Abstract
Exosomes are small vesicles that mediate cell–cell communication. They contain proteins, lipids and RNA, and evidence is accumulating that these molecules are specifically sorted for release via exosomes. We recently showed that endothelial-cell-produced exosomes promote angiogenesis in vivo in a small RNA-dependent manner. Recent deep sequencing studies in exosomes from lymphocytic origin revealed a broad spectrum of small RNAs. However, selective depletion or incorporation of small RNA species into endothelial exosomes has not been studied extensively. With next generation sequencing, we identified all known non-coding RNA classes, including microRNAs (miRNAs), small nucleolar RNAs, yRNAs, vault RNAs, 5p and 3p fragments of miRNAs and miRNA-like fragments. In addition, we mapped many fragments of messenger RNAs (mRNAs) and mitochondrial RNAs (mtRNAs). The distribution of small RNAs in exosomes revealed a considerable overlap with the distribution in the producing cells. However, we identified a remarkable enrichment of yRNA fragments and mRNA degradation products in exosomes consistent with yRNAs having a role in degradation of structured and misfolded RNAs in close proximity to endosomes. We propose that endothelial endosomes selectively sequester cytoplasmic RNA-degrading machineries taking part in gene regulation. The release of these regulatory RNAs via exosomes may have implications for endothelial cell–cell communication.
Keywords: extracellular vesicles, microvesicles, next generation sequencing, quality control
Exosomes are small (50–130 nm) extracellular vesicles generated within multivesicular bodies (MVB). Upon fusion with the plasma membrane, MVB release their intraluminal vesicles into the environment. They were discovered by Johnstone et al. and Harding et al., who described the shedding of transferrin receptors from reticulocytes (1,2). The release of exosomes by a large number of cells has been demonstrated in vitro, and exosomes have been isolated from many body fluids, pointing to the presence and relevance of exosomes in vivo (3). Exosomes were initially assumed to be vehicles for the disposal of superfluous molecules. However, they were later demonstrated to have a possible function in intercellular communication when Raposo et al. showed in 1996 that MHC-II-bearing exosomes released by B-cells are able to activate resting T-cells (4).
Subsequent studies showed that exosomes contain not only proteins and lipids from their cell of origin, but also incorporate functional mRNA molecules (5,6) that can be delivered to and translated in recipient cells (7,8). This finding suggests that exosomes directly influence gene expression of recipient cells upon internalization (6,8,9). However, most recent studies indicate a general enrichment of small RNA species in exosomes. Of these species, so far only the class of mircoRNAs (miRNAs) has been confirmed to sustain gene-regulatory functions upon cell-to-cell transfer (8), which may be exploited therapeutically (9,10).
Apart from miRNAs, the advent of next generation sequencing (NGS) revealed a broad spectrum of additional small non-coding RNAs (ncRNAs) in cells, but these were also found to be incorporated into exosomes. In exosomes from human plasma, saliva and neuronal cells, small RNA sequences derived from transfer RNA (tRNA), ribosomal RNA (rRNA), small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA) have been identified (11–13). Compared to the producing cells, in exosomes small RNAs are differentially distributed, suggesting a selective driving force for incorporation of small RNA species into exosomes. Thus far, several mechanisms for selective incorporation of RNA into exosomes have been described, showing that properties of both RNAs and proteins appear important for cellular retention and exosome incorporation (14–17).
We showed previously that endothelial cell-derived exosomes are functional in vivo and in vitro and that the RNA content depends highly on the physiological condition of the producing cells. Moreover, we demonstrated that individual miRNA species are not equally distributed when comparing cells with exosomes (9,18). We hypothesized that, in endothelial cell-derived exosomes, specific small RNAs are selectively incorporated or depleted. If correct, this point should be reflected in the quantitative as well as the qualitative distribution of cellular and exosomal RNAs. To investigate the identity and nature of small RNAs in endothelial cells and endothelial cell-derived exosomes, we performed a thorough deep sequencing analysis on small RNAs isolated from endothelial cells and their corresponding exosomes. This approach revealed that many small RNAs and fragments from larger RNAs are asymmetrically distributed within cells and exosomes, suggesting a selective driving force for incorporation of these RNA molecules. Because we also observed an unequal distribution for messenger RNA (mRNA) and mitochondrial RNA (mtRNA) fragments, we propose that gene control through mRNA turnover is linked to the exosome biogenesis pathway in endothelial cells.
Material and methods
Cell culture
Human endothelial cell line 1 (HMEC-1) cells (Centers for Disease Control and Prevention, Atlanta, GA, USA) were cultured at 37°C and 5% CO2, in MCDB 131 medium (Life Technologies, Grand Island, NY, USA) supplemented with 10% foetal calf serum (FCS), 100 U/ml penicillin, 100 µg/ml streptomycin, growth factors (10 ng/ml hEGF and 50 nm hydrocortisone) and 10 mM L-glutamine (all from Life Technologies). Cells were grown for up to 27 passages. Before exosome isolation, the cells, with a confluence of about 80%, were grown for 24 h in medium supplemented with exosome-free FCS. This was generated by centrifuging FCS for 1 h at 200,000×g (Beckman LE80K preparation ultracentrifuge, Beckman Coulter, Indianapolis, IN, USA) followed by filtration through a 0.20 µm filter.
Exosome isolation
Exosomes were isolated from harvested medium through differential centrifugation as previously described (9). Briefly, harvested medium was centrifuged for 15 min at 1,500×g to remove apoptotic cells and cellular debris. Subsequently, the supernatant was collected and centrifuged for 30 min at 10,000×g to remove microvesicles and membrane debris. The remaining supernatant was collected and centrifuged for 1 h at 100,000×g to pellet exosomes, which were resuspended in PBS and again centrifuged for 1 h at 100,000×g. Sucrose density gradient analysis was performed as previously described (9), and 400 µl fractions were taken from the top for subsequent immunoblot analysis and RNA isolation.
Immunoblotting
For immunoblotting, exosome and cell samples were diluted 1:1 in exosome sample buffer (5% SDS, 9M urea, 10 mM EDTA, 120 mM Tris-HCl, pH 6.8, 2.5% beta-mercaptoethanol) and heated (95°C, 5 min). For cell samples, cells were scraped from the culture surface, resuspended in lysis buffer [1% SDS and 0.1% Triton X-100 in PBS with protease inhibitors (cOmplete Mini, EDTA-free, Roche, Basel, Switzerland)] and incubated on ice for 30 min. Genomic DNA was sheared through a 27G needle 4 times. SDS-PAGE, protein transfer and blocking were performed as previously described (9). Subsequently, membranes were incubated in one of the following antibodies: rabbit-anti-GAPDH (Cell Signaling, Boston, MA, USA), rabbit anti-flotillin 1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), goat anti-lamin A/C (Santa Cruz Biotechnology), mouse anti-CD9 (Santa Cruz Biotechnology), mouse anti-ATP5A (Abcam, Cambridge, UK), rabbit anti-Tom20 (Santa Cruz Biotechnology) or rabbit anti-histone H2A.X (Merck Millipore, Billerica, MA, USA).
As secondary antibodies, 1:2,000 diluted affinity-purified swine-anti-rabbit, rabbit-anti-mouse, or donkey-anti-goat coupled to horseradish peroxidase (Dako, Glostrup, Denmark) were used. Antigen-antibody reactions were visualized with enhanced chemiluminescence according to the manufacturer's guidelines (chemiluminescent peroxidase substrate, Sigma-Aldrich, St. Louis, MO, USA) and imaged using a GelDoc XR+system (Bio-Rad, Hercules, CA, USA).
Electron microscopy
Transmission electron microscopy was conducted as described (19). In short, Formvar-filmed carbon-coated grids were placed on 5-µl exosome samples for 20 min. After 3 washes with glycine (0.15% in PBS) and 1 with BSA (0.1% in PBS), specimens were fixed in glutaraldehyde (1% in PBS, 5 min) and given 2 washes with PBS. After 4 washes with distilled water, grids were placed on ice-cold 1.8% methylcellulose (25 Ctp)/0.4% uranyl acetate for 5 min. They were then viewed using a FEI Tecnai 12 (FEI, Hillsboro, OR, USA) transmission electron microscope.
Cellular and exosomal RNA isolation
Cellular and exosomal RNA isolation was performed using the mirVana™ miRNA Isolation Kit (Life Technologies), according to the manufacturer's protocol. DNA was digested using the QIAGEN (Venlo, the Netherlands) RNase-Free DNase set as described in the manufacturer's protocol, and DNase was heat-inactivated at 65°C for 10 min. The composition of isolated RNA was analysed using small RNA chips and RNA 6000 LabChip on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA).
Small RNA sequencing
Small RNA sequencing was performed by ServiceXS (Leiden, the Netherlands). Briefly, the Illumina (San Diego, CA, USA) TruSeq Small RNA-Seq Sample Prep Kit was used according to manufacturer's protocol to prepare small RNA for sequencing. The quality and yield after sample preparation was measured with a High Sensitivity DNA Lab-on-a-Chip and corresponded to the expected 150 bp. Clustering and DNA sequencing was accomplished using the Illumina HiSeq 2000 (Solexa) according to manufacturer's protocol. Two sequencing reads were performed of 100 cycles each using Read1 sequencing and Read 2 sequencing primers. Image analysis, basecalling and quality check were performed with the Illumina data analysis pipeline RTA 1.13.48 and/or OLB 1.9 and CASAVA 1.8.2.
Small RNA sequencing data; quantitative analysis
Primary data analysis was also performed by ServiceXS using an in-house analysis pipeline that employs open source bioinformatics tools. Prior to alignment, the reads were trimmed for adapter sequences and filtered for sequence quality. Presumed adapter sequences were removed from the read when the bases matched a sequence in the adapter sequence set (TruSeq adapters) with 2 or fewer mismatches and an alignment score of at least 12.
To remove noise introduced by sequencing errors, the reads were filtered and clipped by quality. The reads were scanned with a sliding window of 4 bases, using an average Phred score of Q20 (corresponding to a chance of 1 error in 100 bases) as a minimum threshold for a 4-base-long section. If such a section had average scores below this level, the bases were removed and the longest remaining fragment was kept. The filtering, trimming and cutting was performed by Trimmomatic 0.22 (20).
The genome reference and annotation file Homo_sapiens.GRCh37.70 was used in FASTA and GTF format. The filtered reads were then aligned to the reference sequence using Tophat 1.4.0 (Baltimore, MD, USA) combined with Bowtie 0.12.7 (21). Alignments were stored in BAM files, which were sorted and indexed by the SAMtools 0.1.18 package (22).
For quantitative analyses, SAM and BAM files were generated and count files were produced showing the aligned read counts per gene, with a minimal match of 6 bases and allowing 2 mismatches (represented in dark grey in the figures). For comparative analysis of the small RNA sequencing results of all 4 samples, normalized read counts were generated for each sample by dividing the amount counts per gene by the number of million mapped reads.
Small RNA sequencing data; qualitative analysis
The produced BAM files, sorted and indexed, were analysed on qualitative differences in the coverage of sequences in exosomes and cells using the Integrative Genomics Viewer (IGV) 2.3 (23,24), using human genome 19 or Ensembl Homo_sapiens.GRCh37.70 as the reference genome. In all figures, coverage tracks are displayed in log scale and set at an automatic range for each track. RNA classes were attributed to the reads using the GeneCards or Ensembl annotation (25,26). Further databases and software used for analysis were the Diana miRPath software, using the DIANA microT 4.0 (beta version) prediction software, for the investigation of clustering of miRNA mRNA targets in cellular pathways and the miRBase database for the identification of 3p and 5p fragments of miRNAs (27,28).
Real-time quantitative PCR
Total RNA was isolated using the mirVana kit as described, and cDNA was synthesized using the NCode miRNA First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) using 100 ng RNA as a template. Subsequently, an equivalent of 500 pg RNA was used in a quantitative RT-PCR amplification in a MyiQ single-color real-time PCR system (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad) in a final reaction volume of 15 µl. Primers: Y51FWD: 5′-AGT TGG TCC GAG TGT TGT GGG-3′; Y52FWD: 5′-CCC CAC AAC CGC GCT TGA C-3′; Y5REV: 5′-GTC AAG CGC GGT TGT GGG G-3′; SNORD93_FWD: 5′-TGG CCA AGG ATG AGA ACT CT-3′; SNORD78_FWD: 5′-ATG AGC ATG TAG ACA AAG GT-3′; SNORA71B_FWD: 5′-CCT GTA TTC GAA AGT GAT CG-3′; J01415.15_FWD: 5′-CAG ATT GTG AAT CTG ACA AC-3′; J01415.18_FWD: 5′-GAG AAA GCT CAC AAG AAC TG-3′; MIR214_FWD: 5′-ACA GCA GGC ACA GAC AGG CA-3′; MIR126_FWD: 5′-CAT TAT TAC TTT TGG TAC GC-3'; MIR10B_FWD: 5′-TAC CCT GTA GAA CCG AAT TTG-3′, and the NCode universal reverse primer. Raw threshold cycle (Ct) values were calculated with the software iQ5 Optical System 2.0 using automatic baseline settings. Thresholds were set in the linear phase of the amplification plots.
Results
RNA profiling of endothelial cells and their exosomes
All sample preparations and analyses were performed in accordance with the recently published ISEV position paper on extracellular vesicle RNA analysis (29). Exosomes from endothelial cells were isolated using a sequential centrifugation protocol (30). Their presence in the final pellet was verified using transmission electron microscopy (Fig. 1a), and potential nuclear and mitochondrial contamination was analysed by immunoblot (Fig. 1b). Total RNA derived from endothelial cells and isolated exosomes was analysed using Agilent Bioanalyzer small RNA chips to compare cellular and exosomal small RNA profiles (Fig. 1c). Next, we characterized the small RNA content of these exosomes and their cells of origin using an NGS approach. Duplicate samples of total RNA, isolated from endothelial cells and their exosomes were subjected to NGS, yielding an average of 2,421,546 and 3,364,970 reads for cellular and exosomal RNAs, respectively. Sequencing data is available at the NCBI SRA, accession number PRJNA272508.
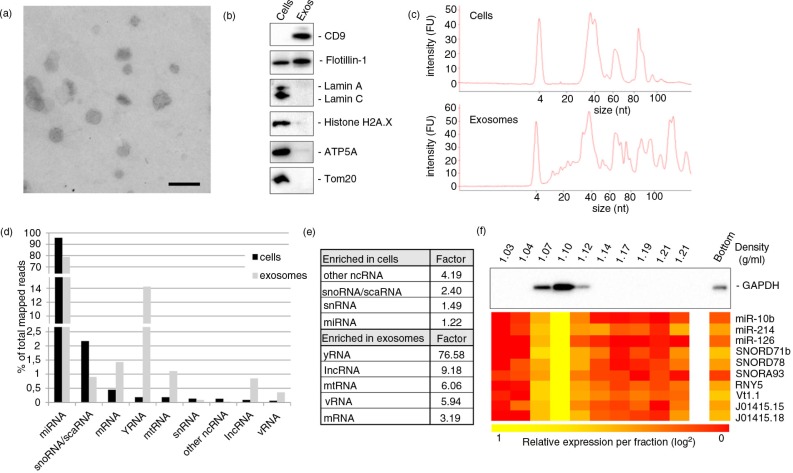
Fig. 1.
General quantitative comparison of small RNAs in cells and exosomes. (a) Exosomes from endothelial cells were isolated by ultracentrifugation and analysed by electron microscopy (scale bar: 250 nm). (b) Assessment of exosome purity by immunoblot for exosome (CD9, flotillin 1), nuclear (lamin A/C and histone H2A.X) and mitochondrial (ATP5A, Tom20) marker proteins. (c) Bioanalyzer messenger RNA and small RNA chips were used to obtain cell (upper panel) and exosome (lower panel) RNA size distribution profiles. (d) Average percentages of mapped reads of the 9 most abundant RNA classes in cells and exosomes ordered by abundance in cells and (e) enrichment factors of these RNAs in either cells or exosomes. (f) Immunoblot for GAPDH and qPCR analysis for selected small RNAs on sucrose density gradient fractions.
The sequences that could be aligned to the reference genome represented on average 71.2% of the cellular RNA reads and 38.4% of the exosomal RNA reads. To concentrate data analysis on the most relevant matches, only aligned sequences with an average read count >10 in any sample, 772 in total, were selected for further analysis (Supplementary Table I). The selected identifications were categorized into different RNA classes based on their GeneCards or Ensembl annotation (25,26).
Different RNA classes show different relative distributions
Most known classes of RNAs were identified in endothelial cells and their exosomes. We determined the relative distribution between cells and exosomes of the ten most abundant classes, which corresponded to 99.7% of all aligned reads. These RNA classes included miRNA, snoRNA, mRNA, yRNA, mtRNA, snRNA, other ncRNA (including pseudogenes and novel intronic sense RNA), long non-coding RNA (lncRNA), vault RNA (vRNA) and small Cajal body-associated RNA (scaRNA) (Fig. 1d). The remaining aligned reads in cells and exosomes represented lincRNA, tRNA and 7SK RNA. For all identified RNA classes, a differential distribution was observable between cells and exosomes. Whereas miRNA, snRNA, scaRNA, other ncRNA and snoRNA are relatively enriched in the cells rather than the exosomes, yRNA, lncRNA, vRNA, mtRNA and mRNA fragments represented a higher percentage in exosomes than in cells. The most distinct difference was visible for yRNA-derived fragments, which were more than 75 times enriched in exosomes (Fig. 1e). This finding provides an unexpected link between yRNA processing at endosomal membranes. To verify that identified exosomal RNAs are indeed associated with exosomes, we performed qPCR analyses on RNA isolated from sucrose density-floated exosomes for a selection of small RNAs, showing that all analysed RNAs have the highest abundance in the 1.10 g/ml fraction, in line with the previously reported density for endothelial cell-derived exosomes (9) (Fig. 1f). Importantly, 84%±10% of the analysed RNAs could be detected in the 3 peak fractions, with only 6%±4% present in the bottom fraction, which may contain vesicles that did not float into the gradient or cellular debris. Overall, we conclude that, within classes, variations in distributions of individual members could be detected, both quantitatively and qualitatively indicating that the sequencing approach is useful for interrogating the small RNA distributions in cells and exosomes.
Distribution of miRNAs in cells and exosomes
Among the aligned sequences, 223 mature miRNAs were identifiable (Supplementary Table I); they were ranked according to their abundance based on the average normalized read count (counts per million, CPM) in cells and exosomes. The most abundant miRNAs in cells corresponded well with the most abundant miRNAs in exosomes. Four miRNAs (miR-92b, miR-30e, miR-125a and miR-30d) were among the most abundant miRNAs in cells, but were present at lower abundance in exosomes, though still at physiologically relevant counts of >6,000 CPM (31). Similarly, 4 miRNAs that are among the most abundant miRNAs in exosomes (miR-25, miR-4485, miR-186 and miR-27a) were not present among the most abundant miRNAs. These observations suggest that exosomal miRNA content largely reflects cellular miRNA content, but exceptions do exist (Table I and II).
Table I.
Top 15 miRNAs in cells
| ID | Rank in cells | Rank in exosomes |
|---|---|---|
| miR-10B | 1 | 1 |
| miR-30A | 2 | 3 |
| miR-27B | 3 | 2 |
| miR-191 | 4 | 4 |
| miR-411 | 5 | 7 |
| miR-92B | 6 | 16 |
| miR-LET7I | 7 | 5 |
| miR-222 | 8 | 15 |
| miR-100 | 9 | 10 |
| miR-30E | 10 | 17 |
| miR-381 | 11 | 13 |
| miR-126 | 12 | 6 |
| miR-125A | 13 | 23 |
| miR-30D | 14 | 18 |
| miR-221 | 15 | 8 |
Table II.
Top 15 miRNAs in exosomes
| ID | Rank in cells | Rank in exosomes |
|---|---|---|
| miR-10B | 1 | 1 |
| miR-27B | 3 | 2 |
| miR-30A | 2 | 3 |
| miR-191 | 4 | 4 |
| miR-LET7I | 7 | 5 |
| miR-126 | 12 | 6 |
| miR-411 | 5 | 7 |
| miR-221 | 15 | 8 |
| miR-25 | 16 | 9 |
| miR-100 | 9 | 10 |
| miR-4485 | 30 | 11 |
| miR-186 | 19 | 12 |
| miR-381 | 11 | 13 |
| miR-27A | 18 | 14 |
| miR-222 | 8 | 15 |
To identify selectively exosome-sorted miRNAs in endothelial cell-derived exosomes, the miRNAs were ordered based on relative enrichment factor in either exosomes or cells. Table III shows that enrichments approaching 100% (i.e. miRNAs that are not detected in either exosomes or cells) can be observed. However, it should be noted that there greater enrichment factors in either cells or exosomes can especially be observed for lower-ranking (i.e. less abundant) miRNAs (Fig. 2a).
Table III.
Enriched and depleted miRNAs
| Higher in exosomes | |
|---|---|
|
| |
| ID | Ratio exosomes/cells |
| MIR486 | 57.93 |
| MIR204 | 25.34 |
| MIR143 | 15.85 |
| MIR1468 | 12.29 |
| MIR148A | 9.19 |
|
| |
| Higher in cells | |
|
| |
| ID | Ratio cells/exosomes |
|
| |
| AC008738.2 | 97.28 |
| MIR29B1 | 18.27 |
| MIR3155A | 13.29 |
| FLJ27365 | 6.87 |
| MIR33B | 6.71 |
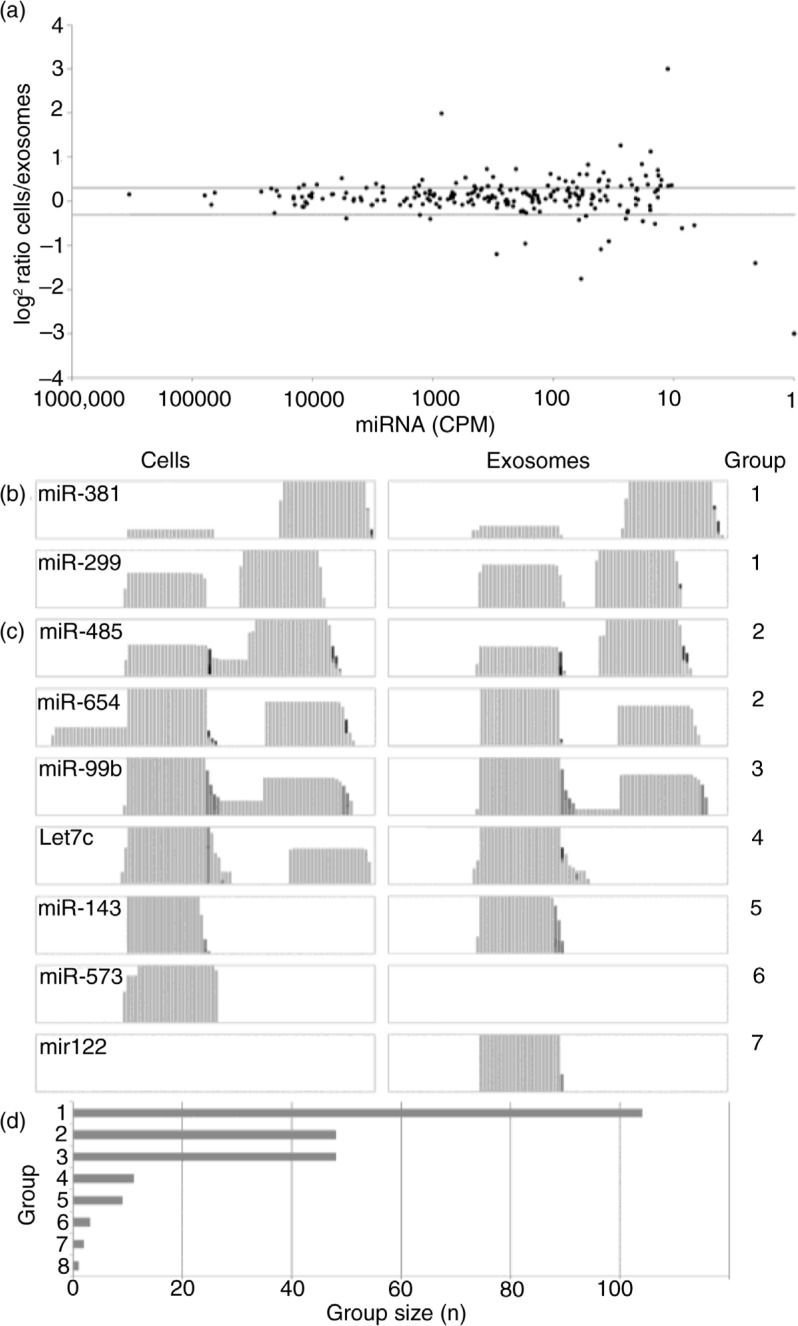
Fig. 2.
Distribution of micro RNAs (miRNAs) in cells and exosomes. (a) Log2 miRNA abundance ratios (cells/exosomes) are displayed with miRNAs ordered by their abundance (CPM). Horizontal grey lines indicate ±1.5-fold ratios. Differences in distribution of miRNA fragments between cells and exosomes as observed in Integrative Genomics Viewer coverage tracks were grouped as follows: (b) group 1: similar distribution; (c) group 2: additional fragment observed in cells; group 3: additional fragment present in both cells and exosomes; group 4: one fragment not observed in exosomes; group 5: only one fragment observed in both cells and exosomes; group 6: miRNA not detected in exosomes and group 7: miRNA not observed in cells. Group 8 encompasses miRNAs not fitting any of the described groups and consists of only 2 miRNAs. All groups are illustrated by representative miRNAs (y-axis in log scale). Comparison of group sizes (d) shows that most miRNA are distributed in a similar manner in cells and exosomes.
For all identified miRNAs, IGV software was used to compare qualitative differences in coverage patterns in exosomes and cells and assigned numbered groups according to their distribution. Group 1, in which the relative abundances of the 5p and 3p fragments were similar in cells and exosomes (Fig. 2b), encompasses the most miRNAs. However, we noted that additional fragments flanking or interconnecting the 5p and/or 3p fragments and representing pre-miRNA stem and loop fragments (Fig. 2c) were often present. These additional fragments were frequently observed in cells (group 2) compared to exosomes (group 3), but were never exclusively found in exosomes. In the fourth group, 2 fragments (5p and 3p) could be identified in cells, whereas one of these fragments was lacking in exosomes. MiRNAs belonging to group 5 showed only one of the 5p or 3p fragments in both cells and exosomes, with the same fragment detected in each sample. In a few cases, a miRNA could exclusively be detected in either cells (group 6) or exosomes (group 7, Fig. 2d). For one (predicted novel) miRNA (32), AC008738.2, fragment distribution patterns in cells and exosomes were completely different (group 8, Supplementary Fig. S1). In conclusion, our data show that quantitatively, miRNAs are predominantly distributed evenly between cells and exosomes. However, more detailed analysis of the distribution of processing fragments revealed that 5p, 3p and stem-loop fragments differ significantly between cells and exosomes in approximately half of the identified miRNAs. The reduced abundance of non-functional stem-loop fragments in exosomes may suggest that functional “mature” products are sorted into exosomes, whereas the non-functional fragments are degraded in the cell's cytoplasm. This finding is biologically relevant as it supports the idea of a mechanistic link between exosome cargo selection machinery and miRNA-dependent RNA degradation by RISC complexes associated with endosomal membranes (17).
snoRNAs and scaRNAs
In our analysis, 97 snoRNAs and 8 scaRNAs could be identified, generally showing higher read counts in cells than in exosomes, representing the single RNA class with the most prominent overall enrichment in cells (2.40-fold). In general, snoRNAs are represented by far more read counts in cells compared to exosomes, and only 19 snoRNAs, appear more abundant in exosomes. These 19 snoRNAs represent 50.8% of all snoRNA reads in exosomes, but only 14.8% of the cellular snoRNA reads. The 15 most abundant snoRNAs in exosomes and cells contain 10 corresponding and 5 different snoRNAs, (Table IV and V).
Table IV.
Top 15 snoRNAs in cells
| ID | Rank in cells | Rank in exosomes |
|---|---|---|
| SNORD78 | 1 | 3 |
| SNORD93 | 2 | 1 |
| SNORD114-22 | 3 | 10 |
| SNORD43 | 4 | 18 |
| SNORD114-9 | 5 | 15 |
| SNORD104 | 6 | 4 |
| SNORD119 | 7 | 8 |
| SNORD114-24 | 8 | 88 |
| SNORD100 | 9 | 6 |
| SNORD82 | 10 | 2 |
| SNORD12 | 11 | 41 |
| SNORD99 | 12 | 7 |
| SNORD69 | 13 | 24 |
| SNORD66 | 14 | 13 |
| SNORD12B | 15 | 17 |
Table V.
Top 15 snoRNAs in exosomes
| ID | Rank in cells | Rank in exosomes |
|---|---|---|
| SNORD78 | 2 | 1 |
| SNORD93 | 10 | 2 |
| SNORD114-22 | 1 | 3 |
| SNORD43 | 6 | 4 |
| SNORD114-9 | 16 | 5 |
| SNORD104 | 9 | 6 |
| SNORD119 | 12 | 7 |
| SNORD114-24 | 7 | 8 |
| SNORD100 | 66 | 9 |
| SNORD82 | 3 | 10 |
| SNORD12 | 18 | 11 |
| SNORD99 | 51 | 12 |
| SNORD69 | 14 | 13 |
| SNORD66 | 68 | 14 |
| SNORD12B | 5 | 15 |
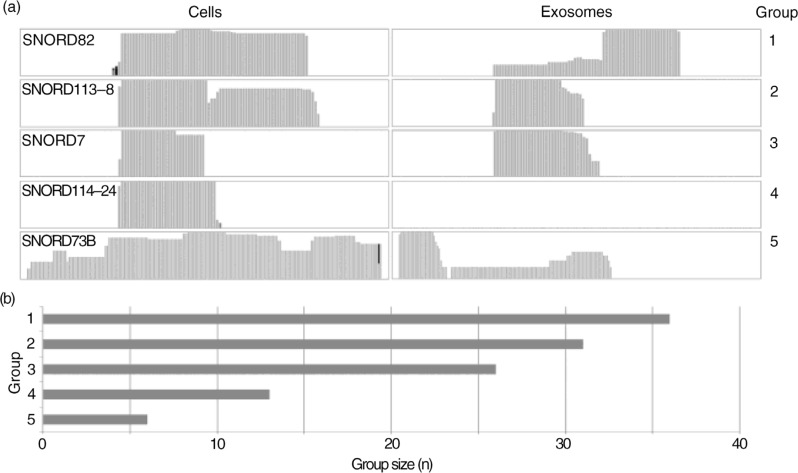
Qualitative differences in the coverage pattern and read length of snoRNA reads in exosomes and cells were analysed. Again, several differences in the distribution of snoRNA fragments between cells and exosomes could be detected, whereas, as observed for the miRNAs, the majority (62 snoRNAs) showed a sequence coverage that was similar in cells and exosomes. In many cases (36/62), the coverage in exosomes appeared more fragmented whereas read lengths in cells were longer, showing more 60–70 bp reads resulting in a longer average read length in cells compared to exosomes (38.7±6.0 nt vs. 30.3±2.6 nt, p=0.0007). It also resulted in a different coverage pattern (group 1), while for the other 26 snoRNAs the reads and coverage pattern appear identical in cells and exosomes (group 2).
The remaining 43 snoRNAs were classified based on the differences in sequence coverage observed in cells and exosomes such as different sequence coverage and absence in either cells or exosomes, and summarized in Fig. 3a and b. We speculate that the skewed distribution of snoRNAs in cells versus exosomes are likely a reflection of their biological function(s) in the cell's nucleus, whereas the observed enrichment of several snoRNA fragments may be attributed to a miRNA-like cytoplasmic role of these fragments (33–35).
Fig. 3.
Distribution and coverage of small nucleolar RNAs (snoRNAs) in cells and exosomes. (a) Different coverage patterns identified in IGV coverage tracks were grouped as follows: group 1: similar coverage but different fragment distribution in cells and exosomes; group 2: similar coverage pattern in cells and exosomes; group 3: additional coverage in cells compared to exosomes; group 4: snoRNA not detected in exosomes and group 5: different coverage and different fragment distribution in cells and exosomes. (b) Bar chart comparing sizes of aforementioned groups.
Exosomal enrichment of vRNAs and yRNAs
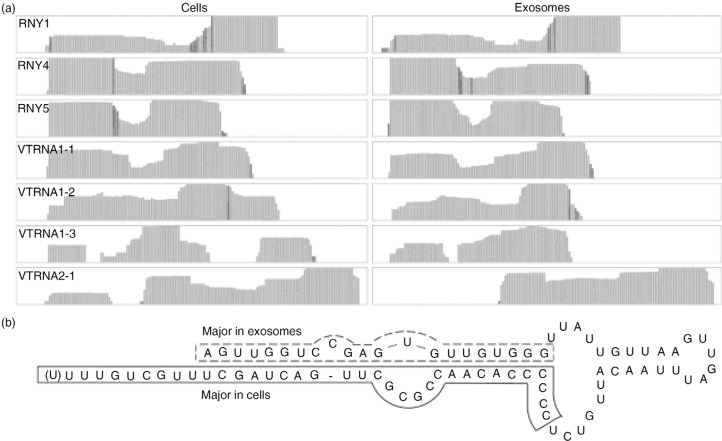
Two separate classes of small RNAs (yRNA and vRNA) were highly enriched in exosomes. Without exception, all identified vRNAs and yRNAs were detected to a much greater extent in exosomes compared to cells, showing enrichment factors of 6 and 80 for vRNAs and yRNAs, respectively. Among these RNAs, RNY5 showed the highest enrichment in exosomes (factor 86) and accounted for the second most identified RNA species in exosomes (rank number 48 in cells). vRNAs, RNY1 and RNY4 ranked number 48 and higher in exosomes and 127 and higher in cells. As observed for miRNAs and snoRNAs, fragments mapping to vRNAs and yRNAs are mostly distributed similarly in cells and exosomes (Fig. 4a). However, for VRNA1-3 and VRNA2-1, an additional fragment was detected in cells, and for RNY5 the 5′ fragment was 3.08±0.08 times more abundant than the 3′ fragment in exosomes compared to cells, which we confirmed by RT-qPCR, showing a 2.49±0.20 higher abundance of the 5′ fragment (n.s.) (Fig. 4b and Supplementary Fig. 2). Altogether, vRNA and yRNA fragments represented the highest enriched RNAs in exosomes, but different fragment distribution patterns were observable compared to cells.
Fig. 4.
yRNA and vault RNA (vRNA). (a) Coverage tracks of identified vRNAs and yRNAs in cells and exosomes and (b) 2D structure of hRNY5, with the predominant fragments identified in cells and exosomes indicated.
mtRNA in cells and exosomes
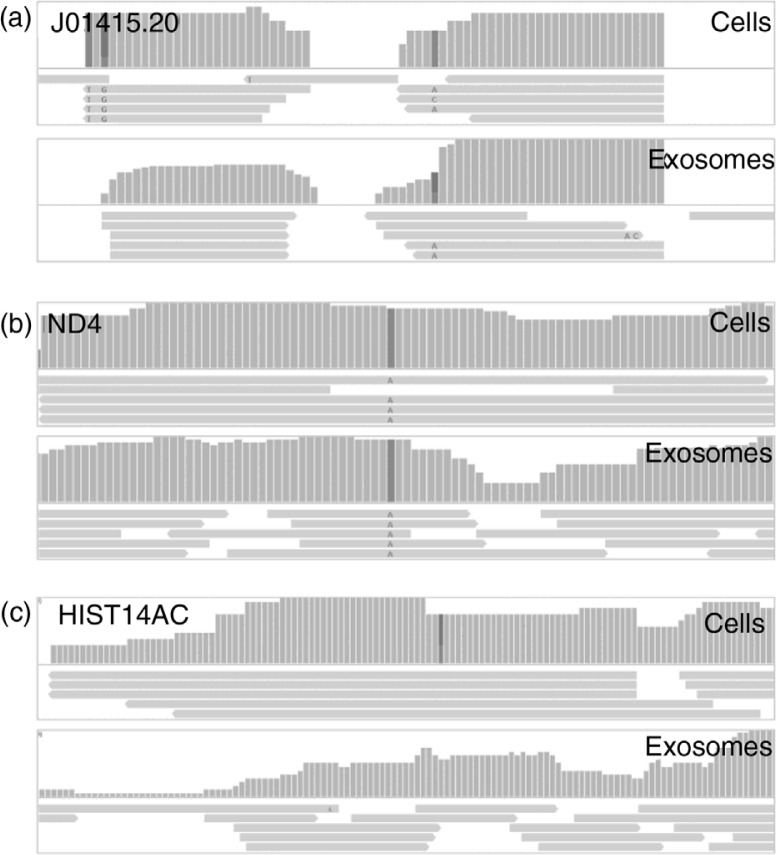
In our data, RNAs representing 28 mitochondrial genes could be identified by 10 or more reads, corresponding to 1.11 and 0.18% of the mapped reads in exosomes and cells, respectively. All but 1 mtRNA (J01415.20) showed higher read counts in exosomes than in cells, resulting in a 6-fold higher abundance of mtRNA reads in exosomes compared to cells. Identified mtRNAs include mitochondrial tRNA and rRNAs and 12 of the 13 mitochondrial mRNAs, with ATP synthase subunit 8 as the only mRNA not identified (Supplementary Table I).
Although a vast enrichment of mtRNAs is observed in exosomes, qualitative analysis reveals that observed fragments in cells are in general longer. These longer reads in cells correspond to protein-coding mRNAs, whereas for mitochondrially encoded tRNAs, read lengths appeared similar in cells and exosomes (Fig. 5a and b). The percentage of full 100 nt reads represents 21.5% of the reads in cells, but only 1.3% in exosomes, and corresponds to an average length of 48.7±30.1 nt in cells and 29.4±10.8 nt in exosomes (p<0.0001). Correspondingly, the total amount of nt sequenced in cells is only 1.66-fold higher in exosomes compared to cells. Summarized, exosomes show a higher abundance of mtRNAs, but identified RNAs are significantly shorter.
Fig. 5.
Mitochondrial RNA (mtRNA) and messenger RNA (mRNA) fragments in cells and exosomes. Analysis of coverage and read lengths reveals similar distributions of (a) mitochondrial transfer RNAs and similar coverage but shorter read lengths in exosomes for (b) protein coding mtRNAs and (c) mRNAs.
Fragments from long coding and non-coding RNAs
Besides small RNAs, (fragments of) 389 different long RNAs (pseudogenes, lincRNAs, lncRNAs and mRNAs) were also identified, accounting for only 0.7 and 2.4% of the total reads in cells and exosomes, respectively. In general, long RNAs are thus represented by low read counts and overall show an enriched abundance in exosomes. This consideration may be of physiological relevance, as some lncRNAs are suspected to have a functional activity at low copy numbers (36) and the demonstrated role of exosome-mediated transfer of mRNA (6,7). Analysis of sequence coverages shows that, within the entire genomic region of identified long RNAs, the few mapped reads are scattered and consequently coverage patterns in cells and exosomes differ, hampering qualitative analysis. In cases where sufficient numbers of reads were mapped to the same region of the RNA in both cells and exosomes for such a comparison, a pattern similar to that observed for mitochondrial mRNAs could be observed, with mapped reads in cells longer than in exosomes, whereas total coverage appeared similar (Fig. 5c). In addition, for a few mRNAs identified with sufficient reads to derive a reliable view of fragment distribution along the coverage track, fragments corresponding to the 3′UTR could be observed (data not shown). In conclusion, apart from small RNAs, fragments from long coding and non-coding RNAs can be observed at a low level in both cells and exosomes. These fragments detected in exosomes do not appear to be derived from a specific RNA processing event, but rather seem to represent RNA degradation products, as length, start and endpoints of these fragments appear random.
Discussion
Individual endothelial cells align into structured tissues that perform highly specialized biological functions and are in constant contact with each other and their surroundings, containing many additional cell types. We have shown previously in mouse models that endothelial cell-derived exosomes contain and transfer functional miRNAs that control key signalling pathways in growing endothelium (9). Here, we performed a first comprehensive analysis on the total small RNA content of endothelial cells and their secreted exosomes. We investigated whether additional small RNA classes are sorted into endothelial exosomes by applying quantitative and qualitative analyses using a small RNA sequencing approach.
Quantitative analysis
Our global quantitative analyses reveals enrichment and selective depletion of particular small RNA classes that are underrepresented (miRNA, sno/scaRNA, snRNA and other ncRNA) or decidedly overrepresented (mtRNA, mRNA, lncRNA, yRNA and vRNA) in exosomes when compared to the producing cells. It should be noted that, in this comparison, percentages of the total read numbers were used, which may not necessarily represent the relative abundance in each total RNA sample, as small RNA served as the input material for sequencing. The effects of potentially contaminating DNA were excluded by a direct comparison by qPCR of DNAse-treated and non-treated RNA samples, showing that after DNAse treatment 99%±1% of the initial signal remained detectable. As reported earlier, we found that the class of miRNAs is more abundant in cells compared to exosomes (9) and, based on Bioanalyzer mRNA chip analyses, small RNAs appear 1.9±0.4 times more abundant in exosomes compared to cells. However, for absolute comparisons, correction for these different abundances or a suitable housekeeping RNA that is equally distributed in cells and exosomes should be performed. Furthermore, it should be realized that, in comparisons between exosomal and cellular RNA, exosomal RNA is derived from a vast amount of exosomes and thus represents the cumulative content of these exosomes, while only RNA from a relatively low amount of cells is analysed. We judged that corrections for either small RNA content (higher in exosomes) or total RNA content (higher in cells) would dramatically skew (semi-) quantitative analyses towards either RNA source. To compensate for the lack of well-established housekeeping RNAs for the comparison of cellular versus exosomal RNAs, we chose to normalize read counts for each individual sample, allowing for the semi-quantitative comparison of RNA abundances.
miRNAs
Functional intercellular transfer of miRNAs through exosomes has been extensively described (8,9,37). In our analysis, 223 miRNAs could be identified in exosomes, representing 96 and 79% of all reads in cells and exosomes, respectively. Analysis of the most abundant miRNAs in exosomes may provide clues as to the function of endothelial cell-derived exosomes. The 10 most abundant miRNAs in exosomes cover 67.92% of all miRNA reads. They have been described to have pro-angiogenic (miR-10b, LET7I, miR-126) (38–40) or anti-angiogenic effects (miR-100) (41), stimulate cell differentiation (miR-27b, miR-30a) (42,43) and affect cell proliferation (miR-25, miR-221) (44,45), whereas for other miRNAs no clear function has been described (miR-191, miR-411). Potential effects of these miRNAs on angiogenesis, cell differentiation and proliferation fit well with the described effects of endothelial cell-derived exosomes (9,46) and may represent additional players in the described processes. Furthermore, these high-abundant miRNA can provide leads for further investigations on the functional effects of endothelial cell-derived exosomes and the molecular mechanisms by which these effects are accomplished, both on endothelial cells and on other cell types. The high-abundant miR-10b, for example, may be a key regulator in the cellular response to hypoxia, explaining the pro-angiogenic effects in endothelial cells (47), although in other cell types the effects on cell differentiation will be more prominent specific identified exosome enriched (48) warranting a broad diversity of further in-depth investigations.
Comparative analysis of miRNAs enriched in either cells or exosomes may provide information about possible mechanisms for selective incorporation of miRNAs into exosomes. Considering only miRNAs with >1,000 reads, miR-486 is the miRNA showing the highest enrichment in exosomes (57.9-fold), followed by miR-143 (15.8-fold). Both miRNAs show extensive 3′ uracil residues in addition to their mature sequence, shown to be important for selective incorporation into exosomes, whereas additional adenosines were detected at the 3′ end of cellular retained miRNAs (miR-193a, miR-181b2) (15). These results are consistent with the function of these modifications on miRNA activity (49).
Small RNA-derived miRNAs (vRNA, yRNA, snoRNA)
Similar to prior exosome-RNA sequencing studies (11,50), snoRNAs, yRNAs and vRNAs were also identifiable in both endothelial cells and exosomes. SnoRNAs represent the second most abundant small RNA class in cells with regard to read counts, and the third largest class when considering IDs, after mRNA fragments and miRNAs. SnoRNA are small ncRNAs that mediate the modification of ribosomal RNAs, and over 350 snoRNAs have been described thus far (51,52). scaRNAs form a subclass of snoRNAs representing snoRNA that specifically bind to the Cajal body in the nucleus (52). yRNA and vRNA are components of ribonucleoprotein complexes and have different functions. In the cytosol, yRNAs interact with the Ro protein, a protein involved in the degradation of misfolded or misprocessed RNAs, thereby preventing a specific binding to (correctly folded) RNAs (53). Besides, yRNAs have a role in the chromosomal DNA replication (54). vRNAs are components of vault complexes, which have been demonstrated to be up-regulated in various cancers, where they have a role in multidrug resistance (55–57). Drug resistance can be conferred either by direct binding to chemotherapeutic compounds (58,59) or by the regulation of expression of multidrug-associated proteins (60).
Our observations that fragments of small non-coding RNAs such as scaRNAs, snoRNAs, vRNAs and yRNAs are more abundant in exosomes compared to cells are interesting in the light of recent reports that such fragments may act as miRNAs, as has previously been reported for several snoRNA-derived RNAs (sdRNAs) (33–35,61). Interestingly, some of these reported functional sdRNAs were identified in our analysis; moreover, many snoRNA-derived fragments with a length of 20–23 nt were identified for snoRNAs without described (functional) sdRNAs. Our finding that, similar to earlier descriptions of exosomal and circulating yRNA (11,62), primarily 18–48 nt fragments were identified in our analysis, supports potential processing of yRNA to miRNA-like molecules (63). Two fragments of 26–29 nt and 32–35 nt were predominantly detected: a 5p fragment started at nt 1 and ended at nt 31, but was also present in slightly longer versions in cells and exosomes, up to nt 39, and a 3p fragment started around nt 50 in cells and exosomes and ended around nt 73 or nt 83. The 3p fragment was most prominent in cells, whereas the 5p was the major fragment in exosomes. Additionally, a fragment starting at nt 64 and ending around nt 83 was present in low amounts.
Thus, the most detected fragments are 26–34 nt in length, which is somewhat longer than the described 22–27 nt for yRNA-derived miRNAs (63).
yRNA-derived fragments have also been described to associate with PNPase and Rsr, forming a RNA-degrading complex for the degradation of structured and misfolded RNAs (64). Moreover, the high amount of yRNAs in exosomes may indicate that the cytoplasmic RNA-degrading machinery localizes to the proximity of MVB, resulting in high focal concentrations of both yRNAs and RNA degradation products (65,66). Fragments of different vRNAs displayed a variety of lengths ranging from miRNA-like sizes (19–22 nt) to up to 40 nt fragments; they included the svRNA molecules described earlier (60).
Long coding and non-coding RNA
Although our goal was to profile small <100 nt RNAs, a low percentage (0.81 and 2.93% of all mapped reads in cells and exosomes, respectively, including mitochondrial encoded mRNAs) of the total reads mapped to long coding and non-coding RNAs. Given the procedure for physical length restriction using agarose gel and the propensity of RNA to fold into compact secondary and tertiary structures, it may be argued that such long, but compactly folded, RNAs migrate in a similar manner as short, linear RNAs, explaining their detection by NGS despite size selection. Additionally, our qualitative analysis revealed that identified long RNAs were represented, in both cells and exosomes, by smaller fragments that nicely corresponded to the size restriction procedure. Nevertheless, in cells 5.3% of the reads mapping to protein-coding RNA were full 100 nt reads, over 10-fold more than in exosomes (0.5%), suggesting that exosomes contain not only fragmented, but also intact RNA, as found by others (6,7).
RNA decay takes place in the cytoplasm, thus enrichment of fragmented RNA in exosomes is consistent with the cytoplasmic origin of exosomes. MiRNA-mediated RNA degradation has been reported to localize to the surface of MVB, leading to high focal concentrations of miRNA and degraded mRNA at the site of intraluminal vesicle budding (67,68). Furthermore, in accordance with observations by Batagov and Kurochkin (66), mRNA 3′ ends were relatively overrepresented in exosomes, which supports the explanation that focal high concentrations of miRNA and miRNA-degraded RNA cause its increased incorporation into exosomes and illustrates that the exosomal pathway plays a role in gene regulation. Additionally, an enrichment of erroneous, degraded RNAs in exosomes may provide an explanation for the finding that only 38.4% of the exosomal reads could be mapped, versus 71.2% of the cellular reads. mtRNAs and proteins have often been observed in exosomes (11,18,69); they likely reflect the intracellular pathways involving late endosome formation, shared by autophagocytosis and exosome synthesis, or the vesicle-mediated transfer of mitochondrial content between mitochondria and lysosomes (70–72). Our observations with regard to the presence of RNA degradation products in exosomes highlight the added value of NGS analysis over RNA expression array analysis. Importantly, the identification of shorter fragments representing long RNAs in exosomes argues for great caution in the interpretation of RNA expression array data, because in these analyses positive signals may represent hybridization of (mRNA) fragments rather than complete mRNAs. Verification of the presence of intact RNAs enriched by PCR – and not qPCR, in which generally ~150 nt fragments are amplified – is thus strongly urged in such analyses. Although it remains to be investigated whether specific identified exosome enriched RNAs reflect the function of exosomes or rather an efficient method for cells to dispose of waste RNA, it is clear that the RNA content of exosomes differs from that of cells in a quantitative and qualitative manner, linking the endosomal-exosomal pathway to subcellular sites of gene control.
Conclusions
In summary, our semi-quantitative and qualitative analysis of cellular and exosomal small RNAs shows that many small RNA classes are present in endothelial cell-derived exosomes; we demonstrated that different RNA classes show different distributions between cells and exosomes with respect to relative amounts and (fragment) distribution patterns. In general, miRNAs are distributed evenly between cells and exosomes, although our qualitative analysis shows that for roughly half of the identified miRNAs, 5p, 3p and stem-loop fragments are differentially distributed. Furthermore, in addition to miRNA-like fragments derived from snoRNAs, vRNAs and yRNAs, the degradation products of long coding and non-coding RNAs appear enriched in exosomes. These findings suggest that, besides possible targeted transport of small RNAs to exosomes, exosomal RNA content reflects a cellular mechanism for localized degradation of RNA in the proximity of MVB and the subsequent disposal of RNA fragments through exosomes.
Supplementary Material
Acknowledgements
BWMvB was supported by the Netherlands Organization for Scientific Research (NGI/ZonMw Horizon project 935190280), a UMC Utrecht Focus and Massa grant (grant DIGD-DGK-DHL), and the Netherlands Institute for Regenerative Medicine (grant FES0908). MCV is supported by the Netherlands Organisation for Scientific Research (ZonMw-TAS grant 116001026; Vidi grant 016.096.359). We thank Olivier G. de Jong (Department of Nephrology and Hypertension, University Medical Center Utrecht, the Netherlands) for excellent technical assistance.
Conflict of interest and funding
The authors have not received any funding or benefits from industry or elsewhere to conduct this study.
References
- 1.Johnstone RM, Adam M, Pan BT. The fate of the transferrin receptor during maturation of sheep reticulocytes in vitro. Can J Biochem Cell Biol. 1984;62:1246–54. doi: 10.1139/o84-159. [DOI] [PubMed] [Google Scholar]
- 2.Harding C, Heuser J, Stahl P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur J Cell Biol. 1984;35:256–63. [PubMed] [Google Scholar]
- 3.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 4.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–56. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 6.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 7.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328–33. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Balkom BW, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. 2013;121:3997–15. doi: 10.1182/blood-2013-02-478925. [DOI] [PubMed] [Google Scholar]
- 10.Almanza G, Anufreichik V, Rodvold JJ, Chiu KT, DeLaney A, Akers JC, et al. Synthesis and delivery of short, noncoding RNA by B lymphocytes. Proc Natl Acad Sci USA. 2013;110:20182–7. doi: 10.1073/pnas.1311145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolte-'t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, 't Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–85. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937–49. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–58. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 16.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–46. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 18.de Jong OG, Verhaar MC, Chen Y, Vader P, Gremmels H, Posthuma G, et al. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012;1 doi: 10.3402/jev.v1i0.18396. 18396, doi: http://dx.doi.org/10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2:2480–91. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- 20.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safran M, Dalah I, Alexander J, Rosen N, Iny ST, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belinky F, Bahir I, Stelzer G, Zimmerman S, Rosen N, Nativ N, et al. Non-redundant compendium of human ncRNA genes in GeneCards. Bioinformatics. 2013;29:255–61. doi: 10.1093/bioinformatics/bts676. [DOI] [PubMed] [Google Scholar]
- 27.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill AF, Pegtel DM, Lambertz U, Leonardi T, O'Driscoll L, Pluchino S, et al. ISEV position paper: extracellular vesicle RNA analysis and bioinformatics. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.22859. 22859, doi: http://dx.doi.org/10.3402/jev.v2i0.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 31.Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012;9:840–6. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–86. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–28. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 35.Falaleeva M, Stamm S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. Bioessays. 2013;35:46–54. doi: 10.1002/bies.201200117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montecalvo A, Larregina AT, Shufesky WJ, Beer SD, Sullivan ML, Karlsson JM, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–66. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Solingen C, Seghers L, Bijkerk R, Duijs JM, Roeten MK, van Oeveren-Rietdijk AM, et al. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J Cell Mol Med. 2009;13:1577–85. doi: 10.1111/j.1582-4934.2008.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plummer PN, Freeman R, Taft RJ, Vider J, Sax M, Umer BA, et al. MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Cancer Res. 2013;73:341–52. doi: 10.1158/0008-5472.CAN-12-0271. [DOI] [PubMed] [Google Scholar]
- 40.Sen CK, Gordillo GM, Khanna S, Roy S. Micromanaging vascular biology: tiny microRNAs play big band. J Vasc Res. 2009;46:527–40. doi: 10.1159/000226221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grundmann S, Hans FP, Kinniry S, Heinke J, Helbing T, Bluhm F, et al. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation. 2011;123:999–1009. doi: 10.1161/CIRCULATIONAHA.110.000323. [DOI] [PubMed] [Google Scholar]
- 42.Biyashev D, Veliceasa D, Topczewski J, Topczewska JM, Mizgirev I, Vinokour E, et al. miR-27b controls venous specification and tip cell fate. Blood. 2012;119:2679–87. doi: 10.1182/blood-2011-07-370635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Q, Lagos-Quintana M, Liu D, Shi Y, Helker C, Herzog W, et al. miR-30a regulates endothelial tip cell formation and arteriolar branching. Hypertension. 2013;62:592–8. doi: 10.1161/HYPERTENSIONAHA.113.01767. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Mao H, Chen JY, Wen S, Li D, Ye M, et al. Increased expression of microRNA-221 inhibits PAK1 in endothelial progenitor cells and impairs its function via c-Raf/MEK/ERK pathway. Biochem Biophys Res Commun. 2013;431:404–8. doi: 10.1016/j.bbrc.2012.12.157. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Aznar E, Barrallo-Gimeno A, Nieto MA. Scratch2 prevents cell cycle re-entry by repressing miR-25 in postmitotic primary neurons. J Neurosci. 2013;33:5095–105. doi: 10.1523/JNEUROSCI.4459-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheldon H, Heikamp E, Turley H, Dragovic R, Thomas P, Oon CE, et al. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. 2010;116:2385–94. doi: 10.1182/blood-2009-08-239228. [DOI] [PubMed] [Google Scholar]
- 47.Nusrin S, Tong SK, Chaturvedi G, Wu RS, Giesy JP, Kong RY. Regulation of CYP11B1 and CYP11B2 steroidogenic genes by hypoxia-inducible miR-10b in H295R cells. Mar Pollut Bull. 2014;85:344–51. doi: 10.1016/j.marpolbul.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Han X, Yan S, Weijie Z, Feng W, Liuxing W, Mengquan L, et al. Critical role of miR-10b in transforming growth factor-beta1-induced epithelial-mesenchymal transition in breast cancer. Cancer Gene Ther. 2014;21:60–7. doi: 10.1038/cgt.2013.82. [DOI] [PubMed] [Google Scholar]
- 49.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–88. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 50.Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol Pharm Bull. 2013;36:66–75. doi: 10.1248/bpb.b12-00607. [DOI] [PubMed] [Google Scholar]
- 51.Ellis JC, Brown DD, Brown JW. The small nucleolar ribonucleoprotein (snoRNP) database. RNA. 2010;16:664–6. doi: 10.1261/rna.1871310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie J, Zhang M, Zhou T, Hua X, Tang L, Wu W. Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs. Nucleic Acids Res. 2007;35:D183–7. doi: 10.1093/nar/gkl873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–39. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang AT, Langley AR, Christov CP, Kheir E, Shafee T, Gardiner TJ, et al. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J Cell Sci. 2011;124:2058–69. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pituch-Noworolska A, Zaremba M, Wieczorek A. Expression of proteins associated with therapy resistance in rhabdomyosarcoma and neuroblastoma tumour cells. Pol J Pathol. 2009;60:168–73. [PubMed] [Google Scholar]
- 56.Valera ET, Scrideli CA, Queiroz RG, Mori BM, Tone LG. Multiple drug resistance protein (MDR-1), multidrug resistance-related protein (MRP) and lung resistance protein (LRP) gene expression in childhood acute lymphoblastic leukemia. Sao Paulo Med J. 2004;122:166–71. doi: 10.1590/S1516-31802004000400007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aronica E, Gorter JA, van Vliet EA, Spliet WG, van Veelen CW, van Rijen PC, et al. Overexpression of the human major vault protein in gangliogliomas. Epilepsia. 2003;44:1166–75. doi: 10.1046/j.1528-1157.2003.22103.x. [DOI] [PubMed] [Google Scholar]
- 58.Mashima T, Kudo M, Takada Y, Matsugami A, Gopinath SC, Kumar PK, et al. Interactions between antitumor drugs and vault RNA. Nucleic Acids Symp Ser (Oxf) 2008:217–18. doi: 10.1093/nass/nrn110. [DOI] [PubMed] [Google Scholar]
- 59.Gopinath SC, Matsugami A, Katahira M, Kumar PK. Human vault-associated non-coding RNAs bind to mitoxantrone, a chemotherapeutic compound. Nucleic Acids Res. 2005;33:4874–81. doi: 10.1093/nar/gki809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11:1268–71. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 61.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Mote P, Martin DI. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics. 2013;45:990–8. doi: 10.1152/physiolgenomics.00129.2013. [DOI] [PubMed] [Google Scholar]
- 63.Verhagen AP, Pruijn GJ. Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. Bioessays. 2011;33:674–82. doi: 10.1002/bies.201100048. [DOI] [PubMed] [Google Scholar]
- 64.Chen X, Taylor DW, Fowler CC, Galan JE, Wang HW, Wolin SL. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell. 2013;153:166–77. doi: 10.1016/j.cell.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL. The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell. 2009;20:1555–64. doi: 10.1091/mbc.E08-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biol Direct. 2013;8:12. doi: 10.1186/1745-6150-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 68.Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–6. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mears R, Craven RA, Hanrahan S, Totty N, Upton C, Young SL, et al. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4:4019–31. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 70.Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22:135–41. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 71.Politi Y, Gal L, Kalifa Y, Ravid L, Elazar Z, Arama E. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev Cell. 2014;29:305–20. doi: 10.1016/j.devcel.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 72.Liou W, Geuze HJ, Geelen MJ, Slot JW. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol. 1997;136:61–70. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.