Abstract
In response to nutrient limitation, budding yeast can undergo filamentous growth by differentiating into elongated chains of interconnected cells. Filamentous growth is regulated by signal transduction pathways that oversee the reorganization of cell polarity, changes to the cell cycle, and an increase in cell adhesion that occur in response to nutrient limitation. Each of these changes can be easily measured. Yeast can also grow colonially atop surfaces in a biofilm or mat of connected cells. Filamentous growth and biofilm/mat formation require cooperation among individuals; therefore, studying these responses can shed light on the origin and genetic basis of multicellular behaviors. The assays introduced here can be used to study analogous behaviors in fungal pathogens, which require filamentous growth and biofilm/mat formation for virulence.
INTRODUCTION
Microbial species use diverse strategies to compete for nutrients. Being nonmotile, fungal microorganisms have developed a unique behavior, called filamentous growth, in which cells change their shape and band together in chains or filaments to scavenge for nutrients. Many fungal species can also grow in interconnected mats of cells called biofilms. The budding yeast Saccharomyces cerevisiae shows these behaviors, providing a genetically tractable system to study the pathways that control nutrient-dependent foraging. Studies on filamentous growth have provided insights into how eukaryotic cells differentiate and cooperate with each other, and how genetic pathways control fungal pathogenesis. Fungal pathogens require filamentous growth and biofilm formation for virulence.
Filamentous Growth
In budding yeast, filamentous growth is triggered by nutrient limitation (Cullen and Sprague 2012). In particular, depletion of glucose or fixed nitrogen induces filamentous growth in both haploid and diploid cells (Cullen and Sprague 2002). The balance of the cell’s nutrient levels is critical for commitment to the filamentous growth program: complete removal of nutrients triggers entry into stationary phase (G0) in both haploids and diploids, and in diploids, depletion of both carbon and nitrogen sources induces sporulation (Neiman 2011). The decision of whether or not to undergo filamentous growth, and the coordination of the response itself, is regulated by signal transduction pathways. Among the pathways that regulate filamentous growth are the RAS protein kinase A (PKA) pathway (Gimeno et al. 1992) and a mitogen-activated protein kinase (MAPK) pathway called the filamentous growth pathway (Roberts and Fink 1994). These pathways regulate changes in gene expression and alter the activity of target proteins, leading to the construction of a new cell type.
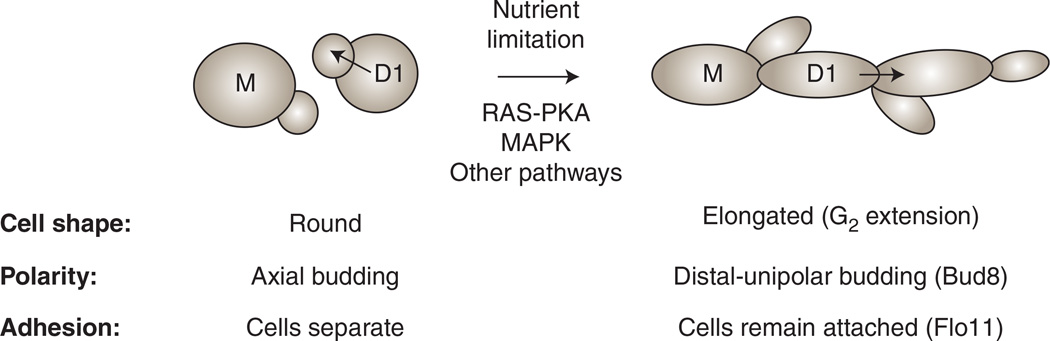
Although filamentous growth is thought to be a complex differentiation response, it includes three easily observable changes in cells (Fig. 1). First, cell polarity is altered. Cell polarity is determined by bud-site-selection proteins, which mark the different poles of the cell (Park and Bi 2007) and direct growth in different directions. Haploid cells bud in an axial pattern, and diploid cells in a bipolar pattern (Chant and Pringle 1995). During filamentous growth, both haploid and diploid cells switch to a distal-unipolar pattern [Fig. 1; (Gimeno et al. 1992; Roberts and Fink 1994; Cullen and Sprague 2002)] by using the distal-pole landmark Bud8 (Harkins et al. 2001). Second, an increase cell length is observed, resulting from a delay in the G2 phase of the cell cycle [Fig. 1 (Kron et al. 1994)]. Finally, cells remain attached to each other (Fig. 1). Unlike yeast-form cells that fully separate after cytokinesis, filamentous cells retain connections between proteins and carbohydrates on the cell wall. One such protein, Flo11, is a mucin-like flocculin and the major cell adhesion molecule that regulates cell–cell adherence (Lambrechts et al. 1996; Lo and Dranginis 1998; Guo et al. 2000; Halme et al. 2004). Together, these changes clearly denote cells undergoing filamentous growth.
FIGURE 1.
Morphological changes that occur during filamentous growth in yeast. Under nutrient-rich conditions (left), yeast-form cells are round in shape and produce daughters that fully separate from their mothers. In haploid cells (shown), daughter cells (D1) bud back toward the mother cell (M) by axial budding (arrow toward left). Under nutrient-limiting conditions (right), cells become elongated and remain attached through Flo11. Daughter cells bud away from the mother cell (arrow toward right) by distal-unipolar budding by using the distal landmark Bud8. Signal transduction pathways (RAS-PKA and MAPK) regulate these changes.
Biofilm/Mat Formation
Budding yeast can also undergo biofilm/mat formation (Reynolds and Fink 2001; Vachova et al. 2011), an ancient microbial response that involves regulating colonial growth at the level of cellular connectivity. In addition to its role in filamentous growth, Flo11 is also required for biofilm/mat formation. Flo11 specifically regulates the complex colony morphology of biofilm/mats (Granek and Magwene 2010), their rim-and-spoke pattern (Reynolds and Fink 2001), and the expansion of mats across surfaces (Reynolds and Fink 2001). Flo11 mediates cellular “sliding” in part because the protein is shed from cells, thereby attenuating cell adhesion and potentially conferring a cellular lubrication property (Karunanithi et al. 2010). Filamentous growth and biofilm formation have distinct regulatory features (Sarode et al. 2011; Ryan et al. 2012), yet are related in that both occur in response to nutrient limitation and require overlapping signaling pathways and target proteins. Under some conditions, filamentous growth and biofilm formation occur in concert, which indicates that these behaviors may represent aspects of a global foraging response (Karunanithi et al. 2012).
TECHNICAL APPROACHES
The associated protocols describe assays to measure and quantitate the changes that occur during filamentous growth and biofilm formation in yeast. The assays are designed to distinguish between phenotypes showed in high- and low-nutrient environments and between wild-type and mutants strains. A key feature of several of these assays is their simplicity. The plate washing assay (see Protocol: The Plate-Washing Assay: A Simple Test for Filamentous Growth in Budding Yeast [Cullen 2015a]) and biofilm/mat assay (see Protocol: Biofilm/Mat Assays for Budding Yeast [Cullen 2015b]) require minimal reagents and measure changes in colony patterns that are visible to the naked eye and interpretable without specialized equipment. In the single-cell invasive growth assay and pseudohyphal growth assay (both presented in Protocol: Evaluating Filamentous Yeast Growth at the Single-Cell Level [Cullen 2015c]), microscopic examination of cells allows quantitation of changes in budding pattern and cell length that occur during filamentous growth.
Three related assays measure the activity of the MAPK pathway that controls filamentous growth (all are presented in Protocol: Evaluating the Activity of the Filamentous Growth MAPK Pathway in Yeast [Cullen 2015d]). First, detecting phosphorylated MAPKs in yeast by western blotting using commercially available antibodies provides a direct measure of MAPK activity. Second, the pectinase assay measures the enzymatic activity of a secreted pectinase that is a target of the filamentous growth pathway. Finally, during filamentous growth, Flo11 (mentioned above) and other mucin-like proteins are shed from cells. Measuring Flo11 shedding provides information about protein levels and biofilm/mat patterning. Another mucin-like protein, Msb2, is the signaling glycoprotein that regulates the filamentous growth pathway (Cullen et al. 2004). Cleavage and release of the extracellular inhibitory domain of Msb2 is required for MAPK activity (Vadaie et al. 2008), which also corresponds to filamentous growth MAPK activity. Secretion profiling of yeast mucin-like proteins provides information about the role of MAPKs in the regulation of filamentous growth.
Finally, the FLO11 gene is regulated by a large and complex promoter where multiple signals converge (Rupp et al. 1999). Measuring changes the expression of FLO 11 (using techniques not described here) can provide a diagnostic readout of changes in the filamentous growth response.
Most yeast strains used in the laboratory do not show filamentous growth because they have acquired mutations as a result of genetic manipulation (Liu et al. 1996). The filamentous (Σ1278b) background is typically used to study filamentous growth (Gimeno et al. 1992). The genome sequence of the Σ1278b background is available (Dowell et al. 2010) as it is a collection of ordered deletion mutants (Ryan et al. 2012). These tools facilitate the genetic analysis of this growth response.
CONCLUSIONS
The current picture of filamentous growth is a complex one, in which multiple pathways and hundreds of targets coordinate a highly integrated response that we are only beginning to understand. Future studies of filamentous growth will aid in the understanding of the genetic basis of cell differentiation, development, and the regulation of multicellularity in eukaryotes. The assays described in the associated protocols are attractive in terms of their simplicity and potential use as teaching tools. Their versatility furthermore allows analysis of filamentous growth and biofilm formation in diverse fungal species including pathogens.
ACKNOWLEDGMENTS
P.J.C. is supported from a U.S. Public Health Service grant (GM098629).
REFERENCES
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ. The plate-washing assay: A simple test for filamentous growth in budding yeast. Cold Spring Harb Protoc. 2015a doi: 10.1101/pdb.prot085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ. Biofilm/mat assays for budding yeast. Cold Spring Harb Protoc. 2015b doi: 10.1101/pdb.prot085076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ. Evaluating filamentous yeast growth at the single-cell level. Cold Spring Harb Protoc. 2015c doi: 10.1101/pdb.prot085084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ. Evaluating the activity of the filamentous growth MAPK pathway in yeast. Cold Spring Harb Protoc. 2015d doi: 10.1101/pdb.prot085092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sabbagh W, Jr, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF., Jr A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF., Jr The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol Biol Cell. 2002;13:2990–3004. doi: 10.1091/mbc.E02-03-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF., Jr The regulation of filamentous growth in yeast. Genetics. 2012;190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, et al. Genotype to phenotype: A complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Granek JA, Magwene PM. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 2010;6:e1000823. doi: 10.1371/journal.pgen.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Harkins HA, Page N, Schenkman LR, De Virgilio C, Shaw S, Bussey H, Pringle JR. Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol Biol Cell. 2001;12:2497–2518. doi: 10.1091/mbc.12.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanithi S, Joshi J, Chavel C, Birkaya B, Grell L, Cullen PJ. Regulation of mat responses by a differentiation MAPK pathway in Saccharomyces cerevisiae. PLoS ONE. 2012;7:e32294. doi: 10.1371/journal.pone.0032294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanithi S, Vadaie N, Chavel CA, Birkaya B, Joshi J, Grell L, Cullen PJ. Shedding of the mucin-like Flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr Biol. 2010;20:1389–1395. doi: 10.1016/j.cub.2010.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts MG, Bauer FF, Marmur J, Pretorius IS. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci. 1996;93:8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Dranginis AM. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189:737–765. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rupp S, Summers E, Lo HJ, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. Embo J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin ZY, Cox MJ, Vizeacoumar F, Cheung D, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337:1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- Sarode N, Miracle B, Peng X, Ryan O, Reynolds TB. Vacuolar protein sorting genes regulate mat formation in Saccharomyces cerevisiae by Flo11p-dependent and -independent mechanisms. Eukaryot Cell. 2011;10:1516–1526. doi: 10.1128/EC.05078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachova L, Stovicek V, Hlavacek O, Chernyavskiy O, Stepanek L, Kubinova L, Palkova Z. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J Cell Biol. 2011;194:679–687. doi: 10.1083/jcb.201103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol. 2008;181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]