Abstract
This review of Brucella–host interactions and immunobiology discusses recent discoveries as the basis for pathogenesis-informed rationales to prevent or treat brucellosis. Brucella spp., as animal pathogens, cause human brucellosis, a zoonosis that results in worldwide economic losses, human morbidity, and poverty. Although Brucella spp. infect humans as an incidental host, 500,000 new human infections occur annually, and no patient-friendly treatments or approved human vaccines are reported. Brucellae display strong tissue tropism for lymphoreticular and reproductive systems with an intracellular lifestyle that limits exposure to innate and adaptive immune responses, sequesters the organism from the effects of antibiotics, and drives clinical disease manifestations and pathology. Stealthy brucellae exploit strategies to establish infection, including i) evasion of intracellular destruction by restricting fusion of type IV secretion system-dependent Brucella-containing vacuoles with lysosomal compartments, ii) inhibition of apoptosis of infected mononuclear cells, and iii) prevention of dendritic cell maturation, antigen presentation, and activation of naive T cells, pathogenesis lessons that may be informative for other intracellular pathogens. Data sets of next-generation sequences of Brucella and host time-series global expression fused with proteomics and metabolomics data from in vitro and in vivo experiments now inform interactive cellular pathways and gene regulatory networks enabling full-scale systems biology analysis. The newly identified effector proteins of Brucella may represent targets for improved, safer brucellosis vaccines and therapeutics.
It is noteworthy that long ago in his publication Epidemics, Hippocrates described brucellosis-type syndromes in humans living in the Mediterranean littoral. Many centuries later, British physician, David Bruce, and Greek physician, Themistokles Zammit, in 1886 would discover the causative agent, Micrococcus melitensis, of brucellosis and would identify milk products of goats as the source of infection for military troops on the island of Malta. Even after more than a century of extensive research, Brucella spp. are still serious animal pathogens that cause brucellosis, a zoonosis that results in substantial economic losses, human morbidity, and perpetuates poverty worldwide.1 These Gram-negative bacteria infect a diverse array of land and aquatic mammals, including swine, cattle, goat, sheep, dogs, dolphins, whales, seals, and desert wood rats. Traditionally, the genus Brucella consisted of six recognized species, grouped according to their primary host preferences, that is, B. abortus, cattle; B. melitensis, sheep and goats; B. suis, pigs; B. ovis, sheep; B. canis, dogs; and B. neotomae, wood desert rats. Recent isolates from human (B. inopinata), aquatic mammals (B. pinnipedialis and B. ceti), and a common vole (B. microti) are recognized as new species, bringing the current number to 10 species in the genus. The basis for host preference remains an open question, although there may be a role for pseudogenes that influence host adaptation. The global disease burden in livestock is enormous. Conservative estimates are that >300 million of the 1.4 billion worldwide cattle population is infected with the pathogen. Brucellosis in animals results in abortion and other disease manifestations.
Brucella spp. infects humans as an incidental host. Human infection usually results from direct contact with tissues or blood from infected animals or by consumption of contaminated animal products, including unpasteurized milk and cheeses. In fact, >500,000 new human infections are estimated to occur annually. Brucellosis in humans typically presents with high, undulating fever. However, chronic brucellosis may affect many host organs, leading to arthritis, orchitis, hepatitis, encephalomyelitis, and endocarditis2,3 (Figure 1). Arthritis represents the most common complication. The diverse manifestations of the disease complicate diagnosis. Brucellosis has eluded systematic attempts at eradication for more than a century, even in most developed countries, and no approved human vaccine is available. The low number of virulent organisms required for infection combined with the capacity for aerosolization renders Brucella spp. as category B pathogens and potential agents for bioterrorism. With an infectious dose of 10 to 100 organisms, the calculated financial risk of such an attack is second only to anthrax and tularemia. In addition, the threat of deliberate release poses a direct risk to public health in an urban population that cannot be mitigated through the normal approach of animal vaccination. Brucellosis in humans and livestock are relatively uncommon in industrialized nations because of routine screening of domestic livestock and animal vaccination. However, brucellosis is endemic in many developing regions of the globe, including the Middle East, Asia, Africa, and South America, and in the United States where foci of disease remain because of persistent infection in wildlife species. This review of Brucella–host interactions and Brucella immunobiology is intended to present recent pathogenetic discoveries as the basis for pathogenesis-informed rationales to prevent and treat brucellosis.
Figure 1.

Hepatic and vertebral histopathology of human brucellosis caused by Brucella melitensis. A: Percutaneous liver biopsy. Mild nonspecific lymphocytic periportal hepatitis (arrow); stained with H&E. B: Percutaneous liver biopsy, culture positive for Brucella melitensis. Early-stage hepatic microgranuloma formation (arrow); stained with H&E. C: Guided needle core biopsy of vertebral body and epidural abscess, culture positive for Brucella melitensis. Lymphohistiocytic discitis osteomyelitis with dense cellular aggregates (arrow); stained with Diff-quik. Panels A and B are reproduced from Young et al2 with permission from Elsevier. Panel C was provided by Drs. Supriya Narasimhan and Michael L. Deftos (Santa Clara Valley Medical Center, San Jose, CA). Original magnification, ×40. H&E, hematoxylin and eosin.
Host Interactions
Pathology of Brucellosis
Brucellae display strong tissue tropism and replicate within vacuoles of macrophages, dendritic cells (DCs), and placental trophoblasts. However, the pathogen has the ability to replicate in a wide variety of mammalian cell types, including microglia, fibroblasts, epithelial cells, and endothelial cells. The intracellular lifestyle of Brucella limits exposure to the host innate and adaptive immune responses,4 sequesters the organism from the effects of some antibiotics, and drives the unique features of pathology in infected hosts, which is typically divided into three distinct phases: the incubation phase before clinical symptoms are evident, the acute phase during which time the pathogen invades and disseminates in host tissue, and the chronic phase that can eventually result in severe organ damage and death of the host organism. Nonspecific influenza-like symptoms observed in humans include pyrexia, diaphoresis, fatigue, anorexia, myalgia, and arthralgia. Furthermore, increasing evidence from endemic regions suggests that an elevated risk of human abortion is associated with exposure.5 Chronic infection results from the ability of the organism to persist in the cells of the host in which brucellae are distributed by way of the lymphoreticular system to eventually cause cardiovascular, hepatic, lymphoreticular, neurologic, and osteoarticular disease (Figure 1). Measurable splenomegaly is associated with increased lymphohistiocytic cells in the spleen, slightly reduced percentage of splenic CD4+ and CD8+ T cells, and major increases in the percentage of splenic macrophages.
Biology of Brucella
Brucellae quickly translocate across the mucosal epithelium layer6 in vivo and are endocytosed by mucosal macrophages and DCs. Brucella survive and replicate inside professional phagocytic cells, evade and modulate the host immune response, and disseminate to preferred tissues through cellular tropism, for example, placental trophoblasts in pregnant females, fetal lung, reticuloendothelial system, and reproductive tract.7 In vitro studies were used as models to understand adhesion, internalization, intracellular trafficking, survival, and replication of Brucella in susceptible hosts. Thus, after attachment to the surface of mucosal epithelial cells, Brucella induces a zipper-like mechanism for internalization.8 As yet incompletely defined binding molecule(s) are activated before and/or on contact with epithelial cells, Brucella bind to epithelial cell surface receptors that contain sialic acid and sulfated residues.9 Binding promotes activation of small GTPases that trigger a signaling cascade that reorganizes the actin cytoskeleton to induce a host cell membrane rearrangement along the surface of the pathogen that enhances invasion. Entry occurs within a few minutes after interaction which requires full activation of a mitogen-activated protein kinase signaling pathway.8 Brucella survive and replicate inside nonprofessional phagocytic cells up to 72 hours in vitro and move across the epithelium in vivo by subverting the mucosal epithelial barrier function to facilitate Brucella transepithelial migration.6 Simultaneously, this interaction initiates a minimal innate immune response with weak proinflammatory activity.8,10 Once translocated through the epithelium, Brucella are engulfed by mucosal phagocytic cells in which <10% of phagocytized bacteria survive an adaptation period. To delay being recognized by the immune system and initiating an immune response, Brucella reduce, modify, or cloak their pathogen-associated molecular patterns10; however, some Toll-like receptors (TLRs; mainly TLR2, TLR4, and TLR9) initiate limited intracellular signaling that activates the transcription factor NF-κB to control expression of inflammatory cytokine genes,11 although at a level that is 10-fold less than enterobacteria.
Inside mononuclear phagocytic cells, Brucella reside in a special vacuole (Brucella-containing vacuole, BCV), modify intracellular trafficking, and transform the vacuole into a replicative compartment or brucellosome.12 Experimental evidence indicates that the microenvironment inside the BCV is one of limited nutrient availability12 to which Brucella adapts soon after invasion. Initially, the pathogen undergoes quantitatively reduced gene expression and protein synthesis involved in anabolic metabolism while increasing amino acid catabolism, switching to alternative energy sources, and altering respiration to adapt to low oxygen tension.13 In an in vitro brucellosis infection model, expression of a type IV secretion system (T4SS) early after infection is essential for intracellular survival and multiplication inside mammalian cells. Yet, in vivo studies found that the T4SS is not necessary for invasion, systemic dissemination, or establishment of initial infection, but it is essential for prolonged persistence14 (C.A. Rossetti, K.L. Drake, S.D. Lawhon, J. Nunes, T. Gull, S. Khare, L.G. Adams, unpublished data) in which expression of T4SS stimulates an inflammatory reaction that was proposed as a mechanism to recruit cells that contribute to persistence.
Over the course of infection, invading brucellae surviving the adaptation period gradually recover the expression of key metabolic process-encoded genes. Transporters, iron metabolism, and cell membranes are primary targets for this transcription–translation reactivation.13 Brucella initiate replication concurrently with the resumption of expression of necessary functions, including virulence genes that in some cases are also tightly regulated by quorum-sensing molecules.15,16 Infected mononuclear phagocytic cells trigger extensive transcriptional changes in response to infection during the adaptation stage and return to normal levels after 12 hours, a time corresponding to the initiation of Brucella replication. Among the early transcription changes that contribute to adaptation, Brucella has several clever strategies to establish and maintain a chronic infection, including inhibition of apoptosis of infected mononuclear cells, prevention of DC maturation, reduced antigen presentation, and reduced activation of naive T cells.17 Once adapted to the intramacrophage environment, Brucella extends its intracellular persistence indefinitely, which contributes to systemic metastasis and infection of preferred targeted cells or tissues, such as placental trophoblasts, fetal lung, male genitalia, skeletal tissues, reticuloendothelial system, and endothelium. Currently, there is minimal information available to describe the interaction of Brucella with these target cells and tissue18,19 to provide a more holistic systems biology analysis of the pathogenesis of brucellosis at the level of the whole host organism.
Intracellular Trafficking
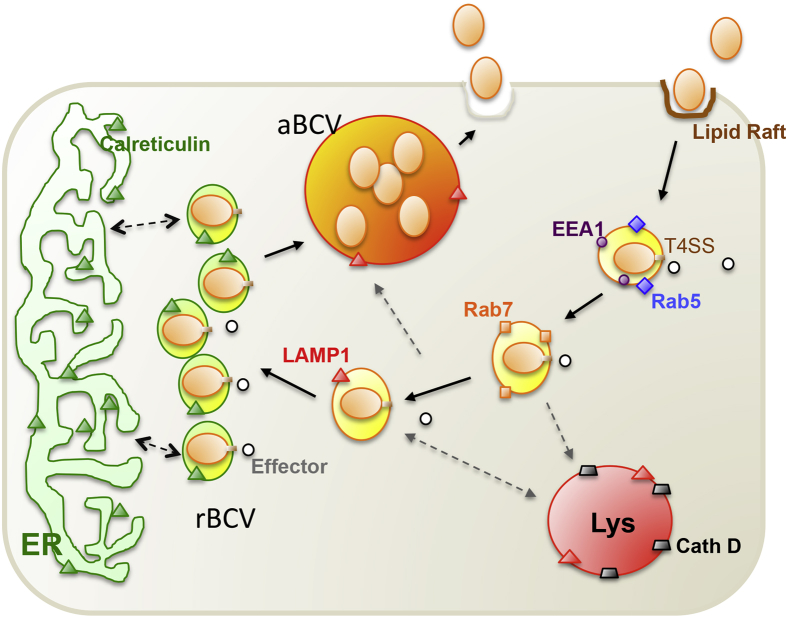
Brucellae evade intracellular destruction by restricting fusion of the BCV with the lysosomal compartment. Some BCVs that harbor internalized Brucella traffic from endocytic compartments to a replicative niche within BCVs that contain markers of the endoplasmic reticulum (ER). BCV seizure of ER membranes and components is accompanied by structural characteristics and functional restructuring of the ER.20 BCVs later accumulate autophagic features and exhibit lysosome-associated membrane protein-1 positivity, constituting a distinctive aspect of the intracellular Brucella lifestyle (Figure 2). The VirB T4SS regulates Brucella intracellular trafficking,21,22 and organisms that lack this system fail to establish an intracellular replicative niche in vitro. The T4SS of Brucella is thought to secrete effector molecules that control the intracellular and stealthy lifestyle of the pathogen.23–25 Table 1 summarizes several of the established interactive host and pathogen elements in the pathogenesis of brucellosis.
Figure 2.
Working model of Brucella intracellular trafficking in macrophage cells. Plasma membrane-associated lipid rafts mediate the internalization of smooth Brucella into macrophage cells. As the BCV matures, it sequentially associates with markers for early (EEA1, purple circle; Rab5, blue diamond) and late (Rab7, orange square) endosomes. The biogenesis and trafficking of BCVs is regulated by bacterial effector proteins (white circles), which are secreted through the Brucella T4SS. BCVs that contain virulent organisms do not fuse with lysosomes (cathepsin D, gray trapezoid), although transient association with LAMP1-positive membranes (orange triangles) is observed. The pathogen replicates in tight rBCVs that are decorated with calreticulin (green triangle), a marker for the ER. At a later point after infection (48 to 72 hours), the pathogen is observed in LAMP1-positive aBCVs that also contain LAMP1. The biogenesis of aBCVs depends on the activities of a subset of autophagy proteins, including ULK1 (not shown) and Beclin1 (not shown). Finally, the pathogen is released from the cell through lytic or nonlytic (shown) mechanisms. aBCV, autophagic Brucella-containing vacuole; BCV, Brucella-containing vacuole; Beclin1, coiled-coil myosin-like BCL2-interacting protein; EEA1, early endosome antigen 1; ER, endoplasmic reticulum; LAMP1, lysosome-associated membrane protein 1; rBCV, replicative Brucella-containing vacuole; T4SS, type IV secretion system; ULK1, Unc-51-like kinase 1.
Table 1.
Pathogenesis-related Brucella spp.–host molecular interacting elements
| Brucella element | Host factor | Reference |
|---|---|---|
| Adhesion and internalization | ||
| SP41 | Sialic acid residues | 9 |
| Hsp60 | PrPc | 26 |
| LPS | Scavenger receptor SR-A | 27 |
| Intracellular trafficking | ||
| RicA | GTPase Rab2 | 25 |
| Intracellular survival | ||
| Cyclic Β1,2-glycan | Cholesterol | 28 |
| Evasion of immunity | ||
| BtpA/Btp1/TcpB | MyD88 MAL |
29,30 |
| BtpB | MyD88 | 24 |
| Proinflammatory reaction | ||
| VceC | BiP/Grp78 | 31 |
Hsp60, heat shock protein 60; LPS, lipopolysaccharide; MAL, MyD88-adaptor like; MyD88, myeloid differentiation response gene 88; PrPc, cellular prion protein; SP41, 41-kDa surface protein.
Two-Component Regulatory Systems
Two-component regulatory systems also play an important role in the stealth program. Among the best studied are the BvrR/BvrS two-component regulatory system and the LuxR-like transcriptional regulator VjbR.16,32 The absence of the BvrR/BvrS sensory-regulatory system results in major changes in the bacterial outer membrane that alters cellular uptake of the organism.33 The BvrR/BvrS regulon also includes carbon and nitrogen metabolic functions and the expression of additional transcriptional regulators among 127 differentially regulated genes.32 In the absence of a functioning BvrR/BvrS, the organism fails to replicate intracellularly and is avirulent in the mouse model. Among the genes regulated by BvrR/BvrS, there are 10 transcriptional regulators, including vjbR. The protein-encoded vjbR was shown to regulate expression of the VirB locus that encodes the T4SS necessary to prevent phagosome–lysosome fusion.21 The absence of VirB alone may explain the attenuated virulence of bvrR/bvrS mutants.
Identification of T4SS-Secreted Substrates
Identification of Brucella virulence factors that facilitate invasion and infection was restricted to the surface O-polysaccharide until the late 1990s.34 At that time, several research groups used multiple approaches to genetically inactivate target genes. These reductionist approaches led to the conclusion that Brucella persists in nutrient-poor BCVs within the host,35 in which the organism replicates and from which infection spreads with minimal activation of the host cell.36 In contrast to the numerous metabolic functions shown to be necessary for intracellular replication, the T4SS stood out as a notable target for further investigation of virulence potential.37,38 Yet despite a decade of research, the complete mechanism of action remains undefined. Although shown to be required to prevent trafficking to the lysosome, the mechanistic steps involved, including interacting partners, enzymatic reactions, protein modifications, and detailed intracellular trafficking, are only now being described after a series of experiments in which Brucella gene reporter fusions were found to be secreted in a T4SS-dependent manner.23,25,39
Putative effector candidates were identified in silico on the basis of several criteria, including shared features with effectors expressed by other bacteria, eukaryotic motifs, GC content, and limited distribution across bacterial genera. Candidate effectors were shown to target secretory pathway compartments when expressed ectopically and impaired host protein secretion.31,39 Genetic studies have found a redundancy of function among effector candidates, consistent with the failure to identify effectors by using early genetic screens. Although several factors are now identified, the list is far from complete, and the mechanism by which replication is enhanced and lysosomal fusion is prevented remains undefined. Effector enzymatic activity, protein–protein interactions, and the identity of effector targets all remain to be identified.
Central Role for the Host UPR
The unfolded protein response (UPR) is an evolutionarily conserved pathway that mediates cellular adaptation to protein-folding stress in the ER. The stress sensors activating transcription factor 6, protein kinase RNA-like ER kinase, and inositol-requiring enzyme 1 α (IRE1α), which are located in the ER membrane, trigger UPR when unfolded proteins accumulate in the lumen of the ER. Activating transcription factor 6 regulates the expression of chaperones that facilitate protein folding. Protein kinase RNA-like ER kinase mediates transient translational attenuation and promotes apoptosis in unresolved stress. IRE1α plays a central role in initiating the UPR by catalyzing the splicing of X box binding protein I (XBP1) mRNA when unfolded proteins accumulate in the ER. The spliced message is then translated to generate active XBP1 transcription factor.40 XBP1 controls the expression of UPR genes that encode ER chaperones, proteins involved in ER-associated degradation, and other proteins that mitigate the harmful consequences of unfolded protein accumulation.41
Subversion of the UPR is critical to the intracellular lifestyle of Brucella.22,42 Murine embryonic fibroblasts that harbor deletions in IRE1α were used to demonstrate that this protein supports intracellular replication of Brucella in vitro. Host phosphatidylinositol 3-kinase (PI3K) activity was confirmed to mediate Brucella uptake; however, replication after uptake was unaffected.42 Celli and colleagues43 showed that Brucella replication occurs in an ER-derived replicative BCVs preceding the acquisition of autophagic markers. The gradual appearance of autophagic BCVs involves the acquisition of several autophagy proteins, including Unc-51-like kinase 1, Beclin 1. The formation of autophagic BCVs is independent of autophagy-elongation proteins, including ATG5 and LC3b,43 and gives rise to lysosome-associated membrane protein-1–positive, calreticulin-negative, pathogen-containing compartments. Importantly, autophagic BCVs were shown to play an important role in cell-to-cell spread of the pathogen.43 Finally, Brucella infection induces XBP1 splicing and UPR gene expression in host macrophages, both in vitro and in vivo.20 Moreover, treatment of host cells with tauroursodeoxycholic acid, a pharmacologic chaperone that ameliorates the UPR, impairs Brucella replication.20 Taken together, these data suggest that Brucella subverts host IRE1α signaling cascades to secure an intracellular niche that supports nutrient acquisition, pathogen replication, or pathogen cell-to-cell spread.
Host PI3K Activity and Brucella TIRAP-Containing Proteins
The role of PI3K in Brucella uptake is especially intriguing. Instrumental in the formation of phosphatidylinositol 3,4,5 trisphosphate, PI3K promotes binding of Toll–IL-1 receptor (TIR)-containing proteins on the plasma membrane and subsequent TLR signaling. Brucella prevents this by expressing at least two TIR-containing proteins to ultimately restrict the proinflammatory response and DC maturation.20,29,30,44,45 Details of the mechanism remain to be established, but evidence suggests that the presence of TIR-containing protein TcpB or BtpA and/or BtpB compete with myeloid differentiation response gene 88 (MyD88) for TIR domain-containing adaptor protein (TIRAP) binding which results in enhanced degradation of TIRAP by and interference with TLR4/TLR2 signaling.30 The presence of phosphoinositol phosphate binding sites in TcpB/BtpA and BtpB is consistent with binding at the plasma membrane and inhibition of downstream events that prevent activation of NF-κB–mediated transcription and development of an effective proinflammatory response.
Systems Biology and Omics Analysis of Brucella and Hosts
The expansion of genomics, next-generation sequencing, and omics technologies has enabled in-depth analysis of the pathogenesis of brucellosis. Large-scale simultaneous Brucella and host global expression data sets can now be combined with proteomics and metabolomics data from in vitro and in vivo experiments in target species and nonhuman primates to generate cellular pathway and gene regulatory networks46–48 that enable full-scale systems biology analysis49 and improved whole system understanding of Brucella pathogenesis. Around the turn of the century, the community of Brucella investigators generated genomes and data sets.7,50,51 Brucella melitensis strain 16M was the first Brucella genome sequenced and published.52 More than 18 complete and 415 whole genome shotgun Brucella genomes were sequenced, published in peer-reviewed journals, and are available online for analysis (Broad Institute, http://www.broadinstitute.org/annotation/genome/brucella_group/Downloads.html; Pathosystems Resource Integration Center, http://patricbrc.org/portal/portal/patric/Taxon?cType=taxon&cId=234; and National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=224914; all last accessed February 10, 2015). These data provide baselines for the analysis of comparative gene structure and homologies, conservation and variability, and gene expression, regulatory networks, and protein synthesis, interaction, and metabolic pathways. DNA sequence availability of several genomes of host species, including domestic animals and humans, together with experimental omics technologies and software tools for data analysis will increasingly facilitate construction of in silico interactome models of the infection biology of Brucella–host relations.
In vitro and in vivo Brucella and/or host gene expression and proteome data sets were generated by several investigators during the past decade, progressing toward a more comprehensive analysis of the host and pathogen.6,8,13,16,18,32,53–60 This broad spectrum of data sets was analyzed to identify candidate genes and biomarkers of Brucella and hosts,61 to analyze62 and predict Brucella antigenic proteins, to identify subunit vaccines,63 to understand gene regulatory networks,54 and to characterize the stringent Brucella stress response64 and modulation of host responses.53
Time-series studies of in vitro or in vivo experimental infection data were performed with a systems-based perspective of complex biological organizations to categorize the interactions of individual proteins within Brucella–host protein–protein networks. Time-series studies of in vivo system have served as a source of Brucella gene expression data interacted computationally with bovine host gene expression data to identify mechanistic genes of interacting cellular pathways as novel biosignatures and potential druggable targets on the basis of predicted protein–protein structural homologies (Figure 3).61,65 In another time-series study, results of systems biology analysis of Brucella and bovine host gene expression data61,65 were combined with reverse vaccinology62 to identify effective candidate subunit vaccines protective against virulent challenge in the mouse model.66 These studies provide provocative examples in support of using systems biology to more effectively integrate and exploit data for model development, for causal discovery, for the prediction of biological activities, for improving the design of in vitro and in vivo experiments, for finding biomarkers for enhanced brucellosis diagnosis, and for druggable targets for more effective treatment of brucellosis.
Figure 3.
Scheme of in vivo and/or in vitro systems biology analysis of host and Brucella interactive pathogenesis. Experimental pathology includes the collection and omics (transcriptomics, proteomics, metabolomics, etc.) data from both mammalian host and Brucella samples from an in vivo time series of Brucella infection of a target organ (eg, Peyer patch, lung, spleen, liver) in a natural target animal (eg, cattle, sheep, goat, pig, nonhuman primate). The resulting omics (eg, transcriptomics, proteomics, metabolomics) data sets are fused and bioinformatically analyzed for known and computed structural modeling of predicted host–pathogen protein–protein interactions to develop an in silico interactome structure learning model. Proteins are inferred from genes if not directly measured. Pairs of predicted candidate Brucella–host protein–protein mechanistic genes from interactive pathways are phenotyped in vitro in standardized gentamicin killing assays by using specific deletion mutants of Brucella, siRNA knockdown of host genes, and confocal microscopy. Brucella–host protein–protein pairs with positive in vitro phenotypes are phenotyped in vivo, and high confidence positive candidate protein pairs undergo pull-down analysis and quantitative selected reaction monitoring mass spectrometry for further confirmation of Brucella–host protein–protein interactions in Brucella pathogenesis. Blue lines and arrows indicate flow of in vivo data and results from the host, and brown lines and arrows indicate flow of data and results from Brucella into in silico analysis to identify and model domain A from a host protein predicted to interact with domain B of Brucella.
Immunobiology of Brucellosis
Central Role for Immunoregulatory Components
The stealthy nature of Brucella was attributed in large part to the structure of the smooth lipopolysaccharide (LPS) on the cell surface.10 The presence of elongated fatty acid molecules on the lipid A portion was shown to reduce the toxicity of Brucella LPS and to reduce the immune response by serving as a poor TLR4 agonist, consistent with the capacity of the organism to invade with minimal activation of the host cell. However, rough brucellae, lacking the O-polysaccharide portion of the LPS, are cytotoxic to macrophage cells. Although a comparative analysis of the lipid A from smooth and rough organisms has not been performed, there is no scientific reason to assume that the length of the fatty acids will be altered. Furthermore, the complete eradication of cytotoxic activity in the absence of the T4SS argues against a role for rough LPS in the cytotoxic activity. The simplest explanation at the current time is that Brucella LPS is a weak TLR4 agonist, whereas the O-polysaccharide is instrumental to the stealthy behavior of the organism. As an inducer of PI3K activity, weak TLR4 activation may be expected to reduce uptake as observed with smooth Brucella, whereas enhanced uptake observed with rough Brucella is consistent with an enhanced PI3K activity and a reduction in stealth.42
In addition to the weak agonist activity of Brucella LPS, the organism expresses novel immunoregulatory factors that suppress the innate immune response (Figure 4). The mechanism has been elaborated for some of these proteins, such as the TIR-containing protein or Brucella TIR protein, TcpB/BtpA.20 Similar to TLRs, TcpB contains a TIR domain through which it interacts with cytoplasmic MyD88 adaptor like/TIRAP. One study concluded that TcpB binding prevents MyD88 binding to MyD88 adaptor like/TIRAP, thereby accelerating MyD88 adaptor like/TIRAP degradation and subverting TLR signaling and proinflammatory cytokine production.30,45 In another study, TcpB was shown to interact with MyD88 to prevent downstream interactions and short-circuiting TLR-mediated activation.29 Although ΔtcpB knockout mutants exhibit minimal change in survival in the wild-type mouse model, replication in macrophages is reduced at late time points (≥48 hours) after infection.20 In addition, current evidence suggests that the absence of tcpB expression results in an elevated state of immune activation that reduces overall organism survival. One implication of this hypothesis is that TcpB may act through protein kinase B to relieve tuberous sclerosis complex 1/2-mediated inhibition of target of rapamycin activity to ultimately suppress the NF-κB–mediated proinflammatory response and induce IL-10 production. To do so, TcpB must enhance the production or stability of phosphatidylinositol-3,4,5 triphosphate, which would deplete the levels of phosphatidylinositol 4,5-bisphosphate and diminish plasma membrane interactions to support TLR signaling.
Figure 4.
Brucella is bipolar. Brucella both inhibits and promotes a proinflammatory immune response. TLR4 signaling during infection is restricted by the presence of elongated fatty acid chains that reduce the toxicity of the LPS (blue studs on Brucella surface) and by blocking downstream IKK phosphorylation via MyD88 binding (blue squares) with Brucella TIR-containing proteins, BtpA and BtpB (red squares), leading to enhanced polyubiquitination and degradation of MAL. However, the T4SS (VjbR-controlled expression) effector VceC (red oval) stimulates an innate immune response via interaction with BiP, an ER molecular chaperone (green squares) to release and phosphorylate IRE1 to promote mRNA splicing of XBP1 and activation of UPR. IRE1 phosphorylation also promotes the proinflammatory response via the release of NF-κB from the complex with IκBα (maroon crescent). The critical distinction between these two pathways may reside in the timing of activation. Inhibiting the host early or MyD88-mediated response may promote acquisition of a replicative niche, whereas the delayed T4SS-mediated VceC effector (VjbR-controlled expression) response may enhance the spread of the organism. Solid arrows indicate Brucella-mediated activation, whereas dotted arrows indicate Brucella-mediated inhibition. ER, endoplasmic reticulum; IκBα, inhibitor of κB protein α; Iκκ, IκB kinase; IRE1, inositol-requiring enzyme 1; LPS, lipopolysaccharide; MAL, MyD88-adaptor like; MYd88, myeloid differentiation response gene 88; T4SS, type IV secretion system; TIRAP, Toll–IL-1 receptor domain-containing adaptor protein; TLR4, Toll-like receptor 4; TNF-α, tumor necrosis factor α; UPR, unfolded protein response; XBP1, X-box binding protein 1.
Recently, a second protein BtpB, containing TIR structural domains, was reported to interfere with TLR signaling through MyD88 to prevent DC maturation.24 Redundancy of TcpB/BtpA and BtpB function may explain the failure to identify these immunoregulatory genes by using simple transposon screens. Interaction between BtpA/BtpB and MyD88 may also explain the substantial increase in survival of Brucella in MyD88−/− knockout mice in contrast to TLR4−/−/TLR2−/− knockout mice in which no substantial increase over wild-type was observed.67 The absence of MyD88 may adversely affect the proinflammatory response that results from a reduction of both ER stress and TLR-induced innate immune responses.31
Protective Immunity against Stealthy Brucella
Knowledge of protection against infection is derived from a number of disparate animal models, including mice, guinea pigs, ruminants, nonhuman primates, and humans. The importance of a T helper cell type 1 (Th1) response against Brucella is supported by numerous studies and is summarized elsewhere,51,68,69 particularly the roles of CD4+ and CD8+ T cells, although these results were contradictory at times. Natural killer cells play an important role in some hosts, but their role was shown to be minimal in the mouse model.70 Passive transfer experiments suggest that antibody to LPS (O-polysaccharide) may contribute to protection,71–75 yet the effectiveness of the T helper cell type 2 (Th2) humoral immune response remains unclear,76 and the efficacy of rough Brucella vaccines contradicts the role of anti-LPS antibodies in protective immunity.
Cytokines are key players in protection against brucellosis, mediating both innate and adaptive immune responses. IL-12 produced by B cells and macrophages leads to a Th1 response and induction of interferon-γ, which activates macrophages. The activity of is interferon-γ is maximized by tumor necrosis factor-α produced by macrophages and natural killer cells. Reports also indicate that IL-1–dependent induction of colony-stimulating factor increases neutrophil and macrophage infiltration into the spleen.77 This phenomenon may also explain a role for IL-6 produced by T cells. Splenocytes of infected hosts express higher levels of mRNA for IL-2, interferon-γ, and IL-10 and reduced levels of mRNA for IL-4, consistent with a Th1 response.78,79 Increased IL-10 observed later in infection may support the capacity of Brucella to avoid immune surveillance, resulting from repression of a protective Th1 response.80 Interestingly, cellular and humoral immune responses against similar Brucella strains vary significantly among susceptible hosts. This confounding aspect of Brucella immunobiology has presented important challenges in the identification of reliable correlates of immune protection in tractable model animal systems.
Resistance to other innate immune system components (eg, complement, opsonins, phagocytic cells, innate lymphocytes, cytokines, and other barriers) was in most cases suspected of being inherent to Brucella and provides passive resistance to intracellular killing mechanisms. However, on the basis of the importance of the T4SS to the long-term success of infection, it is becoming clearer that resistance mechanisms alone are not sufficient for the success of infection. Brucellae, and other intracellular pathogens, alter the innate immune response with the immediate aim of establishing a replicative niche and long-term persistence. To restrict long-term protective immunity, the organism first avoids the innate immune response by stealthy entry into host cells. From there, the organism controls aspects of protein secretion, intracellular trafficking, and bacterial replication,31,39 ultimately altering the course of the innate and adaptive immune responses.81
Failure of long-term protection against Brucella infection is the result of a weakened adaptive immune response controlled in part by the attenuated innate immune response.71,82–84 As a stealth invader, Brucella enters the host cell without apparent activation of the innate immune response through TLR ligand interaction. Knockout mice and animals deficient in either or both TLR2 and TLR4 exhibit little relevant change in the ability to control infection. In contrast, cells deficient in MyD88 sustain a two-log increase in bacterial infection.85 This finding may be best explained by a redundancy in host functions. However, it may also reflect that the primary goal is prevention of long-term adaptive immune response rather than rescue at early stages of infection. Evasion of the host-induced innate immune response may allow the organism to gain a foothold, whereas stimulation at later times aids the spread of infection. Manipulation of the innate immune response was found for at least three factors TcpB/BtpA, BtpB, and VceC (Figure 4). Although numerous other effectors were identified,23–25,31,39 their contribution to pathogen survival remains to be demonstrated. However, it seems clear that the wild-type Brucella has at its disposal a complete battery of effectors and that any delay in the innate immune response induced by these proteins could potentially be manipulated so as to improve the potential for more protective and safer vaccines.
Pathogenesis-Informed Approaches to Therapeutics
Human Brucella infection is typically treated with combination antibiotic therapy. But therapeutic relief from infection is neither certain nor rapid, because the intracellular location reduces antibiotic efficacy. Treatment can be prolonged and associated with undesirable side effects. Moreover, relapse after antibiotic therapy constitutes a substantial risk to the treated patient. As such, the recommended treatment usually entails a combination of drugs for at least 30 days,86 each of which falls into Food and Drug Administration pregnancy categories C (rifampin) or D (tetracycline/doxycycline) that are poor to unsuitable for use in gravid women.
To address these issues, development of novel therapeutic strategies to address brucellosis has been pursued. Recent advances in our understanding of the molecular mechanisms that mediate interactions between Brucella and host cells have facilitated this pursuit. For example, Baron et al87 have spearheaded the development of novel antivirulence compounds that target virulence functions while leaving essential cell functions unharmed. One advantage of the antivirulence approach is that molecules that do not target essential functions will fail to induce selective pressure for antibiotic resistance. To identify novel compounds that inhibit the virulence of Brucella, a high throughput small molecule screen was performed for compounds that inactivated the function of VirB8,88 a central component of the bacterial T4SS, which is essential for virulence and pathogenicity of Brucella,89 and a wide variety of other animal and plant pathogens. This endeavor identified several potent and specific inhibitors of T4SS function. X-ray crystallography and docking studies were used to demonstrate that VirB8 inhibitors bind to a surface groove opposite to the dimerization interface,88,90 thereby providing insight into the mechanism of action of this potential therapeutic.
Pathogenesis-Informed Approaches to Vaccines
Nonviable Brucella vaccines historically have a poor record of success that started with heat-killed Brucella, crude extracts, and then subunit and DNA vaccines. This poor track record is primarily explained by a failure to induce an effective Th1 response comparable with live, attenuated vaccines (LAVs).91 In contrast, LAVs have a long history of successful use against brucellosis and other intracellular pathogens in laboratory animal models and in target species, such as ruminants. However, given the safety concerns associated with human use, including the threat of persistent infection or reversion to virulence extreme, caution must be used in LAV development.
Prevention of animal disease was found to provide substantial protection against human disease. Historically, the focus of immune protection against Brucella infection in animals has been the use of spontaneously attenuated strains.92,93 Their stable and effective use in animals over decades was used to justify support for human trials.94 Three vaccines are used extensively in animals to provide immune protection: B. abortus S19 and RB51 and B. melitensis Rev.1.95–97 S19 and Rev.1 are fortuitously attenuated isolates. One strain was obtained accidentally, and the second was obtained after a stepwise process to identify streptomycin-dependent and then streptomycin-independent isolates. The two strains are considered to be smooth (expressing intact LPS with O-polysaccharide) which distinguishes them from RB51, a rough strain lacking the O-polysaccharide. S19 and Rev.1 provide superior protection but exhibit substantial human virulence and cannot be used in gravid female animals. RB51 is considered to be safe for use in gravid females and does not induce O-polysaccharide antibodies that can be used to distinguish field strain-infected from vaccinated animals. Despite well-defined differences in immune potential among these vaccines, no marker or correlate has been identified that can be used to predict immune protection.
Attenuated virulence may be derived from simple point mutations or from genetic rearrangements, including gene deletion. Although the potential for reversion of a mutant that bears a complete gene deletion is already small, the potential for reversion to virulence can be made infinitesimal by the introduction of a secondary mutation. Optimally, the additional mutation would affect the same pathway as the primary mutation, serving only as a backup to prevent reversion, but inducing no additional reduction in virulence or negative effect on protective immunity.
Obviously, care must be taken so that a balance is struck that supports survival sufficiently to enhance immune protection without posing a risk of inducing disease. One approach to this task is encapsulation of the attenuated vaccine strain to release the organism over time and to provide the added advantage of providing a natural booster response.68,98 This approach uses a vaccine depot from which the attenuated Brucella is gradually released over a 30-day period and significantly enhances immune protection by using highly attenuated LAVs to improve both efficacy and safety.99 In addition, recent cell biology findings have revealed the dependence of Brucella infection on the UPR, specifically IRE1α.20,42 This dependence may be exploited in an effort to provide LAVs that provide enhanced immune protection. ER stress and TLR signaling provide a synergistic stimulation of the proinflammatory response.100 The key to an optimal LAV development strategy is to identify vaccine candidates that fail to restrict the innate immune response and as a result induce an effective adaptive immune response without safety or reversion concerns.
Perspectives
Although Brucella spp. infect humans as an incidental host, 500,000 new human infections occur annually, yet no human vaccine or patient-friendly treatments exist. In addition, the threat of deliberate release poses a direct risk to public health that cannot be mitigated through the usual approach of animal vaccination to protect the public. Nonspecific influenza-like symptoms observed in humans include pyrexia, diaphoresis, fatigue, anorexia, myalgia, and arthralgia. Chronic infection results from the ability of the organism to persist in the cells of the host in which Brucella are distributed through the lymphoreticular system to eventually cause cardiovascular, hepatic, lymphoreticular, neurologic, and osteoarticular disease.
Brucellae quickly translocate across the mucosal epithelium layer and are endocytosed by mucosal macrophages and DCs. Brucellae display strong tissue tropism for the lymphoreticular system and an intracellular lifestyle that limits exposure to the host innate and adaptive immune responses, sequesters the organism from the effects of antibiotics, and drives the unique features of the clinical disease manifestations and pathology. Brucella uses several clever strategies to establish and maintain infection, such as evading intracellular destruction by restricting fusion of the type IV secretion system-dependent BCVs with the lysosome compartment, inhibition of apoptosis of infected mononuclear cells, prevention of DC maturation, inhibition of antigen presentation, and activation of naive T cells. Once adapted to the intramacrophage residence, Brucella prolongs its intracellular lifestyle indefinitely yet may systemically metastasize to infect other preferred targeted cells or tissues, such as placental trophoblasts, fetal lung, male genitalia, skeletal tissues, reticuloendothelial system, and endothelium. The mechanistic steps involved in interacting partners, enzymatic reactions, protein modifications, and detailed process of inhibiting intracellular trafficking to the lysosome are only now being described. Subversion of the host UPR proteins was found to be critical to the intracellular lifestyle of Brucella.101 Recent data suggest that Brucella subverts host IRE1α signaling cascades to secure an intracellular niche that supports nutrient acquisition, pathogen replication, or pathogen cell-to-cell spread.
By strategically converging on the entire range or holistic systems biology with traditional reductionist approaches, remarkable progress is being made toward improved design of brucellosis vaccines, enhanced biomarker diagnostics, and identifying specific druggable targets while avoiding the threat of antimicrobial resistance. Current reductionist approaches have generated extensive information about pathogen and host genes and pathways, yet clearer understanding of the interactions of these genes at the level of the whole host system has yet to emerge.
Despite well-defined differences in immune potential among existing vaccines, no marker or correlate has been identified that can be used to predict immune protection in livestock. LAVs have a long history of successful use against brucellosis and other intracellular pathogens in laboratory animal models and in target species. The key to an optimal LAV development strategy for humans is to identify vaccine candidates that fail to restrict the innate immune response and as a result induce an effective adaptive immune response without safety or reversion concerns. Brucella has a complete battery of effector proteins that delay the innate immune response which could be favorably manipulated to provide safer, more protective brucellosis vaccines and therapeutics.
Summary
After more than a century of research human and animal brucellosis still remains an important challenge to health and well-being. Moreover, no currently available safe, efficacious vaccines or patient-friendly treatments are available for human brucellosis, a disease that also needs low-cost bedside differential diagnostics. However, with the emergence of clearer understandings of the pathology and molecular pathogenesis of brucellosis, opportunities are substantially enhanced to identify and evaluate new druggable therapeutic targets and to apply reverse vaccinology and other postgenomic tools to develop safe, effective subunit or human LAVs.
Acknowledgments
We thank Drs. Angela Arenas, Richard C. Laughlin, and Kenneth L. Drake for their review, discussions, and thoughtful suggestions for this manuscript.
Infectious Disease Theme Issue
Footnotes
Supported by NIH/National Institute of Allergy and Infectious Diseases Western Regional Center of Excellence grant 1U54 AI057156-01 (L.G.A., A.R.-F., and T.A.F.), US Department of Homeland Security National Center of Excellence for Foreign Animal and Zoonotic Disease Defense grant ONR-N00014-04-1-0 (L.G.A.), US Department of Defense grant USAMRMC W81XWH-10-1-125 (A.R.-F.), and an I.N.T.A.-Fulbright Argentina Fellowship (C.A.R.).
Disclosures: None declared.
This article is part of a review series on infectious disease.
References
- 1.Pappas G., Panagopoulou P., Christou L., Akritidis N. Brucella as a biological weapon. Cell Mol Life Sci. 2006;63:2229–2236. doi: 10.1007/s00018-006-6311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young E.J., Hasanjani Roushan M.R., Shafae S., Genta R.M., Taylor S.L. Liver histology of acute brucellosis caused by Brucella melitensis. Hum Pathol. 2014;45:2023–2028. doi: 10.1016/j.humpath.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Dean A.S., Crump L., Greter H., Hattendorf J., Schelling E., Zinsstag J. Clinical manifestations of human brucellosis: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2012;6:e1929. doi: 10.1371/journal.pntd.0001929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martirosyan A., Gorvel J.P. Brucella evasion of adaptive immunity. Future Microbiol. 2013;8:147–154. doi: 10.2217/fmb.12.140. [DOI] [PubMed] [Google Scholar]
- 5.Baud D., Greub G. Intracellular bacteria and adverse pregnancy outcomes. Clin Microbiol Infect. 2011;17:1312–1322. doi: 10.1111/j.1469-0691.2011.03604.x. [DOI] [PubMed] [Google Scholar]
- 6.Rossetti C.A., Drake K.L., Siddavatam P., Lawhon S.D., Nunes J.E., Gull T., Khare S., Everts R.E., Lewin H.A., Adams L.G. Systems biology analysis of Brucella infected Peyer's patch reveals rapid invasion with modest transient perturbations of the host transcriptome. PLoS One. 2013;8:e81719. doi: 10.1371/journal.pone.0081719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams L.G. The pathology of brucellosis reflects the outcome of the battle between the host genome and the Brucella genome. Vet Microbiol. 2002;90:553–561. doi: 10.1016/s0378-1135(02)00235-3. [DOI] [PubMed] [Google Scholar]
- 8.Rossetti C.A., Drake K.L., Adams L.G. Transcriptome analysis of HeLa cells response to Brucella melitensis infection: a molecular approach to understand the role of the mucosal epithelium in the onset of the Brucella pathogenesis. Microbes Infect. 2012;14:756–767. doi: 10.1016/j.micinf.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castaneda-Roldán E.I., Avelino-Flores F., Dall'Agnol M., Freer E., Cedillo L., Dornand J., Girón J.A. Adherence of Brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cell Microbiol. 2004;6:435–445. doi: 10.1111/j.1462-5822.2004.00372.x. [DOI] [PubMed] [Google Scholar]
- 10.Barquero-Calvo E., Chaves-Olarte E., Weiss D.S., Guzmán-Verri C., Chacon-Diaz C., Rucavado A., Moriyón I., Moreno E. Brucella abortus uses a stealthy strategy to avoid activation of the innate immune system during the onset of infection. PLoS One. 2007;2:e631. doi: 10.1371/journal.pone.0000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira S.C., de Oliveira F.S., Macedo G.C., de Almeida L.A., Carvalho N.B. The role of innate immune receptors in the control of Brucella abortus infection: toll-like receptors and beyond. Microbes Infect. 2008;10:1005–1009. doi: 10.1016/j.micinf.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Kohler S., Foulongne V., Ouahrani-Bettache S., Bourg G., Teyssier J., Ramuz M., Liautard J.P. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc Natl Acad Sci U S A. 2002;99:15711–15716. doi: 10.1073/pnas.232454299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamontagne J., Forest A., Marazzo E., Denis F., Butler H., Michaud J.F., Boucher L., Pedro I., Villeneuve A., Sitnikov D., Trudel K., Nassif N., Boudjelti D., Tomaki F., Chaves-Olarte E., Guzman-Verri C., Brunet S., Cote-Martin A., Hunter J., Moreno E., Paramithiotis E. Intracellular adaptation of Brucella abortus. J Proteome Res. 2009;8:1594–1609. doi: 10.1021/pr800978p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roux C.M., Rolan H.G., Santos R.L., Beremand P.D., Thomas T.L., Adams L.G., Tsolis R.M. Brucella requires a functional Type IV secretion system to elicit innate immune responses in mice. Cell Microbiol. 2007;9:1851–1869. doi: 10.1111/j.1462-5822.2007.00922.x. [DOI] [PubMed] [Google Scholar]
- 15.Rambow-Larsen A.A., Rajashekara G., Petersoen E., Splitter G. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the Type IV secretion system and flagella. J Bacteriol. 2008;190:3274–3282. doi: 10.1128/JB.01915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weeks J.N., Galindo C.L., Drake K.L., Adams G.L., Garner H.R., Ficht T.A. Brucella melitensis VjbR and C12-HSL regulons: contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression. BMC Microbiol. 2010;10:167. doi: 10.1186/1471-2180-10-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Billard E., Dornand J., Gross A. Brucella suis prevents human dendritic cell maturation and antigen presentation through regulation of tumor necrosis factor alpha secretion. Infect Immun. 2007;75:4980–4989. doi: 10.1128/IAI.00637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho Neta A.V., Stynen A.P., Paixao T.A., Miranda K.L., Silva F.L., Roux C.M., Tsolis R.M., Everts R.E., Lewin H.A., Adams L.G., Carvalho A.F., Lage A.P., Santos R.L. Modulation of the bovine trophoblastic innate immune response by Brucella abortus. Infect Immun. 2008;76:1897–1907. doi: 10.1128/IAI.01554-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delpino M.V., Fossati C.A., Baldi P.C. Proinflammatory response of human osteoblastic cell lines and osteoblast-monocyte interaction upon infection with Brucella spp. Infect Immun. 2009;77:984–995. doi: 10.1128/IAI.01259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith J.A., Khan M., Magnani D.D., Harms J.S., Durward M., Radhakrishnan G.K., Liu Y.P., Splitter G.A. Brucella induces an unfolded protein response via TcpB that supports intracellular replication in macrophages. PLoS Pathog. 2013;9:e1003785. doi: 10.1371/journal.ppat.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comerci D.J., Martinez-Lorenzo M.J., Sieira R., Gorvel J.P., Ugalde R.A. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. 2001;3:159–168. doi: 10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- 22.Delrue R.M., Martinez-Lorenzo M., Lestrate P., Danese I., Bielarz V., Mertens P., De Bolle X., Tibor A., Gorvel J.P., Letesson J.J. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 2001;3:487–497. doi: 10.1046/j.1462-5822.2001.00131.x. [DOI] [PubMed] [Google Scholar]
- 23.Dohmer P.H., Valguarnera E., Czibener C., Ugalde J.E. Identification of a type IV secretion substrate of Brucella abortus that participates in the early stages of intracellular survival. Cell Microbiol. 2014;16:396–410. doi: 10.1111/cmi.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salcedo S.P., Marchesini M.I., Degos C., Terwagne M., Von Bargen K., Lepidi H., Herrmann C.K., Santos Lacerda T.L., Imbert P.R., Pierre P., Alexopoulou L., Letesson J.J., Comerci D.J., Gorvel J.P. BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front Cell Infect Microbiol. 2013;3:28. doi: 10.3389/fcimb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Barsy M., Jamet A., Filopon D., Nicolas C., Laloux G., Rual J.F., Muller A., Twizere J.C., Nkengfac B., Vandenhaute J., Hill D.E., Salcedo S.P., Gorvel J.P., Letesson J.J., De Bolle X. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell Microbiol. 2011;13:1044–1058. doi: 10.1111/j.1462-5822.2011.01601.x. [DOI] [PubMed] [Google Scholar]
- 26.Watarai M., Kim S., Erdenebaatar J., Makino S., Horiuchi M., Shirahata T., Sakaguchi S., Katamine S. Cellular prion protein promotes Brucella infection into macrophages. J Exp Med. 2003;198:5–17. doi: 10.1084/jem.20021980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S., Watarai M., Suzuki H., Makino S., Kodama T., Shirahata T. Lipid raft microdomains mediate class A scavenger receptor-dependent infection of Brucella abortus. Microb Pathog. 2004;37:11–19. doi: 10.1016/j.micpath.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Arellano-Reynoso B., Lapaque N., Salcedo S., Briones G., Ciocchini A.E., Ugalde R., Moreno E., Moriyon I., Gorvel J.P. Cyclic beta-1,2-glucan is a Brucella virulence factor required for intracellular survival. Nat Immunol. 2005;6:618–625. doi: 10.1038/ni1202. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhary A., Ganguly K., Cabantous S., Waldo G.S., Micheva-Viteva S.N., Nag K., Hlavacek W.S., Tung C.S. The Brucella TIR-like protein TcpB interacts with the death domain of MyD88. Biochem Biophys Res Commun. 2012;417:299–304. doi: 10.1016/j.bbrc.2011.11.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta D., Koblansky A., Gaines J., Brown T., West A.P., Zhang D., Nishikawa T., Park S.G., Roop R.M., 2nd, Ghosh S. Subversion of innate immune responses by Brucella through the targeted degradation of the TLR signaling adapter, MAL. J Immunol. 2010;184:956–964. doi: 10.4049/jimmunol.0902008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jong M.F., Starr T., Winter M.G., den Hartigh A.B., Child R., Knodler L.A., van Dijl J.M., Celli J., Tsolis R.M. Sensing of bacterial type IV secretion via the unfolded protein response. MBio. 2013;4 doi: 10.1128/mBio.00418-12. e00418–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viadas C., Rodriguez M.C., Sangari F.J., Gorvel J.P., Garcia-Lobo J.M., Lopez-Goni I. Transcriptome analysis of the Brucella abortus BvrR/BvrS two-component regulatory system. PLoS One. 2010;5:e10216. doi: 10.1371/journal.pone.0010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manterola L., Guzman-Verri C., Chaves-Olarte E., Barquero-Calvo E., de Miguel M.J., Moriyon I., Grillo M.J., Lopez-Goni I., Moreno E. BvrR/BvrS-controlled outer membrane proteins Omp3a and Omp3b are not essential for Brucella abortus virulence. Infect Immun. 2007;75:4867–4874. doi: 10.1128/IAI.00439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ficht T.A. Discovery of Brucella virulence mechanisms using mutational analysis. Vet Microbiol. 2002;90:311–315. doi: 10.1016/s0378-1135(02)00216-x. [DOI] [PubMed] [Google Scholar]
- 35.Kohler S., Michaux-Charachon S., Porte F., Ramuz M., Liautard J.P. What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. 2003;11:215–219. doi: 10.1016/s0966-842x(03)00078-7. [DOI] [PubMed] [Google Scholar]
- 36.Campos M.A., Rosinha G.M., Almeida I.C., Salgueiro X.S., Jarvis B.W., Splitter G.A., Qureshi N., Bruna-Romero O., Gazzinelli R.T., Oliveira S.C. Role of Toll-like receptor 4 in induction of cell-mediated immunity and resistance to Brucella abortus infection in mice. Infect Immun. 2004;72:176–186. doi: 10.1128/IAI.72.1.176-186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong P.C., Tsolis R.M., Ficht T.A. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect Immun. 2000;68:4102–4107. doi: 10.1128/iai.68.7.4102-4107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Callaghan D., Cazevieille C., Allardet-Servent A., Boschiroli M.L., Bourg G., Foulongne V., Frutos P., Kulakov Y., Ramuz M. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 39.Myeni S., Child R., Ng T.W., Kupko J.J., 3rd, Wehrly T.D., Porcella S.F., Knodler L.A., Celli J. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog. 2013;9:e1003556. doi: 10.1371/journal.ppat.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calfon M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 41.Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., Hurt E.M., Petroulakis E., Sonenberg N., Yewdell J.W., Calame K., Glimcher L.H., Staudt L.M. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Qin Q.M., Pei J., Ancona V., Shaw B.D., Ficht T.A., de Figueiredo P. RNAi screen of endoplasmic reticulum-associated host factors reveals a role for IRE1alpha in supporting Brucella replication. PLoS Pathog. 2008;4:e1000110. doi: 10.1371/journal.ppat.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starr T., Child R., Wehrly T.D., Hansen B., Hwang S., Lopez-Otin C., Virgin H.W., Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaplan-Turkoz B., Koelblen T., Felix C., Candusso M.P., O'Callaghan D., Vergunst A.C., Terradot L. Structure of the Toll/interleukin 1 receptor (TIR) domain of the immunosuppressive Brucella effector BtpA/Btp1/TcpB. FEBS Lett. 2013;587:3412–3416. doi: 10.1016/j.febslet.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 45.Radhakrishnan G.K., Yu Q., Harms J.S., Splitter G.A. Brucella TIR domain-containing protein mimics properties of the Toll-like receptor adaptor protein TIRAP. J Biol Chem. 2009;284:9892–9898. doi: 10.1074/jbc.M805458200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kozhenkov S., Sedova M., Dubinina Y., Gupta A., Ray A., Ponomarenko J., Baitaluk M. BiologicalNetworks–tools enabling the integration of multi-scale data for the host-pathogen studies. BMC Syst Biol. 2011;5:7. doi: 10.1186/1752-0509-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y., Zhao Z., Xu H., Shyr Y., Zhang B. Advances in systems biology: computational algorithms and applications. BMC Syst Biol. 2012;6(Suppl 3):S1. doi: 10.1186/1752-0509-6-S3-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez-Cabrero D., Abugessasia I., Maier D., Teschendorff A., Merkenschlager M., Gisel A., Ballestar E., Bongcam-Rudloff E., Conesa A., Tegner J. Data integration in the era of omics: current and future challenges. BMC Syst Biol. 2014;8(Suppl 2):I1. doi: 10.1186/1752-0509-8-S2-I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conesa A., Mortazavi A. The common ground of genomics and systems biology. BMC Syst Biol. 2014;8(Suppl 2):S1. doi: 10.1186/1752-0509-8-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foulongne V., Bourg G., Cazevieille C., Michaux-Charachon S., O'Callaghan D. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect Immun. 2000;68:1297–1303. doi: 10.1128/iai.68.3.1297-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ko J., Splitter G.A. Molecular host-pathogen interaction in brucellosis: current understanding and future approaches to vaccine development for mice and humans. Clin Microbiol Rev. 2003;16:65–78. doi: 10.1128/CMR.16.1.65-78.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DelVecchio V.G., Kapatral V., Redkar R.J., Patra G., Mujer C., Los T., Ivanova N., Anderson I., Bhattacharya A., Lykidis A., Reznik G., Jablonski L., Larsen N., D'Souza M., Bernal A., Mazur M., Goltsman E., Selkov E., Elzer P.H., Hagius S., O'Callaghan D., Letesson J.J., Haselkorn R., Kyrpides N., Overbeek R. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc Natl Acad Sci U S A. 2002;99:443–448. doi: 10.1073/pnas.221575398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He Y. Analyses of Brucella pathogenesis, host immunity, and vaccine targets using systems biology and bioinformatics. Front Cell Infect Microbiol. 2012;2:2. doi: 10.3389/fcimb.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He Y., Sun S., Sha H., Liu Z., Yang L., Xue Z., Chen H., Qi L. Emerging roles for XBP1, a sUPeR transcription factor. Gene Expr. 2010;15:13–25. doi: 10.3727/105221610x12819686555051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rossetti C.A., Galindo C.L., Garner H.R., Adams L.G. Selective amplification of Brucella melitensis mRNA from a mixed host-pathogen total RNA. BMC Res Notes. 2010;3:244. doi: 10.1186/1756-0500-3-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim H.S., Caswell C.C., Foreman R., Roop R.M., 2nd, Crosson S. The Brucella abortus general stress response system regulates chronic mammalian infection and is controlled by phosphorylation and proteolysis. J Biol Chem. 2013;288:13906–13916. doi: 10.1074/jbc.M113.459305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Q., Han W., Sun C., Zhou L., Ma L., Lei L., Yan S., Liu S., Du C., Feng X. Deep sequencing-based expression transcriptional profiling changes during Brucella infection. Microb Pathog. 2012;52:267–277. doi: 10.1016/j.micpath.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Lin Y., Xiang Z., He Y. Brucellosis Ontology (IDOBRU) as an extension of the Infectious Disease Ontology. J Biomed Semantics. 2011;2:9. doi: 10.1186/2041-1480-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang F., Hu S., Liu W., Qiao Z., Gao Y., Bu Z. Deep-sequencing analysis of the mouse transcriptome response to infection with Brucella melitensis strains of differing virulence. PLoS One. 2011;6:e28485. doi: 10.1371/journal.pone.0028485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rajashekara G., Eskra L., Mathison A., Petersen E., Yu Q., Harms J., Splitter G. Brucella: functional genomics and host-pathogen interactions. Anim Health Res Rev. 2006;7:1–11. doi: 10.1017/S146625230700117X. [DOI] [PubMed] [Google Scholar]
- 61.Adams L.G., Khare S., Lawhon S.D., Rossetti C.A., Lewin H.A., Lipton M.S., Turse J.E., Wylie D.C., Bai Y., Drake K.L. Multi-comparative systems biology analysis reveals time-course biosignatures of in vivo bovine pathway responses to B.melitensis, S.enterica Typhimurium and M.avium paratuberculosis. BMC Proc. 2011;5(Suppl 4):S6. doi: 10.1186/1753-6561-5-S4-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He Y., Xiang Z. Bioinformatics analysis of Brucella vaccines and vaccine targets using VIOLIN. Immunome Res. 2010;6(Suppl 1):S5. doi: 10.1186/1745-7580-6-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomez G., Pei J., Mwangi W., Adams L.G., Rice-Ficht A., Ficht T.A. Immunogenic and invasive properties of Brucella melitensis 16M outer membrane protein vaccine candidates identified via a reverse vaccinology approach. PLoS One. 2013;8:e59751. doi: 10.1371/journal.pone.0059751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanna N., Ouahrani-Bettache S., Drake K.L., Adams L.G., Kohler S., Occhialini A. Global Rsh-dependent transcription profile of Brucella suis during stringent response unravels adaptation to nutrient starvation and cross-talk with other stress responses. BMC Genomics. 2013;14:459. doi: 10.1186/1471-2164-14-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams L.G., Khare S., Lawhon S.D., Rossetti C.A., Lewin H.A., Lipton M.S., Turse J.E., Wylie D.C., Bai Y., Drake K.L. Enhancing the role of veterinary vaccines reducing zoonotic diseases of humans: linking systems biology with vaccine development. Vaccine. 2011;29:7197–7206. doi: 10.1016/j.vaccine.2011.05.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomez G., Adams L.G., Rice-Ficht A., Ficht T.A. Host-Brucella interactions and the Brucella genome as tools for subunit antigen discovery and immunization against brucellosis. Front Cell Infect Microbiol. 2013;3:17. doi: 10.3389/fcimb.2013.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiss D.S., Takeda K., Akira S., Zychlinsky A., Moreno E. MyD88, but not toll-like receptors 4 and 2, is required for efficient clearance of Brucella abortus. Infect Immun. 2005;73:5137–5143. doi: 10.1128/IAI.73.8.5137-5143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arenas-Gamboa A.M., Rice-Ficht A.C., Fan Y., Kahl-McDonagh M.M., Ficht T.A. Extended safety and efficacy studies of the attenuated Brucella vaccine candidates 16 M(Delta)vjbR and S19(Delta)vjbR in the immunocompromised IRF-1-/- mouse model. Clin Vaccine Immunol. 2012;19:249–260. doi: 10.1128/CVI.05321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oliveira S.C., Soeurt N., Splitter G. Molecular and cellular interactions between Brucella abortus antigens and host immune responses. Vet Microbiol. 2002;90:417–424. doi: 10.1016/s0378-1135(02)00225-0. [DOI] [PubMed] [Google Scholar]
- 70.Fernandes D.M., Benson R., Baldwin C.L. Lack of a role for natural killer cells in early control of Brucella abortus 2308 infections in mice. Infect Immun. 1995;63:4029–4033. doi: 10.1128/iai.63.10.4029-4033.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Araya L.N., Elzer P.H., Rowe G.E., Enright F.M., Winter A.J. Temporal development of protective cell-mediated and humoral immunity in BALB/c mice infected with Brucella abortus. J Immunol. 1989;143:3330–3337. [PubMed] [Google Scholar]
- 72.Montaraz J.A., Winter A.J. Comparison of living and nonliving vaccines for Brucella abortus in BALB/c mice. Infect Immun. 1986;53:245–251. doi: 10.1128/iai.53.2.245-251.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Montaraz J.A., Winter A.J., Hunter D.M., Sowa B.A., Wu A.M., Adams L.G. Protection against Brucella abortus in mice with O-polysaccharide-specific monoclonal antibodies. Infect Immun. 1986;51:961–963. doi: 10.1128/iai.51.3.961-963.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Philips M., Deyoe B.L., Canning P.C. Protection of mice against Brucella abortus infection by inoculation with monoclonal antibodies recognizing Brucella O-antigen. Am J Vet Res. 1989;50:2158–2161. [PubMed] [Google Scholar]
- 75.Winter A.J., Duncan J.R., Santisteban C.G., Douglas J.T., Adams L.G. Capacity of passively administered antibody to prevent establishment of Brucella abortus infection in mice. Infect Immun. 1989;57:3438–3444. doi: 10.1128/iai.57.11.3438-3444.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baldwin C.L., Goenka R. Host immune responses to the intracellular bacteria Brucella: does the bacteria instruct the host to facilitate chronic infection? Crit Rev Immunol. 2006;26:407–442. doi: 10.1615/critrevimmunol.v26.i5.30. [DOI] [PubMed] [Google Scholar]
- 77.Doyle A.G., Halliday W.J., Barnett C.J., Dunn T.L., Hume D.A. Effect of recombinant human macrophage colony-stimulating factor 1 on immunopathology of experimental brucellosis in mice. Infect Immun. 1992;60:1465–1472. doi: 10.1128/iai.60.4.1465-1472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baldwin C.L., Jiang X., Fernandes D.M. Macrophage control of Brucella abortus: influence of cytokines and iron. Trends Microbiol. 1993;1:99–104. doi: 10.1016/0966-842x(93)90115-8. [DOI] [PubMed] [Google Scholar]
- 79.Zaitseva M.B., Golding H., Betts M., Yamauchi A., Bloom E.T., Butler L.E., Stevan L., Golding B. Human peripheral blood CD4+ and CD8+ T cells express Th1-like cytokine mRNA and proteins following in vitro stimulation with heat-inactivated Brucella abortus. Infect Immun. 1995;63:2720–2728. doi: 10.1128/iai.63.7.2720-2728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Svetic A., Jian Y.C., Lu P., Finkelman F.D., Gause W.C. Brucella abortus induces a novel cytokine gene expression pattern characterized by elevated IL-10 and IFN-gamma in CD4+ T cells. Int Immunol. 1993;5:877–883. doi: 10.1093/intimm/5.8.877. [DOI] [PubMed] [Google Scholar]
- 81.Xavier M.N., Winter M.G., Spees A.M., Nguyen K., Atluri V.L., Silva T.M., Baumler A.J., Muller W., Santos R.L., Tsolis R.M. CD4+ T cell-derived IL-10 promotes Brucella abortus persistence via modulation of macrophage function. PLoS Pathog. 2013;9:e1003454. doi: 10.1371/journal.ppat.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stevens M.G., Olsen S.C., Pugh G.W., Jr., Brees D. Comparison of immune responses and resistance to brucellosis in mice vaccinated with Brucella abortus 19 or RB51. Infect Immun. 1995;63:264–270. doi: 10.1128/iai.63.1.264-270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stevens M.G., Olsen S.C., Pugh G.W., Jr., Palmer M.V. Immune and pathologic responses in mice infected with Brucella abortus 19, RB51, or 2308. Infect Immun. 1994;62:3206–3212. doi: 10.1128/iai.62.8.3206-3212.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grillo M.J., Bosseray N., Blasco J.M. In vitro markers and biological activity in mice of seed lot strains and commercial Brucella melitensis Rev 1 and Brucella abortus B19 vaccines. Biologicals. 2000;28:119–127. doi: 10.1006/biol.2000.0249. [DOI] [PubMed] [Google Scholar]
- 85.Salcedo S.P., Marchesini M.I., Lelouard H., Fugier E., Jolly G., Balor S., Muller A., Lapaque N., Demaria O., Alexopoulou L., Comerci D.J., Ugalde R.A., Pierre P., Gorvel J.P. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 2008;4:e21. doi: 10.1371/journal.ppat.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Colmenero Castillo J.D., Hernandez Marquez S., Requera Iglesias J.M., Cabrera Franquela F., Rius Diaz F., Alonso A. Comparative trial of doxycycline plus streptomycin versus doxycycline plus rifampin for the therapy of human brucellosis. Chemotherapy. 1989;35:146–152. doi: 10.1159/000238662. [DOI] [PubMed] [Google Scholar]
- 87.Villamil Giraldo A.M., Sivanesan D., Carle A., Paschos A., Smith M.A., Plesa M., Coulton J., Baron C. Type IV secretion system core component VirB8 from Brucella binds to the globular domain of VirB5 and to a periplasmic domain of VirB6. Biochemistry. 2012;51:3881–3890. doi: 10.1021/bi300298v. [DOI] [PubMed] [Google Scholar]
- 88.Paschos A., den Hartigh A., Smith M.A., Atluri V.L., Sivanesan D., Tsolis R.M., Baron C. An in vivo high-throughput screening approach targeting the type IV secretion system component VirB8 identified inhibitors of Brucella abortus 2308 proliferation. Infect Immun. 2011;79:1033–1043. doi: 10.1128/IAI.00993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paschos A., Patey G., Sivanesan D., Gao C., Bayliss R., Waksman G., O'Callaghan D., Baron C. Dimerization and interactions of Brucella suis VirB8 with VirB4 and VirB10 are required for its biological activity. Proc Natl Acad Sci U S A. 2006;103:7252–7257. doi: 10.1073/pnas.0600862103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith M.A., Coincon M., Paschos A., Jolicoeur B., Lavallee P., Sygusch J., Baron C. Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem Biol. 2012;19:1041–1048. doi: 10.1016/j.chembiol.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 91.Avila-Calderon E.D., Lopez-Merino A., Sriranganathan N., Boyle S.M., Contreras-Rodriguez A. A history of the development of Brucella vaccines. Biomed Res Int. 2013;2013:743509. doi: 10.1155/2013/743509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Buck J.M. Studies of vaccination during calfhood to prevent bovine infectious abortion. J Agric Res. 1930;41:667–689. [Google Scholar]
- 93.Alton G.G., Elberg S.S., Crouch D. Rev. 1 Brucella melitensis vaccine. The stability of the degree of attenuation. J Comp Pathol. 1967;77:293–300. doi: 10.1016/0021-9975(67)90038-2. [DOI] [PubMed] [Google Scholar]
- 94.Spink W.W., Hall J.W., 3rd, Finstad J., Mallet E. Immunization with viable Brucella organisms. Results of a safety test in humans. Bull World Health Organ. 1962;26:409–419. [PMC free article] [PubMed] [Google Scholar]
- 95.Cotton W.E., Buck J.M., Smith H.E. Efficacy and safety of abortion vaccines prepared from Brucella abortus strains of different degrees of virulence. J Agric Res. 1933;46:291–314. [Google Scholar]
- 96.Schurig G.G., Roop R.M., 2nd, Bagchi T., Boyle S., Buhrman D., Sriranganathan N. Biological properties of RB51; a stable rough strain of Brucella abortus. Vet Microbiol. 1991;28:171–188. doi: 10.1016/0378-1135(91)90091-s. [DOI] [PubMed] [Google Scholar]
- 97.Elberg S.S., Faunce K., Jr. Immunization against Brucella infection. VI. Immunity conferred on goats by a nondependent mutant from a streptomycin-dependent mutant strain of Brucella melitensis. J Bacteriol. 1957;73:211–217. doi: 10.1128/jb.73.2.211-217.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arenas-Gamboa A.M., Rice-Ficht A.C., Kahl-McDonagh M.M., Ficht T.A. Protective efficacy and safety of Brucella melitensis 16MDeltamucR against intraperitoneal and aerosol challenge in BALB/c mice. Infect Immun. 2011;79:3653–3658. doi: 10.1128/IAI.05330-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu Q., Pei J., Turse C., Ficht T.A. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol. 2006;6:102. doi: 10.1186/1471-2180-6-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martinon F., Chen X., Lee A.H., Glimcher L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Celli J., Tsolis R.M. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nat Rev Microbiol. 2015;13:71–82. doi: 10.1038/nrmicro3393. [DOI] [PMC free article] [PubMed] [Google Scholar]