D, Jaborska A, Chopin C, Boniface B. Multicentric study of monitoring alarms in the adult intensive care unit (icu): a descriptive analysis. Intensive Care Med. 1999;25(12):1360–1366. doi: 10.1007/s001340051082. [DOI] [PubMed] [Google Scholar]
- 6.Giraud-Carrier C. A note on the utility of incremental learning. AI Communications. 2000;13(4):215–223. [Google Scholar]
- 7.Hagenouw RR. Should we be alarmed by our alarms? Curr Opin Anaesthesiol. 2007;20(6):590–594. doi: 10.1097/ACO.0b013e3282f10dff. [DOI] [PubMed] [Google Scholar]
- 8.Hay WW, Jr, Rodden DJ, Collins SM, Melara DL, Hale KA, Fashaw LM. Reliability of conventional and new pulse oximetry in neonatal patients. J Perinatol. 2002;22(5):360–366. doi: 10.1038/sj.jp.7210740. [DOI] [PubMed] [Google Scholar]
- 9.Imhoff M, Kuhls S. Alarm algorithms in critical care monitoring. Anesth Analg. 2006;102(5):1525–1537. doi: 10.1213/01.ane.0000204385.01983.61. [DOI] [PubMed] [Google Scholar]
- 10.Lawless ST. Crying wolf: false alarms in a pediatric intensive care unit. Crit Care Med. 1994;22(6):981–985. [PubMed] [Google Scholar]
- 11.Lepape A, Gillibert RP, Perdrix JP, Grozel JM, Banssillon V. Practical aspects of indirect calorimetry in post-anesthesia recovery [in French] Agressologie. 1990;31(1):74–76. [PubMed] [Google Scholar]
- 12.Mitchell TM. Machine Learning. Boston: WCB McGraw-Hill; 1997. [Google Scholar]
- 13.Moore GE. Craming more components onto integrated circuits. Electronics Magazine. 1965;38(8) [Google Scholar]
- 14.Quinlan JR. C4.5: Programs for Machine Learning. San Mateo, CA: Morgan Kaufmann; 1993. [Google Scholar]
- 15.Shabot MM. Ten commandments for implementing clinical information systems. Baylor University Medical Center Proceedings. 2004;17(3) doi: 10.1080/08998280.2004.11927979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsien C. PhD thesis. Cambridge, MA: Massachusetts Institute of Technology; 2000. TrendFinder: Automated detection of alarmable trends. [Google Scholar]
- 17.Tsien CL, Fackler JC. Poor prognosis for existing monitors in the intensive care unit. Crit Care Med. 1997;25(4):614–619. doi: 10.1097/00003246-199704000-00010. [DOI] [PubMed] [Google Scholar]
- 18.van Gils M, Jansen H, Nieminen K, Summers R, Weller PR. Using artificial neural networks for classifying icu patient states. IEEE Engineering in Medicine and Biology Magazine. 1997;16(6):41–47. doi: 10.1109/51.637116. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y. Master’s thesis. Cambridge, MA: Massachusetts Institute of Technology; 2003. Real-time analysis of physiological data and development of alarm algorithms for patient monitoring in the intensive care unit. [Google Scholar]
- 20.Zhang Y. Real-time development of patient-specific alarm algorithms for critical care. Conf Proc IEEE Eng Med Bio Soc. 2007;1:4351–4354. doi: 10.1109/IEMBS.2007.4353300. [DOI] [PubMed] [Google Scholar]