Abstract
More than twenty years of study has provided a better understanding of hepatitis C virus (HCV) life cycle, including the general properties of viral RNA and proteins. This effort facilitates the development of sensitive diagnostic tools and effective antiviral treatments. At present, serologic screening test is recommended to perform on individuals in the high risk groups and nucleic acid tests are recommended to confirm the active HCV infections. Quantization and genotyping of HCV RNAs are important to determine the optimal duration of anti-viral therapy and predict the likelihood of response. In the early 2000s, pegylated interferon plus ribavirin became the standard anti-HCV treatment. However, this therapy is not ideal. To 2014, boceprevir, telaprevir, simeprevir, sofosbuvir and Harvoni are approved by Food and Drug Administration for the treat of HCV infections. It is likely that the new all-oral, interferon-free, pan-genotyping anti-HCV therapy will be available within the next few years. Majority of HCV infections will be cured by these anti-viral treatments. However, not all patients are expected to be cured due to viral resistance and the high cost of antiviral treatments. Thus, an efficient prophylactic vaccine will be the next challenge in the fight against HCV infection.
Keywords: Hepatitis C virus, Diagnosis, Treatment, Hepatocellular carcinoma, Nucleic acid test, Enzyme immunoassay, Interferon, Direct acting antivirals, Host-targeted agents, Sofosbuvir
Core tip: Understanding the general properties of hepatitis C virus (HCV) viral RNA and proteins facilitates the development of sensitive diagnostic tools and effective antiviral treatments. At present, serologic screening test is recommended to perform on individuals in the high risk groups and nucleic acid tests are recommended to confirm the active HCV infections. To 2014, in addition to pegylated interferon and ribavirin, boceprevir, telaprevir, simeprevir, sofosbuvir and Harvoni are approved by Food and Drug Administration to treat HCV infections. The majority of HCV infections can be cured by these anti-viral treatments. An efficient prophylactic vaccine will be the next challenge in the fight against HCV infection.
INTRODUCTION
Hepatitis C virus (HCV) accounts for approximately 15%-20% cases of acute hepatitis. After acute infection, around 50% to 80% of HCV patients will develop chronic infection. Approximately, HCV infects 170 million individuals worldwide (http://www.who.int). Chronic hepatitis C (CHC) patients are at high risk to develop life-threatening complications, including cirrhosis in 20% of cases and hepatocellular carcinoma (HCC) at an incidence of 4%-5% per year in cirrhotic patients[1-3]. Epidemiological studies also indicate that HCV is associated with a number of extrahepatic manifestations including insulin resistance, type 2 diabetes mellitus, glomerulopathies, oral manifestations, etc.[4-7].
Most HCV-infected patients would develop chronic hepatitis, but approximately 15%-40% of them could clear the virus spontaneously. What are the factors responsible for the different outcomes of HCV infection? The viral evolutionary dynamics and host genetic polymorphisms, e.g., the interleukin 28B (IL28B) gene, are important in determining the outcomes of HCV infection[8,9].
After the discovery of HCV, great nucleotide diversity among isolates was reported[10,11]. Due to the error-prone viral RNA-dependent RNA polymerase (HCV NS5B protein), a closely related but diverse population of viral variants known as quasispecies is produced within HCV-infected patients[12]. There is 1%-5% variation in HCV nucleotide sequence from a single infected patient. Accumulation of nucleotide substitutions in the virus has resulted in diversification into distinct subtypes and even genotypes. Therefore, the HCV RNA genome sequences are highly heterogeneous. At present, HCV is classified into eleven genotypes (designated as 1-11) differing in their nucleotide sequence by 30%-50%, six of them are the major ones (genotypes 1 to 6)[13,14]. Within HCV genotype, several subtypes (designated as a, b, c, etc.) can be defined that differ in their nucleotide sequence by 15%-30%[15,16]. The prevalence of HCV genotypes and subtypes is geographically different[17,18]. At present, genotype 1 is the most prevalent (46%) globally, followed by genotype 3, genotype 2 and genotype 4. Various genotypes have different infectivity and pathogenicity, thereby influencing the rate of progression to cirrhosis and the risk of HCC. HCV heterogeneity would also result in different responses to anti-viral treatments, e.g., genotypes 1 and 4 are more resistant to interferon based therapies than genotypes 2 and 3[13,19,20]. Therefore, HCV heterogeneity poses a challenge to the development of pan-genotypic anti-viral treatments. In addition, HCV heterogeneity hinders the development of a successful vaccine to against all HCV genotypes. Of course, HCV heterogeneity could also affect viral diagnosis.
Though heterogeneous, different HCV genotypes preserve the similarity of life cycle in cells. This review briefly describes the HCV life cycle and the general properties of viral RNA and proteins, which are related to the diagnosis of viral infection and the development of anti-viral therapy. This review also summarizes the current methods to detect and treat HCV infections.
VIROLOGY
HCV life cycle[21-23]
HCV is a small enveloped RNA virus belonging to the family Flaviviridae and genus hepacivirus. HCV genomic RNA was single-stranded with positive polarity, which was packaged by core protein and enveloped by a lipid bilayer containing two viral glycoproteins (E1 and E2) to form the virion[24]. Despite the nucleotide sequence divergence among genotypes, all currently recognized HCV genotypes are hepatotropic and pathogenic.
The HCV lifecycle begins with the attachment of a virion to its specific receptors on hepatocytes[25]. Up to now, the high-density lipoprotein receptor scavenger receptor class B type I, tetraspanin CD81, tight junction protein claudin-1, and occludin are the known cellular receptors initiating the attachment step of HCV infection. It is proposed that the virus, after binding with its receptor complex, is internalized, and that the nucleocapsid is released into the cytoplasm. The virus is then uncoating to free its genomic RNA, and the HCV genomic RNA is used both for polyprotein translation and replication in the cytoplasm. HCV replication takes place within the “replication complex” containing the viral non-structural proteins and cellular proteins[26].
HCV replication is catalyzed by the NS5B protein. However, other viral nonstructural proteins are also important. The NTPase/helicase domain of the NS3 protein has several functions important for viral replication, including RNA stimulated NTPase activity, RNA binding, and unwinding of RNA regions of extensive secondary structure. NS4B initiates the formation of replication complex that supports HCV replication. The NS5A protein also plays an important regulatory role in virus replication. New direct acting antivirals (DAAs), specifically designed to inhibit the NS5B RNA dependent RNA polymerase are now becoming available. Several other newer DAAs (e.g., inhibitors to the NS5A protein) have also shown promise in clinical studies[27,28].
A number of cellular factors are involved in HCV replication, such as, cyclophilin A, required for HCV replication through its interacting with NS5A and the NS5B, and microRNA-122, which helps HCV replication through its binding with the 5’un-translated region (5’UTR) of the HCV genome. Therefore, host factors may also become the potential targets for anti-HCV therapies. At present, at least two host-targeted agents (HTAs) have reached clinical development, including specific inhibitors of cyclophilin A and antagonists of microRNA-122[21,28,29].
The very low-density lipoprotein synthesis/ secretion machinery is involved in the production of the infectious HCV particles. HCV uses this lipoprotein biosynthetic pathway to produce mature viral particles and to export them[30,31].
General properties of HCV RNA and proteins
Viral genomic RNA[22,23,32]: The HCV genomic RNA contains three distinct regions[21,32]: (1) a 5′UTR or non-coding region; (2) a long open reading frame (ORF) of more than 9000 nucleotides (nt); and (3) a short 3′UTR.
The HCV 5’UTR contains 341 nt located upstream of the ORF translation initiation codon. The 5’UTR contains the internal ribosomal entry site which forms a stable pre-initiation complex by direct binding with the 40S ribosomal subunit for the HCV polyprotein translation.
The long ORF encodes a polyprotein of approximately 3000 amino acids, which will be further processed by host and viral proteases.
The 3’UTR is principally involved in minus-strand priming during HCV replication.
The nucleotide sequence variability is distributed throughout the entire viral genome. The 5’UTR is the most conserved region in the genome while the regions encoding envelope proteins (E1, E2) are the most variable ones. Thus, the highly conserved 5’UTR region is usually the target of choice for HCV genome detection across different genotypes (in “Diagnosis” section).
Viral proteins[21-23]: The long ORF encodes a polyprotein which is processed co- and posttranslationally by host and viral proteases into at least 10 different proteins, which are arranged in the order of NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. Host signal peptidase is required for the cleavages at C-E1, E1-E2, E2-p7, and p7-NS2 junctions. Host signal peptide peptidase is also needed for the further maturation of HCV core proteins. Two viral proteases are involved in the processing of HCV nonstructural proteins: NS2, a zinc-dependent metalloprotease that cleaves between NS2 and NS3; and NS3/4A, a serine protease that cleaves between the NS3-NS4A, NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B junctions (Figure 1). These viral proteins remain associated with intracellular membranes after processing.
Figure 1.
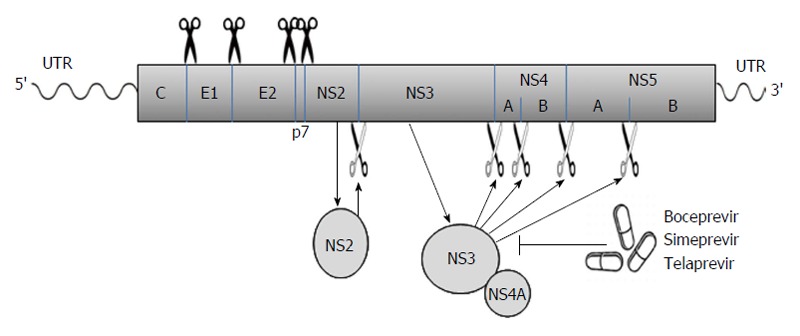
The hepatitis C virus poly-protein is processed co- and posttranslationally by host and viral proteases into at least 10 different proteins, which are arranged in the order of NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. Host signal peptidase is required for the cleavages at C-E1, E1-E2, E2-p7, and p7-NS2 junctions. NS2 cleaves the site between NS2 and NS3; NS3/4A serine protease cleaves the sites at NS3-NS4A, NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B junctions. Several new DAAs (e.g., boceprevir, simeprevir, and telaprevir) specifically designed to inhibit the NS3/4A protease are now becoming available. The wavy lines mark the un-translated regions (UTR) of hepatitis C virus genomic RNA while the rectangle represents the poly-protein derived from the long open reading frame.
Core, a 191-amino acid polypeptide, is a highly conserved basic protein which packages its RNA genome to form the viral nucleocapsid. Core protein may be involved in hepatocarcinogenesis and steatosis[33]. The alternate reading frame protein (ARFP)/core+1/F (frameshift) protein is generated as a result of a -2/+1 ribosomal frameshift in the core-encoding region. The role of ARFP/core+1/F protein in the HCV lifecycle remains unknown[33]. Two envelope glycoproteins, E1 (33-35 kDa) and E2 (70-72 kDa), assemble as non-covalent heterodimers and are necessary for viral entry. Unlike HCV core protein, E2 contains hypervariable regions with amino acid sequences differing up to 80% between different HCV isolates. Therefore, enzyme immuno-assay (EIA) to detect the HCV core antigen rather than the envelope proteins is performed to represent the number of the virions (in “Diagnosis” section).
p7, a 63-amino acid polypeptide, forms ion channels with an essential role in virus infection. NS2, a 21-23 kDa transmembrane protein, is also essential for the completion of the viral replication cycle. The highly hydrophobic N-terminal residues of NS2 protein form three or four transmembrane helices inserting into the endoplasmic reticulum (ER) membrane while the C-terminal domain of NS2 protein plays an important role in NS2/3 auto protease activity together with the N-terminal domain of NS3. NS3, a 67 kDa polypeptide, possesses multifunctional activities. The N-terminus of NS3 has serine protease activity and its C-terminus has NTPase/helicase activity. The NS3 proteins are bound with ER membrane through interacting with NS4A proteins. The enzymatic activity of the NS3 NTPase/helicase activity is indispensable for viral RNA replication. NS4A, a 54-amino acid polypeptide, is a cofactor for NS3 protein. The interaction between NS4A and NS3 proteins stabilizes and facilitates its protease activity. The NS3/4A protease is essential for the cleavages of the viral nonstructural proteins. Thus, it is the target of choice for anti-HCV therapy. Several new DAAs specifically designed to inhibit the NS3/4A protease are now available[21,28,34] (Figure 1). NS4A is also required for the phosphorylation of NS5A and can directly interact with NS5A. NS4B, a small hydrophobic 27 kDa protein, recruits other viral non-structural proteins to form the replication complex. NS5A, a 56-58 kDa hydrophilic phosphoprotein, is also important in viral replication though its exact role is not clear. NS5B, a 65 kDa protein, is an RNA-dependent RNA polymerase (RdRP) responsible for the synthesis of new genomic RNAs. As a central component of the HCV replication complex[26], NS5B has become a major target for antiviral therapy[21,28] (Figure 2).
Figure 2.
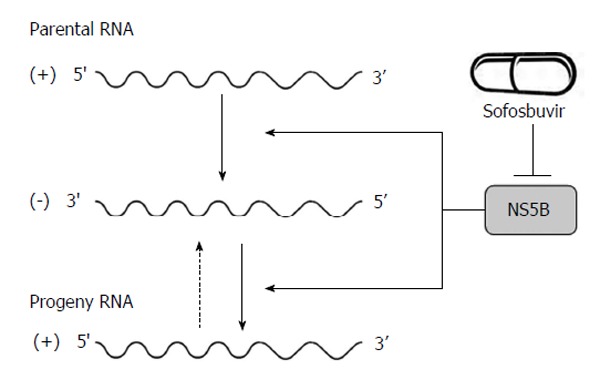
Hepatitis C virus NS5B acts as RNA-dependent RNA polymerase and plays an important role in the synthesis of new RNA genomes. As the central component of the hepatitis C virus replication complex, NS5B has emerged as a major target for antiviral treatment. New direct acting antivirals, specifically designed to inhibit the NS5B are now becoming available (e.g., sofosbuvir).
More than twenty years of study has provided a better understanding of HCV life cycle, including the properties of viral RNA and proteins. This effort facilitates the development of sensitive diagnostic tools and effective antiviral treatments toward HCV infections.
DIAGNOSIS
The purpose of diagnosis of viral infection is to allow the infected persons to be identified and treated. Thus, diagnosis of viral infection is important to prevent disease progression and viral spread. Majority of primary HCV-infected patients are asymptomatic, thus, symptoms could not be used as specific indicators for HCV infection. HCV viremia could still exist despite a normal serum alanine aminotransferase (ALT) level. Therefore, virological methods rather than ALT levels are used to diagnose HCV infection[35].
In general, the virological methods for examining viral infections include indirect and direct tests. The indirect tests are to detect antibody induced by viral infection, including IgM for recent infection and IgG for recent or past infection. The direct tests include virus isolation, detection of viral antigens and viral nucleic acids.
At present, it is difficult to isolate and culture HCV using clinical specimens. Furthermore, anti-HCV IgMs could be detected not only in 50%-93% of patients with acute hepatitis C but also in 50%-70% of CHC patients[36]. Therefore, anti-HCV IgM cannot be used as a reliable marker for the acute HCV infection, and IgM assays have not been used in clinical practice[37,38]. At present, diagnostic assays for anti-HCV total antibody, viral core antigen, and viral genomic RNA are used in clinical practice[19,35,39-44] (Figure 3).
Figure 3.
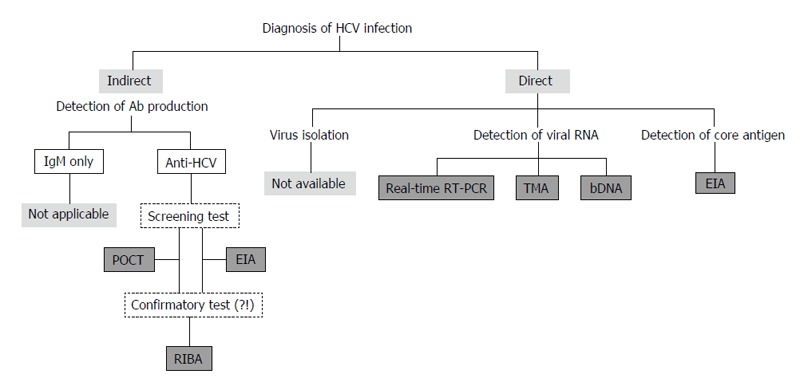
Assays to detect anti-hepatitis C virus antibody, viral core antigen, and viral genomic RNA are used to diagnose HCV infection in clinical practice. HCV: Hepatitis C virus; POCT: Point-of-care test; EIA: Enzyme immuno-assay; RIBA: Recombinant immunoblot assays; RT-PCR: Reverse transcription-polymerase chain reaction; TMA: Transcription-mediated amplification; bDNA: Branched DNA.
Detection of antibody production
In general, serological tests for detecting anti-HCV antibodies include tests for screening and confirmation. Screening tests are used first to screen the antibody positive specimens while confirmatory tests are then used to verify the positive screening specimens.
Screening test: EIA: At present, the third generation test of EIA for the anti-HCV antibody detection is commonly used in the diagnostic laboratory[45]. Conserved antigens from the HCV core, NS3, NS4 and NS5 regions are used in these tests to detect anti-HCV antibodies. The sensitivity of third-generation EIAs was estimated at 98.9% and the specificity was found at 100% in patients with chronic liver disease[46]. EIAs are easy to use and inexpensive. Furthermore, this assay could be fully automated and adapted to large volume testing. Therefore, EIAs to detect anti-HCV antibody are generally recommended for screening the HCV infections[40]. However, this assay should not be used in infants younger than 18 mo due to the possibility of reactivity with maternal antibody[47]. Several Food and Drug Administration (FDA)-approved antibody-based assays are available[47]. However, the time between HCV infection and the appearance of detectable antibodies (serological window period) is generally more than 40 d using the third generation EIAs[48]. In 2008, the fourth generation EIA has become available which could detect the anti-HCV antibody significantly earlier than the other assays (www.microgenbioproducts.com). The antigens utilized in the fourth generation anti-HCV assay are derived from the core (two different epitope clusters), NS3, NS4A, NS4B, as well as the NS5A regions. NS3 and NS4 antigens are derived from genotypes 1a, 1b, 2 and 3.
Screening test: The rapid, point-of-care test[23,49]: Point-of-care tests are used directly at the site of patient care, outside of the diagnostic laboratory. Several point-of-care tests (POCTs) have been developed to detect anti-HCV antibodies with a relatively high sensitivity and specificity[50,51]. The test currently approved by the FDA in 2010 is the OraQuick HCV Rapid Antibody Test (OraSure Technologies, Bethlehem, PA). It is approved for use in patients over 15 years old, for screening persons who are considered at risk for HCV infection. This test detects anti-HCV antibodies in different specimens, e.g., fingerstick and venipuncture whole blood, serum, plasma, or oral fluid. Recombinant proteins or synthetic peptides of core, NS3 and NS4 antigens are immobilized on a nitrocellulose membrane to perform an indirect lateral flow immunoassay, and the results are directly visualized using colloidal gold labeled protein A, which generates a reddish-purple line within 20 to 40 min in the presence of anti-HCV antibodies in the specimens. These rapid tests are suitable for resource-limited settings because they are cheap, simple to perform and fast[52].
Confirmatory tests: Recombinant immunoblot assays[49]: Recombinant immunoblot assays (RIBA) can be used to confirm the presence of anti-HCV antibodies for individuals who have showed positive reactivity by EIAs. This assay is highly specific, as the presence of antibodies against each of the several HCV proteins is assessed as individual bands on a membrane strip[53]. The INNO-LIA™ HCV Score (Fujirebio Europe, previously Innogenetics) assay can be automated. This assay includes recombinant proteins and synthetic peptides from E2 hypervariable region, NS3 helicase, NS4A, NS4B and NS5A regions.
Due to the high sensitivity and specificity of anti-HCV EIAs, RIBA is no longer needed in the diagnostic laboratories for verification[19,45]. Furthermore, nucleic acid tests for viral RNA rather than RIBA are used as a confirmatory test for HCV infection[19].
HCV infection can be detected easily using currently available third generation EIAs[45]. In addition, the use of POCTs can increase HCV screening opportunities. However, these indirect virological tests to detect anti-HCV antibody cannot distinguish current from past infection[37,38]. Active HCV infection must be confirmed by the direct diagnostic methods.
Detection of viral RNA
Based on the items used for amplification, nucleic acid amplification tests (NAT) are divided into target amplification, signal amplification and probe amplification methods[54]. Target amplification methods [e.g., reverse transcription-polymerase chain reaction (RT-PCR) and transcription-mediated amplification (TMA)] and signal amplification methods [e.g., branched DNA (bDNA)] were commonly used to detect the presence of HCV RNA[19,47,49,55]. The presence of HCV RNA in the serum is a reliable marker of viremia. Universal standardization for HCV RNA titer is important. The World Health Organization (WHO) has established an international standard for HCV RNA quantification units[56], i.e., an HCV RNA international unit (IU), which is currently used in all of the commercial HCV RNA quantitative assays no matter what the techniques used[35,57].
Qualitative HCV RNA detection[47]: Qualitative detection assays are based on the principle of target amplification using either RT-PCR or TMA. Several FDA-approved qualitative assays for HCV RNA are available[47]. HCV RNA is extracted and converted into complementary DNA (cDNA) using reverse transcriptase. The cDNA is subsequently processed via cyclic enzymatic reactions leading to the generation of a large number of double-stranded DNAs in PCR-based assays or single-stranded RNAs in TMA. Detection of these amplified products is achieved by hybridizing the produced amplicons onto specific probes. In general, the highly conserved 5’UTR region is the target of choice for HCV genomic RNA detection across different genotypes[49].
Quantitative HCV RNA detection[47]: HCV RNA can be quantified by means of target amplification techniques (real-time RT-PCR or TMA) or signal amplification techniques (bDNA assay). Several FDA-approved quantitative assays to detect HCV RNA are also available[23,39,47]. Real-time RT-PCR is the method of choice for the quantification of HCV RNA levels in clinical practice. This assay is highly sensitive with wide dynamic range of quantification and can prevent carryover contamination.
Fully automated HCV NAT assays have been available in the United States since 2007, and guidelines regarding the requirements for HCV NAT assays were issued in 2010 (http://www.fda.govQBiologicsBloodVaccinesQGuidance- ComplianceRegulatoryInformation/Guidances/default.htm). However, it is necessary to remember that not all HCV genotypes are detected equally by NAT assays, most likely because of nucleotide mismatches which has occurred before[58,59].
HCV RNA in the serum is probably the earliest detectable marker of acute HCV infection, preceding the appearance of anti-HCV antibody by several weeks[35]. CHC infection is defined as the presence of HCV RNA more than 6 mo. HCV RNA levels remain relatively stable over time in CHC patients. Therefore, after a positive reaction screened by the anti-HCV antibody test, NATs to detect HCV RNA is often used as the confirmatory tool to diagnose CHC infection[60]. Detection of HCV RNA is also used to determine the viral load both prior to and during antiviral treatments (www.who.int). On the other hand, the HCV RNA level has no prognostic value[61]. The level of HCV genomic RNA, reflection of HCV replication, does not correlate with the severity of liver disease, not with the risk of liver disease progression to cirrhosis or HCC.
Detection of viral core antigen[44]
Compared to other diagnostic methods like EIA, the advantages of NATs are having higher specificity and sensitivity. However, the disadvantages of these assays are time-consuming and require sophisticated technical equipment, trained technicians, dedicated laboratory space and expensive reagents. In patients with HCV infection, it has been demonstrated that the HCV core antigen level strongly correlates with the HCV RNA level for various genotypes[62]. Thus, due to cheap and easy-to-perform, the HCV core antigen quantification assay can be used as an alternative method to NATs to detect HCV RNA[44]. Currently, core antigen detection by means of a chemiluminescent microparticle immunoassay can be fully automated in the Architect HCV Core antigen test (Abbott Laboratories)[63]. The Architect HCV Ag assay had a specificity of 100%, with a lower limit of detection of 3 fmol/L corresponds to approximately 1000 IU/mL of HCV RNA[62]. Whereas,current HCV RNA assays have a lower level of detection between 5-15 IU/mL[44]. In general, about 90% of HCV RNA positive samples are positive with a viral load above 10000 IU/mL[64], well in the sensitivity range of the HCV core antigen assay[44]. Therefore, HCV antigen detection might be the next step following a positive antibody screening test. Several combination assays for detection of both anti-HCV antibodies and HCV core antigen have been developed[65].
At present, EIA to detect HCV core antigen is too insensitive to replace the NATs to detect HCV RNA in the blood bank setting[66] and in the treatment monitoring according to the current clinical practice guidelines. However, it could be used as a supplemental test in resource-limited settings[67]. The Architect HCV Ag assay has been suggested as a better monitoring tool in the era of new all-oral, interferon-free antiviral treatments that do not require high analytical sensitivity[62].
Interpretations of diagnostic results[19,41,42,47]
The presence of HCV RNA in the absence of anti-HCV antibodies is strongly indicative of acute hepatitis C (AHC), which can be confirmed by seroconversion (i.e., the appearance of anti-HCV antibodies) a few days or weeks later (Figure 4). However, there are still other possibilities for the presence of HCV RNA in the absence of anti-HCV antibodies, e.g., CHC infection in the immunodepressed patients, hemodialysis patients or agammaglobulinemic subjects.
Figure 4.
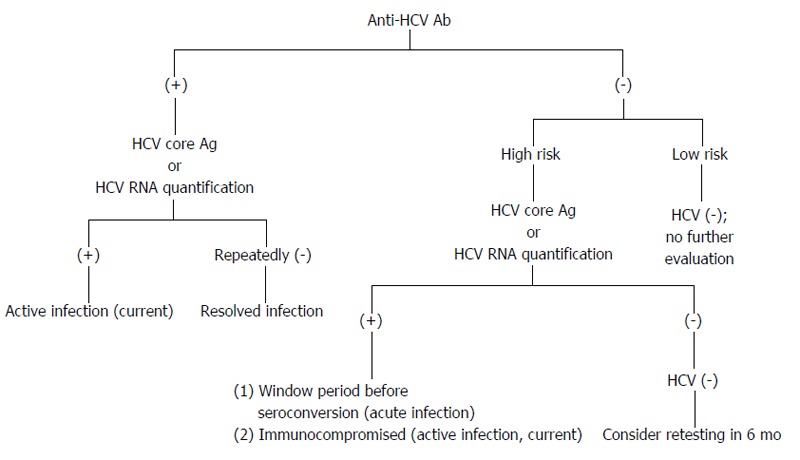
Possible diagnostic results for hepatitis C virus infection. Individuals are in high risk, e.g., persons who have been exposed to HCV; persons with elevated alanine aminotransferase; persons who are immunocompromised. HCV: Hepatitis C virus.
The presence of both anti-HCV and HCV RNA does not allow one to distinguish AHC from an acute exacerbation of CHC. However, the anti-HCV IgG avidity index within the first 8 d following the onset of clinical symptoms may be useful in identifying actual AHC[68].
If the antibody test is positive and the HCV RNA test is negative, this result indicates a resolution of HCV infection or AHC during a period of low-level viremia. If the HCV RNA assay is negative and remains negative for more than 6 mo, then the individuals are recovered from a past HCV infection.
CHC is defined as the persistence of HCV RNA for more than 6 mo. In patients with clinical signs of chronic liver disease, CHC is certain when both anti-HCV antibodies and HCV RNA are present.
Genotyping
Different HCV genotypes would result in different responses to antiviral treatments[35]. Thus, genotyping is important to predict the likelihood of response and determine the optimal duration of therapy[47].
Serological method: The HCV genotype can be determined by detection of antibodies against HCV genotype-specific epitopes using a competitive EIA[69]. The currently available assay (Murex HCV serotyping 1-6 HC02, Abbott Laboratories, North Chicago, Illinois) could identify the six HCV genotypes (1-6) but not subtypes, and provide interpretable results in approximately 90% of chronically infected immunocompetent patients[7].
Molecular techniques: The reference method for HCV genotyping is genome sequencing of the core/E1 or the NS5B regions and subsequent phylogenetic analysis[70]. However, this in-house method is restricted to reference centers. HCV genotyping assays approved for in vitro diagnostic use are also commercially available[23,49]. The Linear Array HCV Genotyping Test (Roche Molecular Systems) targets the 5’UTR[71]. This assay is based on conventional PCR amplification followed by reverse hybridization onto membrane strips containing specific probes. The obtained band pattern can be either visually interpreted or read by a scanner. Assays targeting other regions in addition to the 5′UTR have been recently developed to better discriminate between subtypes 1a and 1b. The Versant HCV genotype 2.0 assay (Siemens) is also based on reverse hybridization and targets the 5’UTR and core regions[72]. On the other hand, the Abbott RealTime HCV Genotype II (Abbott Molecular) targets the 5’UTR and NS5B regions. This assay is based on a single-step real-time RT-PCR with labeled genotype-/subtype-specific probes that minimizes contamination with amplified products[73,74].
Subtyping
HCV subtyping is important for epidemiological studies, especially in the case of outbreaks, but it is not considered to be clinically relevant for the treatment of interferon-α and ribavirin. However, subtyping may be clinically relevant in the era of DAAs. For example, the phase 3 studies of telaprevir, boceprevir, faldaprevir and simeprevir showed lower sustained virologic response (SVR) rates for HCV-subtype 1a than those for subtype 1b[75]. In addition, BILB 1941, a non-nucleoside inhibitor of HCV NS5B, has been shown to have better antiviral efficacy in patients with subtype 1b than in those with subtype 1a[76]. Therefore, methods to determine the HCV subtypes should be important in the era of DAAs. The second-generation line probe assay, a reverse hybridization assay that uses probes targeting both the 5’UTR and core-coding region, correctly identified HCV subtypes 1a and 1b in more than 99% of cases. Thus, this assay could be used to differentiate HCV subtypes 1a and 1b in clinical trials and practice[74].
Screening for HCV-infected patients
According to the WHO (www.who.int), up to 80 percents of HCV-positive patients do not show symptoms. Therefore, most cases of HCV infection are currently undiagnosed. The major way to diagnose HCV infection is to screen high risk groups for anti-HCV antibodies. Humans are the primary HCV reservoir[77]. HCV transmission occurs primarily through direct percutaneous exposure to blood. Therefore, the most common risk factors for HCV infection are persons with history of injection of illicit drugs and with blood transfusion prior to July 1992. The populations with less common risk factors for HCV infection are persons with organ transplant prior to July 1992, receiving clotting factor concentrate prior to 1987, being born to an HCV-infected mother, and with a history of chronic hemodialysis, intranasal use of illicit drugs, acquiring a tattoo, incarceration, having sex with an HCV-infected partner, needlestick or other mucosal exposure, with persistently elevated levels of ALT[40]. Therefore, WHO recommends that anti-HCV EIA be performed on individuals who are part of a population with high HCV seroprevalence or who have a history of HCV risk exposure and/or behavior, rather than at the time of presentation with symptomatic diseases. In addition, it is suggested that NATs for the detection of HCV RNA be performed directly following a seropositive test result to establish a definitive diagnosis of HCV infection (www.who.int). The Center for Disease Control (CDC) has also recommended screening high-risk individuals for HCV since 1998. The CDC further modified the HCV screening guidelines in 2012 to include a one-time HCV test for all US residents born during 1945-1965, independent of risk factors[40].
TREATMENT
Around 50%-80% of persons with acute hepatitis C will develop CHC infection, and 5%-25% of them reportedly progress to cirrhosis after 20-25 years[78]. Persons with cirrhosis are at risk for developing end-stage liver disease as well as HCC[79]. The goal of antiviral treatment for CHC is to halt disease progression, prevent cirrhosis decompensation and reduce the risk of HCC[80]. However, it is really difficult to design and carry out clinical trials to provide direct evidence related to these outcomes (www.ahrq.gov). SVR is defined as undetectable levels of HCV RNA at least 24 wk after completion of therapy. At present, SVR is the primary endpoint of successful therapy and is associated with durable clearance of virus[81]. CHC patients with a SVR after antiviral therapy had a lower risk of all-cause mortality than patients with no SVR[82]. Therefore, SVR is the standard marker of the successful antiviral treatment in clinical trials.
In the early 2000s, the combination of pegylated interferon plus ribavirin (PR) became the standard anti-HCV treatment[19,28,83]. However, the anti-HCV interferon therapy is not ideal because it requires weekly injections and is associated with numerous systemic side effects (e.g., flu-like symptoms, fatigue, etc.). Therefore, other anti-HCV therapies are needed. In principle, every step of the HCV lifecycle, including receptor attachment, endocytosis, uncoating, translation, polyprotein processing, RNA replication, virion assembly, maturation and release, can be a target for new anti-HCV drugs[21]. Advances in understanding of the HCV lifecycle have led to the development of numerous highly effective, well-tolerated oral DAAs[84-86]. In 2011, the United States FDA approved the first DAAs, boceprevir (trade name Victrelis™) (www.fda.gov/ForConsumers/ByAudience/ForPatientAdvocates/ucm255413.htm) and telaprevir (trade name Incivek®) (www.fda.gov/ForConsumers/ByAudience/ForPatientAdvocates/ucm256328.htm), for the treatment of chronic HCV genotype 1 infection (Figure 1). Both drugs are classified as NS3/4A protease inhibitors, with a potential advantage of shorter therapy duration (24 to 28 wk) compared with standard PR treatment for genotype 1 infection (48 wk)[87,88]. Either drug is administered in combination with PR[89]. In 2013, FDA approved another NS3/4A protease inhibitor: simeprevir (http://www.olysio.com/). The HCV NS5B protein is an essential enzyme (RNA-dependent RNA polymerase) in HCV viral replication and has been a prime target in the search for antiviral therapies. In 2013, the FDA approved sofosbuvir (an inhibitor of NS5B) in combination with ribavirin for oral dual therapy of HCV genotypes 2 and 3, and for triple therapy with PR for treatment-naive patients with HCV genotypes 1 and 4 (Figure 2). Sofosbuvir treatment regimens last 12 wk for genotypes 1, 2 and 4, and 24 wk for treatment of genotype 3. This is typically half the time as with prior treatments. Thus, to December 2013, licensed treatments for HCV infection include pegylated and standard interferon alpha, ribavirin, the NS3/4A protease inhibitors boceprevir, telaprevir and simeprevir; and the NS5B nucleotide polymerase inhibitor sofosbuvir. Without taking resource used into consideration, WHO provides the following guidelines (http://www.who.int/hiv/pub/hepatitis/hepatitis-c-guides): (1) Pegylated interferon in combination with ribavirin is recommended for the treatment of CHC rather than standard non-pegylated interferon with ribavirin; (2) Treatment with the DAAs telaprevir or boceprevir, given in combination with PR, is suggested for genotype 1 chronic HCV infection rather than PR alone; (3) Sofosbuvir, given in combination with ribavirin with or without pegylated interferon (depending on the HCV genotype), is recommended in genotypes 1, 2, 3 and 4 HCV infection rather than PR alone (or no treatment for persons who cannot tolerate interferon); and (4) Simeprevir, given in combination with PR, is recommended for persons with subtype 1b HCV infection and for persons with subtype 1a HCV infection without the Q80K polymorphism rather than PR alone.
Although interferon-free anti-HCV therapies will be available in the near future, before then, peginterferon will be still required with either the protease inhibitor simeprevir, or the nucleotide analogue polymerase inhibitor, sofosbuvir, for the treatment of genotype 1 infection. Peginterferon also appears to be a useful adjunct to sofosbuvir and ribavirin for patients with genotype 3 infection, particularly those with cirrhosis. Therefore, pretreatment assessments are needed for the anti-HCV treatments containing interferon, including HCV genotype determination, liver disease staging (e.g., fibrosis), psychiatric assessment (e.g., depression and suicide risk), assessments for alcohol or substance use disorders; adherence, evaluation for HIV co-infection, pregnancy, testing for IL28B genotype, and concomitant medical conditions (e.g., autoimmune disorders). Adverse effects will be still checked for the anti-HCV treatments containing interferon, including anemia, neutropenia, rash and skin reactions, anorectal signs and symptoms, elevated uric acid, bilirubin levels, etc. (http://www.who.int).
HCV resistance is defined as the selection of viral variants, reducing the susceptibility to the drug’s inhibitory activity in the presence of anti-HCV drugs. Resistance-associated variants are naturally produced during the HCV replication. At present, there is no commercially available assay to detect the presence of resistant viruses before or during antiviral treatments[75]. The only way to check if a patient has developed a resistant virus is to monitor for HCV RNA rebound (more than 10 fold increase from the nadir HCV RNA) during anti-HCV treatment.
During treatment, NATs to quantitate HCV RNA should be performed at weeks 4, 8 (with boceprevir-containing regimens), 12, and 24 of treatment, at the end-of-treatment, and 24 wk after treatment to monitor the viral titers. The determination of the viral titers helps to detect drug-resistant viruses and to adjust the dose and duration of the anti-HCV treatment.
The factors influencing the efficacy of anti-HCV treatments based on interferon are divided into two categories: viral-related and host-related factors[90,91]. The viral-related factors include the HCV genotype, baseline viral load, and virological response during treatment. The host-related factors include age, gender, race-ethnicity, fibrosis stage, obesity, hepatic steatosis, low-density lipoprotein cholesterol, insulin resistance, and IL28B gene polymorphisms. In particular, IL28B gene polymorphisms are associated with the SVR. Thus, host genetic factors are important to determine the effect of anti-HCV therapy based on interferon. The anti-HCV treatment will change significantly over the next few years as therapeutic regimens based on interferon-free are rapidly evolving. Thus, it is necessary to determine the effects of these viral-related and host-related factors on the efficacy of anti-HCV therapy based on DAAs therapy without interferon.
In early 2014, several reports regarding novel DAAs have been published: (1) Combined simeprevir and sofosbuvir was efficacious and well tolerated for patients with HCV genotype 1[92]; (2) Combined daclatasvir (NS5A replication complex inhibitor) and asunaprevir (NS3/4A protease inhibitor) could be used as an all-oral, PR-free treatment option for patients with HCV subtype 1b infection, including those with cirrhosis[93]; (3) In combinations with other oral DAAs, dasabuvir (a non-nucleoside inhibitor of NS5B) results in very high rates of SVR (about 95%) in patients with HCV genotype 1 infection with a good tolerability and safety[94]; (4) The sustained response rate of ABT-450, a potent inhibitor of NS3/4A protease, plus other direct antiviral drugs reaches 90%-95% in both naïve and treatment-experienced genotype 1 patients, and tolerability is good[95]; (5) Sofosbuvir was also effective in patients co-infected with HCV and HIV[96]; (6) Sofosbuvir plus PR achieved high SVR rates in patients with HCV genotype 1 infection, and also appeared effective in patients with HCV genotype 4, 5 or 6 infection. Oral sofosbuvir was generally well tolerated in CHC patients[97]; (7) In combinations with other oral DAAs, ombitasvir (an inhibitor of the HCV NS5A) achieves very high rates of SVR (about 95%) in patients with HCV genotype 1 infection with a good tolerability[98]; and (8) Combination of daclatasvir and asunaprevir results in a very high rate of viral eradication in both treatment-naïve and treatment-experienced patients, with a SVR rate of 80%-90%[99]. In October of 2014, FDA approved Harvoni (ledipasvir and sofosbuvir) to treat chronic HCV genotype 1 infection (http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm418365.htm). Ledipasvir is an inhibitor of HCV NS5A protein (www.gilead.com). Harvoni is the first combination pill approved to treat chronic HCV genotype 1 infection.
Not only DAAs, HTAs as anti-HCV therapy are also developed. HTAs block HCV production by interacting with cellular factors. Because they target conserved host proteins, not variable viral proteins, HTAs have the potential for pangenotypic antiviral activity and a high barrier to resistance[29]. Until now, only two HTAs have reached clinical trial, including specific inhibitors to cyclophilin A peptidyl-prolyl cis/trans isomerase activity and antagonists of microRNA-122[21,28].
The pace of DAAs and/or HTAs entering clinical trials is breathtaking[84]. The optimal combination of DAAs (and/or HTAs) that maximizes potency, minimizes resistance, and limits toxicity will be available soon[100]. Once combination DAA therapies are available, peginterferon will serve a smaller and smaller role[101]. Indeed, DAAs trump interferon-alpha in their capacity to rescue exhausted T cells upon HCV clearance[102]. From 2015, interferon-free anti-HCV regimens with short treatment duration and fewer side effects will be available[84,85,103]. However, peginterferon may still have a role in resource-limited regions due to high cost of DAAs[104].
CHC patients achieved a SVR after anti-HCV treatments exhibited a reduction in all-cause mortality > 50% compared with those who are non-responders[82]. However, in such a nonrandom clinical trial, an improved outcome could be biased by parameters, such as the good health conditions of the responders. Indeed, it has been reported that some patients who achieve SVRs still go on to develop end-stage liver disease[105]. Thus, the concept which cure rests solely on SVR may not be always correct. Actually, despite improving SVR, there is no evidence that PR beneficially affects patient-relevant outcomes such as mortality and liver morbidity[106,107]. Therefore, it is better to remember that SVR might not work as a surrogate for patient-relevant outcomes[108].
The cure rate for HCV infection is expected to be over 95% with the new all-oral, interferon-free regimens within the next few years. However, due to drug resistance[109,110], suboptimal activity against certain HCV genotypes and the extremely high cost[100,104], not all patients can be cured. Currently, no effective vaccine is available for HCV infection. Therefore, an efficient prophylactic vaccine will be the next challenge in the combat against HCV infection[111-113].
Footnotes
P- Reviewer: Hung LY, Sipos F S- Editor: Tian YL L- Editor: A E- Editor: Liu SQ
Supported by Grants from the National Science Council of Taiwan, No. NSC 101-2320-B-320-011-MY3 (Dr. Shih-Yen Lo); and from the Tzu Chi University to Dr. Shih-Yen Lo, No. TCIRP 103002-03; and to Dr. Hui-Chun Li, No. TCIRP 103002-02.
Conflict-of-interest: The authors declare no conflicting interests (including but not limited to commercial, personal, political, intellectual, or religious interests) in this work.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: August 28, 2014
First decision: December 17, 2014
Article in press: April 2, 2015
References
- 1.Alberti A, Chemello L, Benvegnù L. Natural history of hepatitis C. J Hepatol. 1999;31 Suppl 1:17–24. doi: 10.1016/s0168-8278(99)80369-9. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 3.Chen SL, Morgan TR. The natural history of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:47–52. doi: 10.7150/ijms.3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrozzo M, Scally K. Oral manifestations of hepatitis C virus infection. World J Gastroenterol. 2014;20:7534–7543. doi: 10.3748/wjg.v20.i24.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozkok A, Yildiz A. Hepatitis C virus associated glomerulopathies. World J Gastroenterol. 2014;20:7544–7554. doi: 10.3748/wjg.v20.i24.7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimbert S, Valensi P, Lévy-Marchal C, Perret G, Richardet JP, Raffoux C, Trinchet JC, Beaugrand M. High prevalence of diabetes mellitus in patients with chronic hepatitis C. A case-control study. Gastroenterol Clin Biol. 1996;20:544–548. [PubMed] [Google Scholar]
- 7.Montenegro L, De Michina A, Misciagna G, Guerra V, Di Leo A. Virus C hepatitis and type 2 diabetes: a cohort study in southern Italy. Am J Gastroenterol. 2013;108:1108–1111. doi: 10.1038/ajg.2013.90. [DOI] [PubMed] [Google Scholar]
- 8.Rau M, Baur K, Geier A. Host genetic variants in the pathogenesis of hepatitis C. Viruses. 2012;4:3281–3302. doi: 10.3390/v4123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito T, Ueno Y. Transmission of hepatitis C virus: self-limiting hepatitis or chronic hepatitis? World J Gastroenterol. 2013;19:6957–6961. doi: 10.3748/wjg.v19.i41.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timm J, Roggendorf M. Sequence diversity of hepatitis C virus: implications for immune control and therapy. World J Gastroenterol. 2007;13:4808–4817. doi: 10.3748/wjg.v13.i36.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Sequence diversity of hepatitis C viral genomes. Mol Biol Med. 1990;7:495–501. [PubMed] [Google Scholar]
- 12.Gómez J, Martell M, Quer J, Cabot B, Esteban JI. Hepatitis C viral quasispecies. J Viral Hepat. 1999;6:3–16. doi: 10.1046/j.1365-2893.1999.t01-1-6120131.x. [DOI] [PubMed] [Google Scholar]
- 13.Chayama K, Hayes CN. Hepatitis C virus: How genetic variability affects pathobiology of disease. J Gastroenterol Hepatol. 2011;26 Suppl 1:83–95. doi: 10.1111/j.1440-1746.2010.06550.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y. Scotomas in molecular virology and epidemiology of hepatitis C virus. World J Gastroenterol. 2013;19:7910–7921. doi: 10.3748/wjg.v19.i44.7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 16.Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 17.Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61:S45–S57. doi: 10.1016/j.jhep.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 18.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chevaliez S, Pawlotsky JM. Hepatitis C virus: virology, diagnosis and management of antiviral therapy. World J Gastroenterol. 2007;13:2461–2466. doi: 10.3748/wjg.v13.i17.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Shamy A, Hotta H. Impact of hepatitis C virus heterogeneity on interferon sensitivity: an overview. World J Gastroenterol. 2014;20:7555–7569. doi: 10.3748/wjg.v20.i24.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemon SM, Walker C, Alter MJ, Yi M. Hepatitis C virus. In: Knipe DM, Howley PM, editors. Field Virology. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1253–1304. [Google Scholar]
- 23.Chevaliez S, Pawlotsky JM. Virology of hepatitis C virus infection. Best Pract Res Clin Gastroenterol. 2012;26:381–389. doi: 10.1016/j.bpg.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci USA. 2013;110:9505–9510. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu YZ, Qian XJ, Zhao P, Qi ZT. How hepatitis C virus invades hepatocytes: the mystery of viral entry. World J Gastroenterol. 2014;20:3457–3467. doi: 10.3748/wjg.v20.i13.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu M, Rice CM. Structures of hepatitis C virus nonstructural proteins required for replicase assembly and function. Curr Opin Virol. 2013;3:129–136. doi: 10.1016/j.coviro.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol. 2013;11:482–496. doi: 10.1038/nrmicro3046. [DOI] [PubMed] [Google Scholar]
- 28.Pawlotsky JM. Hepatitis C virus: standard-of-care treatment. Adv Pharmacol. 2013;67:169–215. doi: 10.1016/B978-0-12-405880-4.00005-6. [DOI] [PubMed] [Google Scholar]
- 29.Pawlotsky JM. What are the pros and cons of the use of host-targeted agents against hepatitis C? Antiviral Res. 2014;105:22–25. doi: 10.1016/j.antiviral.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartenschlager R, Penin F, Lohmann V, André P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Lin CC, Tsai P, Sun HY, Hsu MC, Lee JC, Wu IC, Tsao CW, Chang TT, Young KC. Apolipoprotein J, a glucose-upregulated molecular chaperone, stabilizes core and NS5A to promote infectious hepatitis C virus virion production. J Hepatol. 2014;61:984–993. doi: 10.1016/j.jhep.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 33.Li HC, Ma HC, Yang CH, Lo SY. Production and pathogenicity of hepatitis C virus core gene products. World J Gastroenterol. 2014;20:7104–7122. doi: 10.3748/wjg.v20.i23.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakulin I, Pasechnikov V, Varlamicheva A, Sannikova I. NS3 protease inhibitors for treatment of chronic hepatitis C: Efficacy and safety. World J Hepatol. 2014;6:326–339. doi: 10.4254/wjh.v6.i5.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chevaliez S, Pawlotsky JM. How to use virological tools for optimal management of chronic hepatitis C. Liver Int. 2009;29 Suppl 1:9–14. doi: 10.1111/j.1478-3231.2008.01926.x. [DOI] [PubMed] [Google Scholar]
- 36.Sagnelli E, Coppola N, Marrocco C, Coviello G, Rossi G, Battaglia M, Sagnelli C, Messina V, Tonziello A, Scolastico C, et al. Diagnosis of HCV related acute hepatitis by serial determination of IgM to HCV: a preliminary observation. J Biol Regul Homeost Agents. 2003;17:207–210. [PubMed] [Google Scholar]
- 37.Farci P, Alter HJ, Govindarajan S, Wong DC, Engle R, Lesniewski RR, Mushahwar IK, Desai SM, Miller RH, Ogata N. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 38.Yuki N, Hayashi N, Ohkawa K, Hagiwara H, Oshita M, Katayama K, Sasaki Y, Kasahara A, Fusamoto H, Kamada T. The significance of immunoglobulin M antibody response to hepatitis C virus core protein in patients with chronic hepatitis C. Hepatology. 1995;22:402–406. [PubMed] [Google Scholar]
- 39.Albertoni G, Castelo Girão MJ, Schor N. Mini review: current molecular methods for the detection and quantification of hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1. Int J Infect Dis. 2014;25:145–149. doi: 10.1016/j.ijid.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 40.Campos-Outcalt D. Hepatitis C: new CDC screening recommendations. J Fam Pract. 2012;61:744–746. [PubMed] [Google Scholar]
- 41.Chevaliez S, Pawlotsky JM. Diagnosis and management of chronic viral hepatitis: antigens, antibodies and viral genomes. Best Pract Res Clin Gastroenterol. 2008;22:1031–1048. doi: 10.1016/j.bpg.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Chevaliez S, Pawlotsky JM. Virological techniques for the diagnosis and monitoring of hepatitis B and C. Ann Hepatol. 2009;8:7–12. [PubMed] [Google Scholar]
- 43.Gupta E, Bajpai M, Choudhary A. Hepatitis C virus: Screening, diagnosis, and interpretation of laboratory assays. Asian J Transfus Sci. 2014;8:19–25. doi: 10.4103/0973-6247.126683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tillmann HL. Hepatitis C virus core antigen testing: role in diagnosis, disease monitoring and treatment. World J Gastroenterol. 2014;20:6701–6706. doi: 10.3748/wjg.v20.i22.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alborino F, Burighel A, Tiller FW, van Helden J, Gabriel C, Raineri A, Catapano R, Stekel H. Multicenter evaluation of a fully automated third-generation anti-HCV antibody screening test with excellent sensitivity and specificity. Med Microbiol Immunol. 2011;200:77–83. doi: 10.1007/s00430-010-0171-0. [DOI] [PubMed] [Google Scholar]
- 46.Colin C, Lanoir D, Touzet S, Meyaud-Kraemer L, Bailly F, Trepo C. Sensitivity and specificity of third-generation hepatitis C virus antibody detection assays: an analysis of the literature. J Viral Hepat. 2001;8:87–95. doi: 10.1046/j.1365-2893.2001.00280.x. [DOI] [PubMed] [Google Scholar]
- 47.Mack CL, Gonzalez-Peralta RP, Gupta N, Leung D, Narkewicz MR, Roberts EA, Rosenthal P, Schwarz KB. NASPGHAN practice guidelines: Diagnosis and management of hepatitis C infection in infants, children, and adolescents. J Pediatr Gastroenterol Nutr. 2012;54:838–855. doi: 10.1097/MPG.0b013e318258328d. [DOI] [PubMed] [Google Scholar]
- 48.Barrera JM, Francis B, Ercilla G, Nelles M, Achord D, Darner J, Lee SR. Improved detection of anti-HCV in post-transfusion hepatitis by a third-generation ELISA. Vox Sang. 1995;68:15–18. doi: 10.1111/j.1423-0410.1995.tb02538.x. [DOI] [PubMed] [Google Scholar]
- 49.Saludes V, González V, Planas R, Matas L, Ausina V, Martró E. Tools for the diagnosis of hepatitis C virus infection and hepatic fibrosis staging. World J Gastroenterol. 2014;20:3431–3442. doi: 10.3748/wjg.v20.i13.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scalioni Lde P, Cruz HM, de Paula VS, Miguel JC, Marques VA, Villela-Nogueira CA, Milagres FA, Cruz MS, Bastos FI, Andrade TM, et al. Performance of rapid hepatitis C virus antibody assays among high- and low-risk populations. J Clin Virol. 2014;60:200–205. doi: 10.1016/j.jcv.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Shivkumar S, Peeling R, Jafari Y, Joseph L, Pant Pai N. Accuracy of rapid and point-of-care screening tests for hepatitis C: a systematic review and meta-analysis. Ann Intern Med. 2012;157:558–566. doi: 10.7326/0003-4819-157-8-201210160-00006. [DOI] [PubMed] [Google Scholar]
- 52.Lee SR, Kardos KW, Schiff E, Berne CA, Mounzer K, Banks AT, Tatum HA, Friel TJ, Demicco MP, Lee WM, et al. Evaluation of a new, rapid test for detecting HCV infection, suitable for use with blood or oral fluid. J Virol Methods. 2011;172:27–31. doi: 10.1016/j.jviromet.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Martin P, Fabrizi F, Dixit V, Quan S, Brezina M, Kaufman E, Sra K, DiNello R, Polito A, Gitnick G. Automated RIBA hepatitis C virus (HCV) strip immunoblot assay for reproducible HCV diagnosis. J Clin Microbiol. 1998;36:387–390. doi: 10.1128/jcm.36.2.387-390.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Persing DH, Landry ML. In vitro amplification techniques for the detection of nucleic acids: new tools for the diagnostic laboratory. Yale J Biol Med. 1989;62:159–171. [PMC free article] [PubMed] [Google Scholar]
- 55.Irshad M, Mankotia DS, Irshad K. An insight into the diagnosis and pathogenesis of hepatitis C virus infection. World J Gastroenterol. 2013;19:7896–7909. doi: 10.3748/wjg.v19.i44.7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiffman ML, Ferreira-Gonzalez A, Reddy KR, Sterling RK, Luketic VA, Stravitz RT, Sanyal AJ, Garrett CT, De Medina M, Schiff ER. Comparison of three commercially available assays for HCV RNA using the international unit standard: implications for management of patients with chronic hepatitis C virus infection in clinical practice. Am J Gastroenterol. 2003;98:1159–1166. doi: 10.1111/j.1572-0241.2003.07449.x. [DOI] [PubMed] [Google Scholar]
- 57.Pawlotsky JM. Use and interpretation of virological tests for hepatitis C. Hepatology. 2002;36:S65–S73. doi: 10.1053/jhep.2002.36815. [DOI] [PubMed] [Google Scholar]
- 58.Chevaliez S, Bouvier-Alias M, Castéra L, Pawlotsky JM. The Cobas AmpliPrep-Cobas TaqMan real-time polymerase chain reaction assay fails to detect hepatitis C virus RNA in highly viremic genotype 4 clinical samples. Hepatology. 2009;49:1397–1398. doi: 10.1002/hep.22767. [DOI] [PubMed] [Google Scholar]
- 59.Chevaliez S, Bouvier-Alias M, Rodriguez C, Soulier A, Poveda JD, Pawlotsky JM. The Cobas AmpliPrep/Cobas TaqMan HCV test, version 2.0, real-time PCR assay accurately quantifies hepatitis C virus genotype 4 RNA. J Clin Microbiol. 2013;51:1078–1082. doi: 10.1128/JCM.02004-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butcher A, Aslam S, Hemyari P, Cowen U, Heilek G. HCV RNA detection in HCV antibody-positive patients with the COBAS AmpliPrep/COBAS TaqMan HCV test, v2.0 in comparison with FDA-approved nucleic acid tests. J Clin Virol. 2014;60:336–340. doi: 10.1016/j.jcv.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 61.Pawlotsky JM. More sensitive hepatitis C virus RNA detection: what for? J Hepatol. 2010;52:783–785. doi: 10.1016/j.jhep.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 62.Chevaliez S, Soulier A, Poiteau L, Bouvier-Alias M, Pawlotsky JM. Clinical utility of hepatitis C virus core antigen quantification in patients with chronic hepatitis C. J Clin Virol. 2014;61:145–148. doi: 10.1016/j.jcv.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 63.Morota K, Fujinami R, Kinukawa H, Machida T, Ohno K, Saegusa H, Takeda K. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J Virol Methods. 2009;157:8–14. doi: 10.1016/j.jviromet.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Fytili P, Tiemann C, Wang C, Schulz S, Schaffer S, Manns MP, Wedemeyer H. Frequency of very low HCV viremia detected by a highly sensitive HCV-RNA assay. J Clin Virol. 2007;39:308–311. doi: 10.1016/j.jcv.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Laperche S, Le Marrec N, Girault A, Bouchardeau F, Servant-Delmas A, Maniez-Montreuil M, Gallian P, Levayer T, Morel P, Simon N. Simultaneous detection of hepatitis C virus (HCV) core antigen and anti-HCV antibodies improves the early detection of HCV infection. J Clin Microbiol. 2005;43:3877–3883. doi: 10.1128/JCM.43.8.3877-3883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waldenström J, Konar J, Ekermo B, Norder H, Lagging M. Neonatal transfusion-transmitted hepatitis C virus infection following a pre-seroconversion window-phase donation in Sweden. Scand J Infect Dis. 2013;45:796–799. doi: 10.3109/00365548.2013.797601. [DOI] [PubMed] [Google Scholar]
- 67.Vermehren J, Susser S, Berger A, Perner D, Peiffer KH, Allwinn R, Zeuzem S, Sarrazin C. Clinical utility of the ARCHITECT HCV Ag assay for early treatment monitoring in patients with chronic hepatitis C genotype 1 infection. J Clin Virol. 2012;55:17–22. doi: 10.1016/j.jcv.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 68.Coppola N, Pisapia R, Marrocco C, Martini S, Vatiero LM, Messina V, Tonziello G, Sagnelli C, Filippini P, Piccinino F, et al. Anti-HCV IgG avidity index in acute hepatitis C. J Clin Virol. 2007;40:110–115. doi: 10.1016/j.jcv.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 69.Pawlotsky JM, Prescott L, Simmonds P, Pellet C, Laurent-Puig P, Labonne C, Darthuy F, Remire J, Duval J, Buffet C, et al. Serological determination of hepatitis C virus genotype: comparison with a standardized genotyping assay. J Clin Microbiol. 1997;35:1734–1739. doi: 10.1128/jcm.35.7.1734-1739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy DG, Willems B, Deschênes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5’ untranslated region sequences. J Clin Microbiol. 2007;45:1102–1112. doi: 10.1128/JCM.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lam TH, Cheng RS, Lai ST, Tsang TY, Cheng VC, Ho SL, Yam WC. Evaluation of in-house and commercial genotyping assays for molecular typing of hepatitis C virus in Hong Kong. Br J Biomed Sci. 2010;67:82–85. doi: 10.1080/09674845.2010.11730296. [DOI] [PubMed] [Google Scholar]
- 72.Verbeeck J, Stanley MJ, Shieh J, Celis L, Huyck E, Wollants E, Morimoto J, Farrior A, Sablon E, Jankowski-Hennig M, et al. Evaluation of Versant hepatitis C virus genotype assay (LiPA) 2.0. J Clin Microbiol. 2008;46:1901–1906. doi: 10.1128/JCM.02390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shinol RC, Gale HB, Kan VL. Performance of the Abbott RealTime HCV Genotype II RUO assay. J Clin Microbiol. 2012;50:3099–3101. doi: 10.1128/JCM.01249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chevaliez S, Bouvier-Alias M, Brillet R, Pawlotsky JM. Hepatitis C virus (HCV) genotype 1 subtype identification in new HCV drug development and future clinical practice. PLoS One. 2009;4:e8209. doi: 10.1371/journal.pone.0008209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schneider MD, Sarrazin C. Antiviral therapy of hepatitis C in 2014: do we need resistance testing? Antiviral Res. 2014;105:64–71. doi: 10.1016/j.antiviral.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 76.Erhardt A, Deterding K, Benhamou Y, Reiser M, Forns X, Pol S, Calleja JL, Ross S, Spangenberg HC, Garcia-Samaniego J, et al. Safety, pharmacokinetics and antiviral effect of BILB 1941, a novel hepatitis C virus RNA polymerase inhibitor, after 5 days oral treatment. Antivir Ther. 2009;14:23–32. [PubMed] [Google Scholar]
- 77.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36:S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 79.Fattovich G, Giustina G, Degos F, Diodati G, Tremolada F, Nevens F, Almasio P, Solinas A, Brouwer JT, Thomas H, et al. Effectiveness of interferon alfa on incidence of hepatocellular carcinoma and decompensation in cirrhosis type C. European Concerted Action on Viral Hepatitis (EUROHEP) J Hepatol. 1997;27:201–205. doi: 10.1016/s0168-8278(97)80302-9. [DOI] [PubMed] [Google Scholar]
- 80.Toshikuni N, Arisawa T, Tsutsumi M. Hepatitis C-related liver cirrhosis - strategies for the prevention of hepatic decompensation, hepatocarcinogenesis, and mortality. World J Gastroenterol. 2014;20:2876–2887. doi: 10.3748/wjg.v20.i11.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swain MG, Lai MY, Shiffman ML, Cooksley WG, Zeuzem S, Dieterich DT, Abergel A, Pessôa MG, Lin A, Tietz A, et al. A sustained virologic response is durable in patients with chronic hepatitis C treated with peginterferon alfa-2a and ribavirin. Gastroenterology. 2010;139:1593–1601. doi: 10.1053/j.gastro.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 82.Backus LI, Boothroyd DB, Phillips BR, Belperio P, Halloran J, Mole LA. A sustained virologic response reduces risk of all-cause mortality in patients with hepatitis C. Clin Gastroenterol Hepatol. 2011;9:509–516.e1. doi: 10.1016/j.cgh.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 83.Garg G, Kar P. Management of HCV infection: current issues and future options. Trop Gastroenterol. 2009;30:11–18. [PubMed] [Google Scholar]
- 84.Au JS, Pockros PJ. Novel therapeutic approaches for hepatitis C. Clin Pharmacol Ther. 2014;95:78–88. doi: 10.1038/clpt.2013.206. [DOI] [PubMed] [Google Scholar]
- 85.Pawlotsky JM. New hepatitis C virus (HCV) drugs and the hope for a cure: concepts in anti-HCV drug development. Semin Liver Dis. 2014;34:22–29. doi: 10.1055/s-0034-1371007. [DOI] [PubMed] [Google Scholar]
- 86.Pawlotsky JM. New hepatitis C therapies. Semin Liver Dis. 2014;34:7–8. doi: 10.1055/s-0034-1371178. [DOI] [PubMed] [Google Scholar]
- 87.Poordad F, McCone J, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, McNair L, Alam J, Muir AJ. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 89.Yee HS, Chang MF, Pocha C, Lim J, Ross D, Morgan TR, Monto A. Update on the management and treatment of hepatitis C virus infection: recommendations from the Department of Veterans Affairs Hepatitis C Resource Center Program and the National Hepatitis C Program Office. Am J Gastroenterol. 2012;107:669–89; quiz 690. doi: 10.1038/ajg.2012.48. [DOI] [PubMed] [Google Scholar]
- 90.Gatselis NK, Zachou K, Saitis A, Samara M, Dalekos GN. Individualization of chronic hepatitis C treatment according to the host characteristics. World J Gastroenterol. 2014;20:2839–2853. doi: 10.3748/wjg.v20.i11.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu Y, Chen S. Antiviral treatment of hepatitis C virus infection and factors affecting efficacy. World J Gastroenterol. 2013;19:8963–8973. doi: 10.3748/wjg.v19.i47.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lawitz E, Poordad FF, Pang PS, Hyland RH, Ding X, Mo H, Symonds WT, McHutchison JG, Membreno FE. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet. 2014;383:515–523. doi: 10.1016/S0140-6736(13)62121-2. [DOI] [PubMed] [Google Scholar]
- 93.Manns M, Pol S, Jacobson IM, Marcellin P, Gordon SC, Peng CY, Chang TT, Everson GT, Heo J, Gerken G, et al. All-oral daclatasvir plus asunaprevir for hepatitis C virus genotype 1b: a multinational, phase 3, multicohort study. Lancet. 2014;384:1597–1605. doi: 10.1016/S0140-6736(14)61059-X. [DOI] [PubMed] [Google Scholar]
- 94.Gentile I, Buonomo AR, Borgia G. Dasabuvir: A Non-Nucleoside Inhibitor of NS5B for the Treatment of Hepatitis C Virus Infection. Rev Recent Clin Trials. 2014:Epub ahead of print. doi: 10.2174/1574887109666140529222602. [DOI] [PubMed] [Google Scholar]
- 95.Gentile I, Borgia F, Buonomo AR, Zappulo E, Castaldo G, Borgia G. ABT-450: a novel protease inhibitor for the treatment of hepatitis C virus infection. Curr Med Chem. 2014;21:3261–3270. doi: 10.2174/0929867321666140706125950. [DOI] [PubMed] [Google Scholar]
- 96.Bichoupan K, Dieterich DT, Martel-Laferrière V. HIV-hepatitis C virus co-infection in the era of direct-acting antivirals. Curr HIV/AIDS Rep. 2014;11:241–249. doi: 10.1007/s11904-014-0217-9. [DOI] [PubMed] [Google Scholar]
- 97.Keating GM. Sofosbuvir: a review of its use in patients with chronic hepatitis C. Drugs. 2014;74:1127–1146. doi: 10.1007/s40265-014-0247-z. [DOI] [PubMed] [Google Scholar]
- 98.Gentile I, Buonomo AR, Borgia G. Ombitasvir: a potent pan-genotypic inhibitor of NS5A for the treatment of hepatitis C virus infection. Expert Rev Anti Infect Ther. 2014;12:1033–1043. doi: 10.1586/14787210.2014.940898. [DOI] [PubMed] [Google Scholar]
- 99.Gentile I, Buonomo AR, Zappulo E, Minei G, Morisco F, Borrelli F, Coppola N, Borgia G. Asunaprevir, a protease inhibitor for the treatment of hepatitis C infection. Ther Clin Risk Manag. 2014;10:493–504. doi: 10.2147/TCRM.S66731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.deLemos AS, Chung RT. Hepatitis C treatment: an incipient therapeutic revolution. Trends Mol Med. 2014;20:315–321. doi: 10.1016/j.molmed.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 101.Feld JJ. The beginning of the end: what is the future of interferon therapy for chronic hepatitis C? Antiviral Res. 2014;105:32–38. doi: 10.1016/j.antiviral.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 102.Maini MK, Schurich A. Direct-acting antivirals trump interferon-alpha in their capacity to rescue exhausted T cells upon HCV clearance. J Hepatol. 2014;61:459–461. doi: 10.1016/j.jhep.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 103.Manns MP, von Hahn T. Novel therapies for hepatitis C - one pill fits all? Nat Rev Drug Discov. 2013;12:595–610. doi: 10.1038/nrd4050. [DOI] [PubMed] [Google Scholar]
- 104.Bichoupan K, Martel-Laferriere V, Sachs D, Ng M, Schonfeld EA, Pappas A, Crismale J, Stivala A, Khaitova V, Gardenier D, et al. Costs of telaprevir-based triple therapy for hepatitis C: $189,000 per sustained virological response. Hepatology. 2014;60:1187–1195. doi: 10.1002/hep.27340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gurusamy KS, Wilson E, Koretz RL, Allen VB, Davidson BR, Burroughs AK, Gluud C. Is sustained virological response a marker of treatment efficacy in patients with chronic hepatitis C viral infection with no response or relapse to previous antiviral intervention? PLoS One. 2013;8:e83313. doi: 10.1371/journal.pone.0083313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brok J, Gluud LL, Gluud C. Ribavirin plus interferon versus interferon for chronic hepatitis C. Cochrane Database Syst Rev. 2010;(1):CD005445. doi: 10.1002/14651858.CD005445.pub2. [DOI] [PubMed] [Google Scholar]
- 107.Hauser G, Awad T, Brok J, Thorlund K, Štimac D, Mabrouk M, Gluud C, Gluud LL. Peginterferon plus ribavirin versus interferon plus ribavirin for chronic hepatitis C. Cochrane Database Syst Rev. 2014;2:CD005441. doi: 10.1002/14651858.CD005441.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gluud C, Koretz R, Gurusamy K. Hepatitis C: a new direction, but an old story? Lancet. 2014;383:2122–2123. doi: 10.1016/S0140-6736(14)61025-4. [DOI] [PubMed] [Google Scholar]
- 109.Poveda E, Wyles DL, Mena A, Pedreira JD, Castro-Iglesias A, Cachay E. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral Res. 2014;108:181–191. doi: 10.1016/j.antiviral.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 110.Donaldson EF, Harrington PR, O’Rear JJ, Naeger LK. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology. 2015;61:56–65. doi: 10.1002/hep.27375. [DOI] [PubMed] [Google Scholar]
- 111.Colombatto P, Brunetto MR, Maina AM, Romagnoli V, Almasio P, Rumi MG, Ascione A, Pinzello G, Mondelli M, Muratori L, et al. HCV E1E2-MF59 vaccine in chronic hepatitis C patients treated with PEG-IFNα2a and Ribavirin: a randomized controlled trial. J Viral Hepat. 2014;21:458–465. doi: 10.1111/jvh.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Houghton M, Law JL, Tyrrell DL. An inactivated hepatitis C virus vaccine on the horizon? Gastroenterology. 2013;145:285–288. doi: 10.1053/j.gastro.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 113.Law JL, Chen C, Wong J, Hockman D, Santer DM, Frey SE, Belshe RB, Wakita T, Bukh J, Jones CT, et al. A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS One. 2013;8:e59776. doi: 10.1371/journal.pone.0059776. [DOI] [PMC free article] [PubMed] [Google Scholar]


