Abstract
Influenza A virus infections in humans generally cause self-limited infections, but can result in severe disease, secondary bacterial pneumonias, and death. Influenza viruses can replicate in epithelial cells throughout the respiratory tree and can cause tracheitis, bronchitis, bronchiolitis, diffuse alveolar damage with pulmonary edema and hemorrhage, and interstitial and airspace inflammation. The mechanisms by which influenza infections result in enhanced disease, including development of pneumonia and acute respiratory distress, are multifactorial, involving host, viral, and bacterial factors. Host factors that enhance risk of severe influenza disease include underlying comorbidities, such as cardiac and respiratory disease, immunosuppression, and pregnancy. Viral parameters enhancing disease risk include polymerase mutations associated with host switch and adaptation, viral proteins that modulate immune and antiviral responses, and virulence factors that increase disease severity, which can be especially prominent in pandemic viruses and some zoonotic influenza viruses causing human infections. Influenza viral infections result in damage to the respiratory epithelium that facilitates secondary infection with common bacterial pneumopathogens and can lead to secondary bacterial pneumonias that greatly contribute to respiratory distress, enhanced morbidity, and death. Understanding the molecular mechanisms by which influenza and secondary bacterial infections, coupled with the role of host risk factors, contribute to enhanced morbidity and mortality is essential to develop better therapeutic strategies to treat severe influenza.
Influenza viruses are important pathogens from a public health perspective, because they are common causes of human respiratory illness. Significantly, influenza infections are associated with high morbidity and mortality, especially in elderly persons, infants, and those with chronic diseases.1 Human influenza virus infections are associated with endemically circulating strains causing annual or seasonal epidemic outbreaks (usually in the winter months) and occasional and unpredictably emerging pandemic strains, as well as zoonotic infections from avian and mammalian animal hosts.
Epidemic influenza infection results in up to 49,000 deaths and 200,000 hospitalizations each year in the United States alone.2 However, pandemic strains of influenza can cause even higher mortality. For example, the 1918 Spanish influenza pandemic resulted in approximately 675,000 deaths in the United States in a population approximately one-third the size of the current population and approximately 50 million deaths globally.3,4 Since 1997, there has been heightened concern that avian influenza viruses associated with zoonotic outbreaks, such as H5N1 and H7N9, could adapt to achieve efficient transmissibility in humans and cause a pandemic.5
Influenza virus infections are generally acute, self-limited infections.6 Clinically, influenza manifests as an acute respiratory disease characterized by the sudden onset of high fever, coryza, cough, headache, prostration, malaise, and inflammation of the upper respiratory tree and trachea. Acute symptoms and fever often persist for 7 to 10 days, and in most cases, the infection is self-limited. Generally, pneumonic involvement is not clinically prominent, although weakness and fatigue may linger for weeks. People of all ages are afflicted, but the prevalence is greatest in school-aged children; disease severity is greatest in infants, elderly persons, and those with underlying illnesses. Influenza A viral replication peaks approximately 48 hours after infection of the nasopharynx and declines thereafter, with little virus shed after approximately 6 days. The virus replicates in both the upper and lower respiratory tract. The diagnosis of influenza can be established by viral culture, demonstration of viral antigens, demonstration of viral genetic material (in clinical specimens), or changes in specific antibody titers in serum or respiratory secretions.7
Influenza is the leading cause of respiratory viral disease in all hospitalized patients >16 years.8 People with underlying comorbidities, including chronic pulmonary or cardiac disease, immunosuppression, or diabetes mellitus, are at high risk of developing severe complications from influenza A viruses (IAVs). These may include hemorrhagic bronchitis, laryngotracheitis in young children, pneumonia (primary viral or secondary bacterial), and death.6,9 Obesity has recently been identified as an independent risk factor, and pregnancy has long been associated with increased risk.10,11
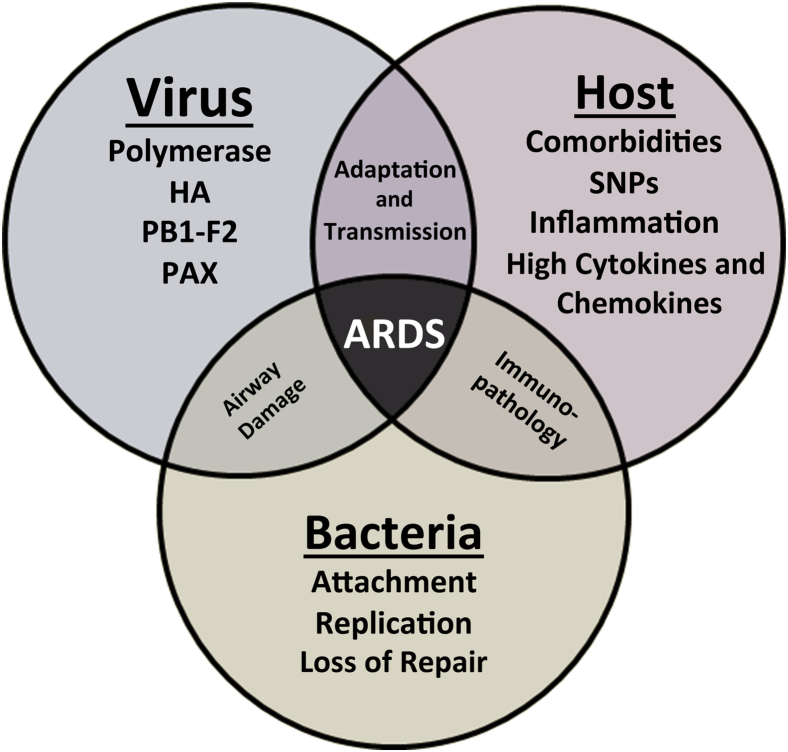
Complications of influenza, including hemorrhagic bronchitis, diffuse alveolar damage, and pneumonia, can develop within hours. Fulminant fatal influenza viral pneumonia occasionally occurs, but most pneumonias are caused by secondary bacterial infections.12,13 Development of pneumonia, dyspnea, cyanosis, hemoptysis, pulmonary edema leading to acute respiratory distress syndrome, and death may proceed in as little as 48 hours after the onset of symptoms. Progression to severe disease, including development of acute respiratory distress, pneumonia, and death, is likely a multifactorial process involving viral, host, and bacterial factors (Figure 1). In this review, we will examine each of these underlying factors to discuss their contribution to influenza pathogenesis.
Figure 1.
Diagram showing interrelationships between viral genes, host factors, and secondary bacterial infections associated with development of severe pathology and disease. ARDS, acute respiratory distress syndrome; HA, hemagglutinin; PAX, polymerase acidic frameshift protein; PB1, polymerase basic protein 1 frame 2 protein; SNP, single-nucleotide polymorphism.
Influenza A Virus Ecobiology
Overview
Influenza viruses (of the family Orthomyxoviridae) are enveloped, negative-sense, single-stranded RNA viruses with segmented genomes. There are five genera, including Influenzavirus A, Influenzavirus B, Influenzavirus C, Thogotovirus, and Isavirus (infectious salmon anemia virus).1,14,15
The Influenzavirus genera differ in host range and pathogenicity and diverged evolutionarily at least several thousand years ago.16 Influenza A and B viruses have a similar structure, whereas influenza C is more divergent. Influenza A and B type viruses contain eight discrete single-stranded RNA gene segments, each encoding at least one protein. Influenza B and C viruses are predominantly human-adapted viruses, whereas IAVs naturally infect hundreds of warm-blooded animal hosts, including both avian and mammalian species. Wild aquatic birds are the major reservoir of IAV, where it causes predominantly asymptomatic gastrointestinal tract infections.17 Mixed IAV infections and gene segment reassortment are common in wild aquatic birds. These data suggest that IAVs in wild birds form transient genome constellations without the strong selective pressure to be maintained as linked genomes, leading to the continual emergence of novel genotypes.18 This genetic and antigenic diversity of IAVs thus poses a significant risk of zoonotic infection, host switch events, and the generation of pandemic IAV strains. IAVs encode at least 13 proteins via alternative open reading frames, splicing, or ribosomal frame shifting.19
Viral Surface and Structural Proteins
IAVs express three surface proteins: hemagglutinin (HA), neuraminidase (NA), and matrix 2. IAVs are classified, or subtyped, by antigenic or genetic characterization of the HA and NA glycoproteins. Eighteen different HA and 11 different NA subtypes are known,18,20 with 16 of the HAs and 9 of the NAs consistently found in avian hosts in various combinations (eg, H1N1 or H3N2).18,20 HA is a glycoprotein that functions both as the viral receptor-binding protein and as the fusion protein. HA binds to sialic acid (N-acetyl neuraminic acid) bound to underlying sugars on the tips of host cell glycoproteins. IAVs have HAs with varying specificities for the disaccharide consisting of sialic acids (SAs) and the penultimate sugar (galactose or N-acetylgalactosamine) with different glycosidic bonds. IAVs adapted to birds typically have HA receptor binding specificity for α2,3 SA, whereas HAs from IAVs adapted to humans have higher specificity for α2,6 SA.1,14
After receptor binding, the virus is internalized. The endosomal compartment's acidic pH leads to an HA conformational change, facilitating fusion of the viral and endosomal membranes, release of viral ribonucleoproteins (RNPs) into the cytoplasm, and their subsequent transport to the nucleus. Viral HA is a homotrimer, and each monomer undergoes proteolytic cleavage to generate HA1 and HA2 polypeptide chains before activation. IAV does not encode a protease and requires exogenous serine proteases (trypsin-like enzymes) for activation that recognize a conserved Q/E-X-R motif found at the HA cleavage site.21 In humans and other mammals, Clara tryptase, produced by cells of the bronchiolar epithelium, likely serves this role. IAVs of H5 and H7 subtypes can acquire insertional mutations at the HA cleavage site, which change their protease recognition site to a furin-like recognition sequence R-X-R/K-R. This polybasic cleavage site broadens protease specificity, allowing for intracellular cleavage activation, and systemic replication of such viruses in poultry, resulting in the emergence of highly pathogenic avian influenza.
NA is a glycoprotein with neuraminidase (sialidase) enzymatic activity required for cleavage of these host cell SAs, allowing newly produced virions to be released and to cleave SAs from viral glycoproteins to prevent aggregation of nascent viral particles. The surface glycoproteins HA and NA are the major antigenic targets of the humoral immune response to IAV, and NA is the target of the antiviral drugs oseltamivir and zanamivir. The matrix 1 protein is the most abundant structural protein. Localized beneath the viral membrane, it interacts with the cytoplasmic domains of the surface glycoproteins HA and NA and also with the viral RNP complexes. The small protein matrix 2 is a proton channel necessary for viral replication and is the target of the adamantane class of antiviral drugs.22 Matrix 2 functions as a low pH gated ion channel that lowers the pH of the virion after internalization and leads to membrane fusion with the lysogenic vacuole. The viral RNPs are released into the cytoplasm and are then translocated to the nucleus to initiate viral RNA synthesis.
Viral RNA Polymerase
IAVs are segmented, negative-stranded viruses, and viral RNPs consist of each RNA gene segment, encapsidated by the single-stranded RNA binding protein nucleoprotein (NP) and associated with three viral polymerase proteins that comprise the RNA-dependent RNA polymerase-polymerase basic protein 2 (PB2), polymerase basic protein 1 (PB1), and polymerase acidic protein (PA). The polymerase proteins form a heterotrimer bound to short hairpin structures formed by the complementary terminal 5′ and 3′ untranslated regions of each RNA segment. PB1 is the RNA-dependent RNA polymerase, and PB2 functions in mRNA synthesis by binding host mRNA caps. Although PA is necessary for a functional polymerase complex, including endonucleolytic cleavage of host pre-mRNAs, its biological roles remain less well understood. NP plays important roles in transcription, and in the trafficking of RNPs between the cytoplasm and nucleus. IAV is dependent on the RNA processing machinery of the host cell, and transcription and replication occur in the host nucleus.
Non-Structural Proteins
Several non-structural proteins play important roles in the viral replicative cycle. The non-structural protein 1 has a variety of functions, including double-stranded RNA binding and antagonism of host cell type I interferon responses.14 The NS2 (or NEP) protein enables nuclear export of viral RNP complexes. PB1-F2 targets the mitochondrial inner membrane and may play a role in apoptosis during IAV infection.23 PAX is a newly discovered protein that functions to repress cellular gene expression and modulates IAV pathogenicity by an unknown mechanism.24
Polymerase Infidelity and Viral Evolution
The viral RNA-dependent RNA polymerase lacks proofreading, and IAVs are thus evolutionarily dynamic viruses with high mutation rates that range from approximately 1 × 10−3 to 8 × 10−3 substitutions per site per year.25 Mutations that alter amino acids in the antigenic portions of the surface glycoproteins HA and NA may allow IAVs to evade preexisting immunity. These mutations are especially important in human seasonal IAV strains, which are subjected to strong population immunological pressures. Anti-HA antibodies can prevent receptor binding, can neutralize infection, and are effective at preventing reinfection with the same strain. This selective mutation in the antigenic domains of HA and NA has been termed antigenic drift26 and is the basis for the need to yearly update the annual influenza vaccine formulation.1
The high mutation rate can also result in the rapid establishment of antiviral drug-resistant populations, including resistance to NA inhibitors and adamantanes. Clinical studies have shown that NA-resistant viruses can rapidly acquire mutations in NA after initiation of treatment,27,28 without apparent loss of fitness. Global circulation of such human IAVs bearing antiviral resistance mutations has been observed for different strains of influenza viruses in the past decade.29 Because the IAV genome consists of eight RNA segments, coinfection of one host cell with two different IAVs can result in progeny viruses containing gene segments derived from both parental viruses. When the process of genetic reassortment involves the gene segments encoding the HA and/or NA genes, it is termed antigenic shift. Reassortment is an important feature in IAV evolution18,30 and host switch.15
Viral Factors Contributing to Influenza Pathogenesis
Considering viral factors specifically, the molecular basis of virulence is also usually polygenic. Because IAV strains can move between hosts to cause either self-limited spillover infections or stably adapt to new hosts, viral pathogenesis varies by host and must be considered when examining viral virulence factors.
The processes by which IAV switch hosts, particularly those changes associated with adaptation of avian IAV to mammalian and ultimately human hosts, have been extensively studied, but are still only partially understood.15 Ultimately, influenza viruses must be able to infect target cells, replicate, and be transmitted efficiently to be adapted to a particular host. However, influenza pathogenesis does not require transmissibility, and zoonotic infections of nonadapted, or partially host-adapted, viruses can cause severe disease, such as observed in human infections with avian H5N1 and H7N9 viruses.5,31 It is likely that viral adaptive mutations enhancing replication, pathogenicity, and transmissibility in a particular host are polygenic traits driven by mutational pressures that are independent and possibly even competing.15,32,33
Virulence Factors Encoded by the Viral HA Gene
In seasonal influenza, rapid mutation of the antigenic regions of the HA results in antigenic drift and the necessity to continually reformulate the vaccine strains selected for inclusion. Emergence of antigenically drifted variants has been associated with more severe annual epidemics (eg, the 1947 H1N1 epidemic) and more recent H3N2 variants in 1997 and 2003 to 2004.30,34,35 Lack of protective immunity to pandemic IAV with antigenically novel HAs is also a likely factor contributing to enhanced morbidity and mortality in pandemics.36,37
Certain H5 or H7 strains of IAV can develop an insertional polybasic mutation at the HA cleavage site in poultry to become highly pathogenic avian influenza. These viruses have enhanced pathogenicity because HA activation can use intracellular furin-like proteases, allowing virus to replicate systemically.1 These viruses typically cause severe illness and high mortality in gallinaceous poultry, such as chickens and turkeys, but may be nonpathogenic in ducks, further supporting the species specificity of viral pathogenesis.38 Human zoonotic infections with highly pathogenic avian influenza H5N1 viruses have been documented in 650 patients in 16 countries since 2003, with 386 deaths (59% case fatality rate; World Health Organization, http://www.who.int/influenza/human_animal_interface/EN_GIP_20140124CumulativeNumberH5N1cases.pdf?ua=1, last accessed August 13, 2014).
The HA proteins encoded by the last four pandemic viruses (1918 H1N1, 1957 H2N2, 1968 H3N2, and 2009 H1N1) cause enhanced disease in mice compared to seasonal influenza H1N1 or H3N2 subtypes when expressed on a chimeric IAV in which the other seven genes were derived from a seasonal H1N1 human virus. These pandemic HA-expressing viruses had an expanded cellular tropism to infect alveolar epithelial cells and macrophages, and this was correlated with an inability of these zoonotically derived HAs to be neutralized by lung surfactant protein D in vitro.39 In addition, viral constructs expressing the 1918 pandemic HA on a seasonal H1N1 or H3N2 human virus40,41 showed murine respiratory tract pathology similar to that induced by the fully reconstructed 1918 virus, with a prominent infiltrate of alveolar neutrophils and macrophages.39
Interestingly, in a recent study, a chimeric 1918 virus in which its HA was replaced with a wild bird–derived avian H1 HA gene without any prior mammalian adaptation was not attenuated and induced a pattern of disease and histopathology indistinguishable from the fully reconstructed 1918 virus.42 This observation suggests that the HA virulence factor associated with the 1918 pandemic might be shared with avian influenza-derived H1 HAs.
Virulence Factors Encoded by the Viral Polymerase Genes
The IAV polymerase genes have also been shown to contain virulence factors in animal models. Chimeric IAVs encoding the four RNP gene segments of the 1918 virus (PB2, PB1, PA, and NP) in the context of the remaining genes from seasonal influenza viruses showed enhanced pathogenicity in mice and ferrets.43,44 Chimeric seasonal H1N1 viruses encoding the PB1 gene of the 1918 virus enhanced viral replication in mice and ferrets,41,44 whereas the PB1 gene from a seasonal influenza virus attenuated the 1918 virus in mice.41,44
Experimental data suggest that the most efficient influenza transcriptional activity in vitro was seen with an avian-derived PB1 gene45; given that the 1918, 1957, and 1968 pandemic viruses had avian influenza-like PB1 genes, there may be fitness advantages for human-adapted IAV strains to encode an avian influenza-derived PB1 polymerase.46 The IAV PB1 segment also variably encodes a second protein in a second reading frame, PB1-F2. The 1918 virus PB1-F2 was shown to be a virulence factor related to a delay in innate immune responses.47,48 The 1918 PB1-F2 was also shown to enhance the frequency and severity of secondary bacterial pneumonias in mice.49 The PA segment encodes a second protein produced by ribosomal frame shifting. The 1918 PAX protein modulated virulence in mice by decreasing pathogenicity. The 1918 viral constructs with loss of PAX expression led to changes in the kinetics of the host inflammatory response.24
Host Factors Contributing to Influenza Pathogenesis
Differences in the response of the host to influenza virus infection can have profound effects on disease severity and outcome. These can arise from preexisting medical conditions and viral gene-dependent differential activation of host inflammatory responses. Although most influenza virus infections result in acute, self-limited infections with minimal respiratory pathology, severe and fatal infections are associated with hemorrhagic bronchitis, bronchiolitis, and alveolitis with pulmonary edema and hemorrhage.6 As shown in Figure 2 (and the references therein), the pathological changes associated with fatal primary viral and secondary bacterial pneumonias were similar in both 1918 and 2009 pandemic autopsies.9,50,51
Figure 2.
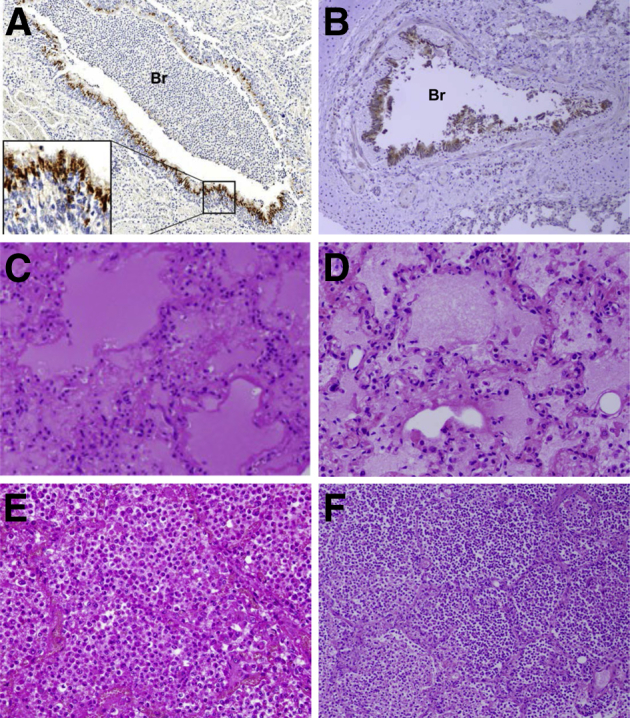
Representative pathological changes during fatal primary influenza and secondary bacterial infections in human autopsies from the 1918 (A, C, and E) and 2009 (B, D, and F) pandemics. A: Immunohistochemical stained section of lung from a 1918 pandemic influenza fatal case showing acute influenza viral bronchiolitis with infiltration of neutrophils and other inflammatory cells in the lumen of a bronchiole (Br). Influenza viral antigen (reddish-brown stain) is readily apparent in the apical cells of the bronchiolar respiratory epithelium (inset), on a hematoxylin-stained background.50B: Immunohistochemical stained section of lung from a 2009 pandemic influenza fatal case showing acute influenza viral bronchiolitis (Br). Influenza viral antigen (reddish-brown stain) is readily apparent in the apical cells of the bronchiolar respiratory epithelium, on a hematoxylin-stained background.51C: Hematoxylin and eosin (H&E)–stained section of lung from a 1918 pandemic influenza fatal case showing diffuse alveolar damage with hyaline membranes lining alveoli. The alveolar airspaces contain edema fluid, strands of fibrin, desquamated epithelial cells, and inflammatory cells.50D: H&E-stained section of lung from a 2009 pandemic influenza fatal case showing diffuse alveolar damage with hyaline membranes lining alveoli. The alveolar air spaces contain edema fluid, strands of fibrin, desquamated epithelial cells, and inflammatory cells.51E: H&E-stained section of lung from a 1918 pandemic influenza fatal case showing a massive infiltrate of neutrophils that fills the alveolar air spaces associated with a secondary bacterial bronchopneumonia. Alveolar capillary congestion is prominent.6F: H&E-stained section of lung from a 2009 pandemic influenza fatal case showing a massive infiltration of neutrophils in the airspaces of alveoli associated with a secondary bacterial bronchopneumonia.51A and C were modified from Sheng et al (published by College of American Pathologists),50E from Taubenberger and Morens (published by Annual Reviews),6 and B, D, and F from Gill et al (published by National Academy of Sciences of the United States of America).51 All images used with permission of the publishers. Original magnifications: ×40 (A and B); ×100 (E and F); ×200 (C and D).
Role of Comorbidities and Genetic Predisposition
Essential epidemiological attributes of seasonal influenza and pandemics are risk factors for severe complications and death. These include age, underlying chronic respiratory and cardiovascular conditions, diabetes mellitus, pregnancy, and immunosuppression. Both the young and old show increased morbidity and mortality to both seasonal and pandemic influenza viral infections, resulting in the typical U-shaped age-specific mortality curves for pneumonia and influenza.4 Immunocompromised patients also have an increased risk of enhanced disease after influenza infection and can shed virus for many months, despite treatment with antiviral drugs, such as NA inhibitors.52 Treatment of this patient population with antiviral drugs, however, may also result in the rapid accumulation of drug-resistance mutations. Immunocompromised patients are significantly more likely to have severe disease/complications and radiological imaging abnormalities while paradoxically demonstrating fewer clinical symptoms and signs, such as dry cough, chills, sweats, myalgia, shortness of breath, chemosis, and neurological symptoms.52
Recently, a single-nucleotide polymorphism (rs12252-C) in the interferon-inducible transmembrane 3 (IFITM3) gene has been identified in hospitalized patients who were diagnosed with seasonal or 2009 pandemic H1N1 infections that were associated with increased disease severity.53 Presence of this single-nucleotide polymorphism was also shown to be associated with increased cytokine expression and plasma and death in patients diagnosed with H7N9 infection.54 Although the relationship of genetic predispositions leading to enhanced disease is just beginning to emerge,55 numerous studies have shown a clear correlation between influenza viral pathogenesis and robust immune and inflammatory responses.
Role of Inflammatory Responses in Disease Severity
Numerous studies have shown an association between fulminant influenza virus infections with robust activation of host inflammatory responses.56–58 These responses include high levels of expression of cytokines, chemokines, and acute phase response reactants. These aberrant inflammatory responses lead to activation of immune cells and cell death responses and result in immunopathologies that contribute to disease severity. Significantly, transcriptomic analysis of a 1918 pandemic autopsy sample59 showed a marked concordance with studies in mice57 and cynomolgus macaques,58 revealing that significant activation of inflammatory and cell death responses, as seen in experimental animals, was also observed in a clinical specimen. The killing of infected cells by immune cells, such as macrophages, natural killer cells, cytotoxic T cells, and neutrophils, is a key component of the inflammatory response.
Immune cell-mediated killing occurs through a variety of mechanisms, including secretion of perforin, proteases, activation of cell death via death receptor ligands like tumor necrosis factor and FasL, and the production of reactive oxygen species (ROS). The in situ generation of ROS by neutrophils and macrophages, catalyzed by the NADPH-oxidase system, produces hydrogen peroxide and other ROS.60–63 ROS cause oxidation of cellular proteins, lipids, and DNA and result in cellular dysfunction or death.64,65 In animal models, highly pathogenic influenza viruses, such as 1918 virus, cause significant activation of antiviral, proinflammatory, ROS, and cell death responses during infection. In the case of 1918 virus, these exaggerated immune responses lead to severe necrotizing bronchitis, bronchiolitis, neutrophil-predominant alveolitis, acute edema, and pulmonary hemorrhage.57 Recently, we reported that treatment of mice infected with a lethal dose of the 1918 influenza virus with a catalytic ROS scavenging organometallic drug with superoxide dismutase and catalase mimetic activity increased survival.66 In this study, drug treatment resulted in decreased severity of lung pathology and greatly reduced ROS damage, but did not affect viral replication.66
Bacterial Factors Contributing to Influenza Pathogenesis
Primary influenza viral infections have been associated with secondary bacterial infections since the late 19th century.13 Although the use of antibiotics has had an impact on secondary bacterial infections, bacterial pneumonia after a primary influenza infection remains a significant threat to public health. During the 1918 influenza pandemic, most of the 50 million worldwide deaths were associated with concurrent or secondary bacterial infections, primarily with Gram-positive bacteria.13 Secondary bacterial infections also increased morbidity and mortality during the 1957 H2N2 and 1968 H3N2 pandemics. Even in the era of widespread antibiotic use, bacterial infections were identified in up to 34% of 2009 H1N1 pandemic influenza cases managed in intensive care units and up to 55% of fatal cases.12,51 The bacteria most frequently associated with secondary infections after influenza are the pneumopathogens Staphylococcus aureus, including methicillin-resistant strains, Streptococcus pneumoniae, and S. pyogenes.12,13 Thus, the increased susceptibility to the complication of secondary bacterial infections should be considered as an intrinsic virulence property of influenza viruses.
Numerous mechanisms have been proposed and experimentally shown to participate in the development of secondary bacterial pneumonias. These include increased colonization of the upper respiratory tract due to loss of physical barriers that inhibit the introduction of bacteria into the lung, such as necrosis of airway epithelial cells that can result in loss of mucociliary clearance, which facilitates colonization of the lungs with bacteria present in the oronasopharynx.67 The damaged surfaces of the airways with associated fibrin deposition, tissue repair, regenerative processes, and the sialidase activity of the viral NA can result in the exposure of bacterial attachment sites.68,69
Studies in mice have shown that primary influenza viral infection can inhibit the Th17 pathway and suppress macrophage function, including inhibition of NADPH-oxidase–dependent phagocytic bacterial clearance, leading to more severe secondary bacterial pneumonias.70,71 Our group has shown that another pathogenic mechanism of bacterial secondary pneumonia is the loss of lung repair responses and loss of basal epithelial cells required for reestablishment of the airway epithelium, including type I and II alveolar epithelial cells. Loss of basal cells was associated with bacterial attachment and apoptosis, suggesting a direct role of the bacteria in death of this essential cell population.72
Once a secondary bacterial infection is established, the blockage and loss of small airway function from cell debris and proteinaceous edema fluid and the development of exudative transmural pleuritis can severely compromise the respiratory system.67,73 Increased lung damage and loss of repair processes would also lead to loss of tissue integrity and contribute to the development of bacteremia. Thus, the risk of bacterial pneumonia may relate to increasing influenza viral pathogenicity, resulting in primary viral epithelial cell damage, bacterial invasion, and decreased lung repair due to loss of respiratory epithelial progenitor basal cells.72
Conclusions
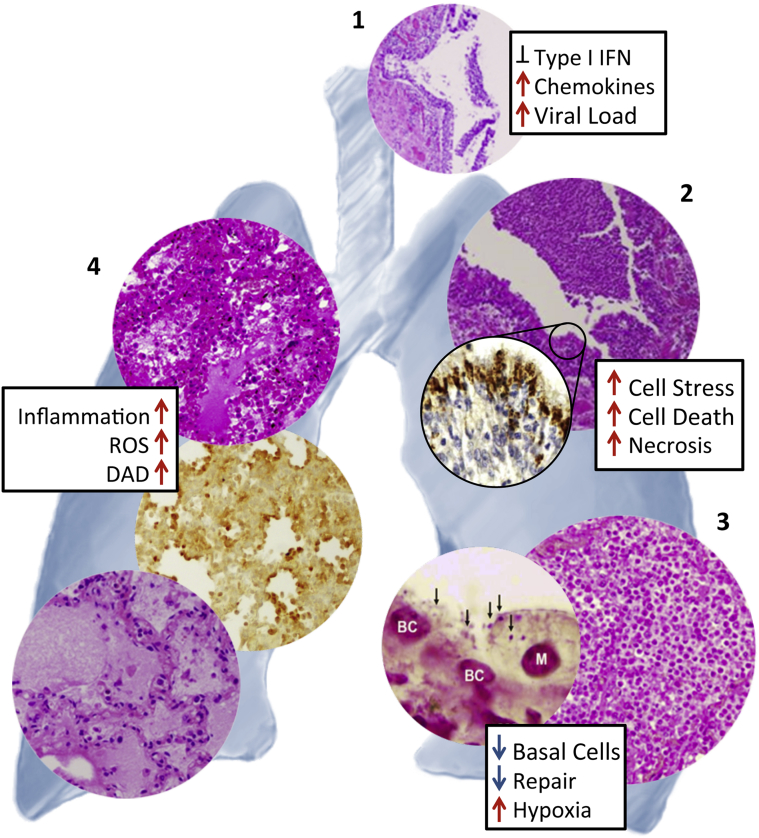
Influenza viruses pose significant public health threats whether examining the impact of seasonal influenza epidemics, zoonotic outbreaks, or the unpredictable emergence of novel pandemic viruses. Although most influenza infections are acute and self-limited, a set of multifactorial processes involving interrelated host, viral, and bacterial factors is involved in disease progression, leading to the development of pneumonia and death in a subset of patients (Figure 3).
Figure 3.
Relationship between pathological changes and host responses during severe primary influenza and secondary bacterial infections in human autopsies and experimental animal infections. Necrotizing tracheitis (1) in a 2009 pandemic H1N1 fatality.51Inset shows the relative expression of inflammatory and type I interferon (IFN) gene expression responses in fatal 1918 influenza infection compared to seasonal virus in cynomolgus macaques.58 Severe necrotizing bronchitis/bronchiolitis (2) in a 1918 influenza autopsy sample showing intense viral antigen staining in the ciliated and goblet cells of the respiratory epithelium, but not in basal cells.50Inset shows the relative expression of inflammatory response and cell death genes in lungs of mice infected with 1918 compared to seasonal H1N1.58 Secondary bacterial pneumonias (3) with massive infiltration of neutrophils into alveolar airspaces in a 1918 pandemic influenza13 fatal case (right panel). Left panel: Gram stain showing presence of Streptococcus pneumoniae bacteria (arrows) on remaining basal epithelial cells (nuclei labeled BC) and a macrophage (M) in mouse lung after coinfection of 2009 pandemic H1N1 and S. pneumoniae.72Inset shows loss of lung repair and regeneration gene expression responses in mice infected with 2009 pandemic H1N1 with a secondary S. pneumoniae infection compared to primary 2009 pandemic H1N1 infection.66 Development of diffuse alveolar damage (DAD) (4) with alveolar hyaline membrane formation, varying degrees of acute intra-alveolar edema, acute hemorrhage, interstitial and airspace inflammatory infiltrates, and small-vessel thromboses in 1918 pandemic influenza (top panel)6,9,51 and 2009 pandemic influenza (bottom panel)51 autopsies, along with pronounced alveolar epithelial immunostaining for reactive oxygen species (ROS) damage in mouse tissue infected with 1918 influenza virus.66Inset shows relative expression of inflammatory and ROS response genes in lungs of mice infected with 1918 compared to seasonal H1N1.66 Photomicrograph in 1 was modified from Gill et al (published by National Academy of Sciences of the United States of America)51; in 2 from Sheng et al (published by College of American Pathologists)50; in 3 from Morens et al (published by Oxford University Press)13 (right panel) and Kash et al (published by American Society for Microbiology)72 (left panel); and in 4 from Taubenberger and Morens (published by Annual Reviews)6 (top panel), Kash et al (published by Elsevier B.V.)66 (middle panel), and Gill et al (published by National Academy of Sciences of the United States of America)51 (bottom panel). All images used with permission of the publishers.
Influenza virus infections can involve all levels of the respiratory tree, and infection of the respiratory epithelium corresponds to the clinical signs and symptoms of tracheitis and bronchitis.6 Pathologically, influenza infection may be associated with severe, necrotizing tracheitis and bronchitis/bronchiolitis (Figure 3), where highly virulent infections are associated with lack of activation of many antiviral responses, such as expression of type I interferons, and marked expression of proinflammatory cytokines and chemokines (Figure 3).
Secondary or coincident bacterial pneumonias frequently occur and complicate the pathological picture. In such cases, a massive infiltration of neutrophils into alveolar airspaces is usually observed, resulting in significant tissue damage that is enhanced by the concomitant loss of lung repair and consequently results in severe respiratory compromise (Figure 3).
Autopsy studies in humans and necropsy studies in experimental animals have helped elucidate the characteristic changes of severe influenza viral pneumonia, including the development of diffuse alveolar damage with alveolar hyaline membrane formation, varying degrees of acute intra-alveolar edema and/or acute hemorrhage, interstitial and airspace inflammatory infiltrates, and small-vessel thromboses.6 These changes correspond clinically to acute respiratory distress syndrome and are associated with increased ROS damage and cell stress responses (Figure 3). In later stages, organizing diffuse alveolar damage and changes associated with repair (eg, fibrosis, epithelial regeneration, and squamous metaplasia) are commonly observed.9
Although the multifactorial processes cannot, in general, be separated in a patient with severe influenza, experimental animal models have greatly contributed to our understanding of how different factors interact to enhance influenza disease. A better understanding of these processes is critically important for our ability to develop new therapeutic modalities to improve outcomes in high-risk patients and patients with severe disease.
Infectious Disease Theme Issue
Footnotes
Supported by the National Institute of Allergy and Infectious Diseases, NIH, Intramural Research Program grant 1-ZIA-AI000986-08.
Disclosures: None declared.
This article is part of a review series on infectious disease.
References
- 1.Wright P.F., Neumann G., Kawaoka Y. Orthomyxoviruses. In: Knipe D.M., Howley P.M., editors. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1691–1740. [Google Scholar]
- 2.Thompson M.G., Shay D.K., Zhou H., Bridges C.B., Cheng P.Y., Burns E., Bresee J.S., Cox N.J. Estimates of deaths associated with seasonal influenza: United States, 1976-2007. MMWR Morb Mortal Wkly Rep. 2010;59:1057–1062. [PubMed] [Google Scholar]
- 3.Johnson N.P., Mueller J. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull Hist Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 4.Taubenberger J.K., Morens D.M. 1918 Influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris J.S.M. Avian influenza viruses in humans. Rev Sci Tech. 2009;28:161–173. doi: 10.20506/rst.28.1.1871. [DOI] [PubMed] [Google Scholar]
- 6.Taubenberger J.K., Morens D.M. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taubenberger J.K., Layne S.P. Diagnosis of influenza virus: coming to grips with the molecular era. Mol Diagn. 2001;6:291–305. doi: 10.1054/modi.2001.28063. [DOI] [PubMed] [Google Scholar]
- 8.Gaunt E.R., Harvala H., McIntyre C., Templeton K.E., Simmonds P. Disease burden of the most commonly detected respiratory viruses in hospitalized patients calculated using the disability adjusted life year (DALY) model. J Clin Virol. 2011;52:215–221. doi: 10.1016/j.jcv.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuiken T., Taubenberger J.K. The pathology of human influenza revisited. Vaccine. 2008;26:D59–D66. doi: 10.1016/j.vaccine.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson E.A., Marcelin G., Webby R.J., Schultz-Cherry S. Review on the impact of pregnancy and obesity on influenza virus infection. Influenza Other Respir Viruses. 2012;6:449–460. doi: 10.1111/j.1750-2659.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memoli M.J., Harvey H., Morens D.M., Taubenberger J.K. Influenza in pregnancy. Influenza Other Respir Viruses. 2013;7:1033–1039. doi: 10.1111/irv.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chertow D.S., Memoli M.J. Bacterial coinfection in influenza: a grand rounds review. JAMA. 2013;309:275–282. doi: 10.1001/jama.2012.194139. [DOI] [PubMed] [Google Scholar]
- 13.Morens D.M., Taubenberger J.K., Fauci A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palese P., Shaw M.L. Orthomyxoviridae: The Viruses and Their Replication. In: Knipe D.M., Howley P.M., editors. Lippincott, Williams & Wilkins; Philadelphia: 2007. pp. 1647–1690. [Google Scholar]
- 15.Taubenberger J.K., Kash J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki Y., Nei M. Origin and evolution of influenza virus hemagglutinin genes. Mol Biol Evol. 2002;19:501–509. doi: 10.1093/oxfordjournals.molbev.a004105. [DOI] [PubMed] [Google Scholar]
- 17.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dugan V.G., Chen R., Spiro D.J., Sengamalay N., Zaborsky J., Ghedin E., Nolting J., Swayne D.E., Runstadler J.A., Happ G.M., Senne D.A., Wang R., Slemons R.D., Holmes E.C., Taubenberger J.K. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wise H.M., Hutchinson E.C., Jagger B.W., Stuart A.D., Kang Z.H., Robb N., Schwartzman L.M., Kash J.C., Fodor E., Firth A.E., Gog J.R., Taubenberger J.K., Digard P. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012;8:e1002998. doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y., Tefsen B., Shi Y., Gao G.F. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 2014;22:183–191. doi: 10.1016/j.tim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Lee K.H., Steinhauer D.A., Stevens D.J., Skehel J.J., Wiley D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 22.Hayden F.G. Antivirals for influenza: historical perspectives and lessons learned. Antiviral Res. 2006;71:372–378. doi: 10.1016/j.antiviral.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Conenello G.M., Palese P. Influenza A virus PB1-F2: a small protein with a big punch. Cell Host Microbe. 2007;2:207–209. doi: 10.1016/j.chom.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Jagger B.W., Wise H.M., Kash J.C., Walters K.A., Wills N.M., Xiao Y.L., Dunfee R.L., Schwartzman L.M., Ozinsky A., Bell G.L., Dalton R.M., Lo A., Efstathiou S., Atkins J.F., Firth A.E., Taubenberger J.K., Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen R., Holmes E.C. Avian influenza virus exhibits rapid evolutionary dynamics. Mol Biol Evol. 2006;23:2336–2341. doi: 10.1093/molbev/msl102. [DOI] [PubMed] [Google Scholar]
- 26.Murphy B.R., Clements M.L. The systemic and mucosal immune response of humans to influenza A virus. Curr Top Microbiol Immunol. 1989;146:107–116. doi: 10.1007/978-3-642-74529-4_12. [DOI] [PubMed] [Google Scholar]
- 27.Memoli M.J., Hrabal R.J., Hassantoufighi A., Eichelberger M.C., Taubenberger J.K. Rapid selection of oseltamivir- and peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin Infect Dis. 2010;50:1252–1255. doi: 10.1086/651605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Memoli M.J., Hrabal R.J., Hassantoufighi A., Jagger B.W., Sheng Z.M., Eichelberger M.C., Taubenberger J.K. Rapid selection of a transmissible multidrug-resistant influenza A/H3N2 virus in an immunocompromised host. J Infect Dis. 2010;201:1397–1403. doi: 10.1086/651610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samson M., Pizzorno A., Abed Y., Boivin G. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res. 2013;98:174–185. doi: 10.1016/j.antiviral.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Rambaut A., Pybus O.G., Nelson M.I., Viboud C., Taubenberger J.K., Holmes E.C. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morens D.M., Taubenberger J.K., Fauci A.S. H7N9 avian influenza A virus and the perpetual challenge of potential human pandemicity. MBio. 2013;4 doi: 10.1128/mBio.00445-13. e00445–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herfst S., Schrauwen E.J., Linster M., Chutinimitkul S., de Wit E., Munster V.J., Sorrell E.M., Bestebroer T.M., Burke D.F., Smith D.J., Rimmelzwaan G.F., Osterhaus A.D., Fouchier R.A. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai M., Watanabe T., Hatta M., Das S.C., Ozawa M., Shinya K., Zhong G., Hanson A., Katsura H., Watanabe S., Li C., Kawakami E., Yamada S., Kiso M., Suzuki Y., Maher E.A., Neumann G., Kawaoka Y. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilbourne E.D., Smith C., Brett I., Pokorny B.A., Johansson B., Cox N. The total influenza vaccine failure of 1947 revisited: major intrasubtypic antigenic change can explain failure of vaccine in a post-World War II epidemic. Proc Natl Acad Sci U S A. 2002;99:10748–10752. doi: 10.1073/pnas.162366899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmes E.C., Ghedin E., Miller N., Taylor J., Bao Y., St George K., Grenfell B.T., Salzberg S.L., Fraser C.M., Lipman D.J., Taubenberger J.K. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 2005;3:e300. doi: 10.1371/journal.pbio.0030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morens D.M., Taubenberger J.K., Fauci A.S. The persistent legacy of the 1918 influenza virus. N Engl J Med. 2009;361:225–229. doi: 10.1056/NEJMp0904819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonsen L., Clarke M.J., Schonberger L.B., Arden N.H., Cox N.J., Fukuda K. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J Infect Dis. 1998;178:53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- 38.Sturm-Ramirez K.M., Hulse-Post D.J., Govorkova E.A., Humberd J., Seiler P., Puthavathana P., Buranathai C., Nguyen T.D., Chaisingh A., Long H.T., Naipospos T.S., Chen H., Ellis T.M., Guan Y., Peiris J.S., Webster R.G. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi L., Kash J.C., Dugan V.G., Jagger B.W., Lau Y.F., Sheng Z.M., Crouch E.C., Hartshorn K.L., Taubenberger J.K. The ability of pandemic influenza virus hemagglutinins to induce lower respiratory pathology is associated with decreased surfactant protein D binding. Virology. 2011;412:426–434. doi: 10.1016/j.virol.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kobasa D., Takada A., Shinya K., Hatta M., Halfmann P., Theriault S., Suzuki H., Nishimura H., Mitamura K., Sugaya N., Usui T., Murata T., Maeda Y., Watanabe S., Suresh M., Suzuki T., Suzuki Y., Feldmann H., Kawaoka Y. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- 41.Pappas C., Aguilar P.V., Basler C.F., Solorzano A., Zeng H., Perrone L.A., Palese P., Garcia-Sastre A., Katz J.M., Tumpey T.M. Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc Natl Acad Sci U S A. 2008;105:3064–3069. doi: 10.1073/pnas.0711815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi L., Davis A.S., Jagger B.W., Schwartzman L.M., Dunham E.J., Kash J.C., Taubenberger J.K. Analysis by single-gene reassortment demonstrates that the 1918 influenza virus is functionally compatible with a low-pathogenicity avian influenza virus in mice. J Virol. 2012;86:9211–9220. doi: 10.1128/JVI.00887-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jagger B.W., Memoli M.J., Sheng Z.M., Qi L., Hrabal R.J., Allen G.L., Dugan V.G., Wang R., Digard P., Kash J.C., Taubenberger J.K. The PB2-E627K mutation attenuates viruses containing the 2009 H1N1 influenza pandemic polymerase. MBio. 2010;1 doi: 10.1128/mBio.00067-10. e00067–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe T., Watanabe S., Shinya K., Kim J.H., Hatta M., Kawaoka Y. Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc Natl Acad Sci U S A. 2009;106:588–592. doi: 10.1073/pnas.0806959106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naffakh N., Massin P., Escriou N., Crescenzo-Chaigne B., van der Werf S. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J Gen Virol. 2000;81:1283–1291. doi: 10.1099/0022-1317-81-5-1283. [DOI] [PubMed] [Google Scholar]
- 46.Taubenberger J.K., Reid A.H., Lourens R.M., Wang R., Jin G., Fanning T.G. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 47.Conenello G.M., Zamarin D., Perrone L.A., Tumpey T., Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alymova I.V., Samarasinghe A., Vogel P., Green A.M., Weinlich R., McCullers J.A. A novel cytotoxic sequence contributes to influenza A viral protein PB1-F2 pathogenicity and predisposition to secondary bacterial infection. J Virol. 2014;88:503–515. doi: 10.1128/JVI.01373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McAuley J.L., Hornung F., Boyd K.L., Smith A.M., McKeon R., Bennink J., Yewdell J.W., McCullers J.A. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2:240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng Z.M., Chertow D.S., Ambroggio X., McCall S., Przygodzki R.M., Cunningham R.E., Maximova O.A., Kash J.C., Morens D.M., Taubenberger J.K. Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc Natl Acad Sci U S A. 2011;108:16416–16421. doi: 10.1073/pnas.1111179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gill J.R., Sheng Z.M., Ely S.F., Guinee D.G., Beasley M.B., Suh J., Deshpande C., Mollura D.J., Morens D.M., Bray M., Travis W.D., Taubenberger J.K. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med. 2010;134:235–243. doi: 10.5858/134.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Memoli M.J., Athota R., Reed S., Czajkowski L., Bristol T., Proudfoot K., Hagey R., Voell J., Fiorentino C., Ademposi A., Shoham S., Taubenberger J.K. The natural history of influenza infection in the severely immunocompromised vs nonimmunocompromised hosts. Clin Infect Dis. 2014;58:214–224. doi: 10.1093/cid/cit725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., Wise H.M., Kane L., Goulding D., Digard P., Anttila V., Baillie J.K., Walsh T.S., Hume D.A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S.B., Smyth R.L., Openshaw P.J., Dougan G., Brass A.L., Kellam P. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z., Zhang A., Wan Y., Liu X., Qiu C., Xi X., Ren Y., Wang J., Dong Y., Bao M., Li L., Zhou M., Yuan S., Sun J., Zhu Z., Chen L., Li Q., Zhang Z., Zhang X., Lu S., Doherty P.C., Kedzierska K., Xu J. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc Natl Acad Sci U S A. 2014;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albright F.S., Orlando P., Pavia A.T., Jackson G.G. Cannon Albright LA: Evidence for a heritable predisposition to death due to influenza. J Infect Dis. 2008;197:18–24. doi: 10.1086/524064. [DOI] [PubMed] [Google Scholar]
- 56.de Jong M.D., Simmons C.P., Thanh T.T., Hien V.M., Smith G.J., Chau T.N., Hoang D.M., Chau N.V., Khanh T.H., Dong V.C., Qui P.T., Cam B.V., Ha do Q., Guan Y., Peiris J.S., Chinh N.T., Hien T.T., Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kash J.C., Tumpey T.M., Proll S.C., Carter V., Perwitasari O., Thomas M.J., Basler C.F., Palese P., Taubenberger J.K., Garcia-Sastre A., Swayne D.E., Katze M.G. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobasa D., Jones S.M., Shinya K., Kash J.C., Copps J., Ebihara H., Hatta Y., Kim J.H., Halfmann P., Hatta M., Feldmann F., Alimonti J.B., Fernando L., Li Y., Katze M.G., Feldmann H., Kawaoka Y. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 59.Xiao Y.L., Kash J.C., Beres S.B., Sheng Z.M., Musser J.M., Taubenberger J.K. High-throughput RNA sequencing of a formalin-fixed, paraffin-embedded autopsy lung tissue sample from the 1918 influenza pandemic. J Pathol. 2013;229:535–545. doi: 10.1002/path.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheson B.D., Curnette J.T., Babior B.M. The oxidative killing mechanisms of the neutrophil. Prog Clin Immunol. 1977;3:1–65. [PubMed] [Google Scholar]
- 61.DeLeo F.R., Allen L.A., Apicella M., Nauseef W.M. NADPH oxidase activation and assembly during phagocytosis. J Immunol. 1999;163:6732–6740. [PubMed] [Google Scholar]
- 62.DeLeo F.R., Quinn M.T. Assembly of the phagocyte NADPH oxidase: molecular interaction of oxidase proteins. J Leukoc Biol. 1996;60:677–691. doi: 10.1002/jlb.60.6.677. [DOI] [PubMed] [Google Scholar]
- 63.Segel G.B., Halterman M.W., Lichtman M.A. The paradox of the neutrophil's role in tissue injury. J Leukoc Biol. 2011;89:359–372. doi: 10.1189/jlb.0910538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stadtman E.R. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 65.Stadtman E.R., Levine R.L. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 66.Kash J.C., Xiao Y., Davis A.S., Walters K.A., Chertow D.S., Easterbrook J.D., Dunfee R.L., Sandouk A., Jagger B.W., Schwartzman L.M., Kuestner R.E., Wehr N.B., Huffman K., Rosenthal R.A., Ozinsky A., Levine R.L., Doctrow S.R., Taubenberger J.K. Treatment with the reactive oxygen species scavenger EUK-207 reduces lung damage and increases survival during 1918 influenza virus infection in mice. Free Radic Biol Med. 2014;67:235–247. doi: 10.1016/j.freeradbiomed.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCullers J.A. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeLeo F.R., Musser J.M. Axis of coinfection evil. J Infect Dis. 2010;201:488–490. doi: 10.1086/650304. [DOI] [PubMed] [Google Scholar]
- 69.McCullers J.A., Bartmess K.C. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J Infect Dis. 2003;187:1000–1009. doi: 10.1086/368163. [DOI] [PubMed] [Google Scholar]
- 70.Robinson K.M., McHugh K.J., Mandalapu S., Clay M.E., Lee B., Scheller E.V., Enelow R.I., Chan Y.R., Kolls J.K., Alcorn J.F. Influenza A virus exacerbates Staphylococcus aureus pneumonia in mice by attenuating antimicrobial peptide production. J Infect Dis. 2014;209:865–875. doi: 10.1093/infdis/jit527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun K., Metzger D.W. Influenza infection suppresses NADPH oxidase-dependent phagocytic bacterial clearance and enhances susceptibility to secondary methicillin-resistant Staphylococcus aureus infection. J Immunol. 2014;192:3301–3307. doi: 10.4049/jimmunol.1303049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kash J.C., Walters K.A., Davis A.S., Sandouk A., Schwartzman L.M., Jagger B.W., Chertow D.S., Li Q., Kuestner R.E., Ozinsky A., Taubenberger J.K. Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. MBio. 2011;2 doi: 10.1128/mBio.00172-11. e00172–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kostrzewska K., Massalski W., Narbutowicz B., Zielinski W. Pulmonary staphylococcal complications in patients during the influenza epidemic in 1971-1972. Mater Med Pol. 1974;6:207–212. [PubMed] [Google Scholar]