Significance
PINK1 protein kinase and PARKIN UB ligase are mutated in inherited forms of Parkinson’s disease and several cancers. Thus, it is of great significance to understand normal functions that could be disrupted in disease. A role for PARKIN and PINK1 is in mediating autophagy of damaged mitochondria (mitophagy) through polyubiquitylation of numerous mitochondrial outer membrane proteins in a reaction that involves phosphorylation of both PARKIN and ubiquitin (UB) by PINK1. The mechanism remains unclear, however, due to challenges in defining individual steps in the pathway. Here, we use a UB replacement system to elucidate steps in the pathway that require PARKIN and/or UB phosphorylation by PINK1 and provide evidence of a PINK1- and UB-driven feed-forward mechanism important for efficient mitochondrial ubiquitylation and mitophagy.
Keywords: PARKIN, ubiquitin, phosphorylation, PINK1, mitochondria
Abstract
The PTEN-induced putative kinase protein 1 (PINK1) and ubiquitin (UB) ligase PARKIN direct damaged mitochondria for mitophagy. PINK1 promotes PARKIN recruitment to the mitochondrial outer membrane (MOM) for ubiquitylation of MOM proteins with canonical and noncanonical UB chains. PINK1 phosphorylates both Ser65 (S65) in the UB-like domain of PARKIN and the conserved Ser in UB itself, but the temporal sequence and relative importance of these events during PARKIN activation and mitochondria quality control remain poorly understood. Using “UBS65A-replacement,” we find that PARKIN phosphorylation and activation, and ubiquitylation of Lys residues on a cohort of MOM proteins, occur similarly irrespective of the ability of the UB-replacement to be phosphorylated on S65. In contrast, polyubiquitin (poly-UB) chain synthesis, PARKIN retention on the MOM, and mitophagy are reduced in UBS65A-replacement cells. Analogous experiments examining roles of individual UB chain linkage types revealed the importance of K6 and K63 chain linkages in mitophagy, but phosphorylation of K63 chains by PINK1 did not enhance binding to candidate mitophagy receptors optineurin (OPTN), sequestosome-1 (p62), and nuclear dot protein 52 (NDP52) in vitro. Parallel reaction monitoring proteomics of total mitochondria revealed the absence of p-S65-UB when PARKIN cannot build UB chains, and <0.16% of the monomeric UB pool underwent S65 phosphorylation upon mitochondrial damage. Combining p-S65-UB and p-S65-PARKIN in vitro showed accelerated transfer of nonphosphorylated UB to PARKIN itself, its substrate mitochondrial Rho GTPase (MIRO), and UB. Our data further define a feed-forward mitochondrial ubiquitylation pathway involving PARKIN activation upon phosphorylation, UB chain synthesis on the MOM, UB chain phosphorylation, and further PARKIN recruitment and enzymatic amplification via binding to phosphorylated UB chains.
The RING-Between-RING ubiquitin (UB) ligase PARKIN and the PTEN-induced putative kinase protein 1 (PINK1), both of which are mutated in Parkinson’s disease, promote ubiquitylation of numerous outer membrane proteins on damaged mitochondria, which facilitates mitophagy (1). When mitochondria are healthy, cytoplasmic PARKIN is thought to be in an unphosphorylated and auto-inhibited state (2–6). Upon mitochondrial damage, PINK1 is stabilized on the mitochondrial outer membrane (MOM) and promotes phosphorylation of both Ser65 (S65) on PARKIN’s UB-like (UBL) domain and the conserved S65 residue in UB itself, which is 62% similar to PARKIN’s UBL (1, 7–14). PARKIN is also retained on the mitochondrial surface and ubiquitylates numerous MOM proteins through the formation of both canonical and noncanonical UB chains. The order of events, as well as the precise roles of UB and PARKIN phosphorylation, is poorly defined, and multiple models have been proposed. All models agree that PARKIN activation reverses auto-inhibition but differ in the mechanism and role of p-S65-UB (8, 11–15). In one model, binding of monomeric p-S65-UB to unphosphorylated PARKIN activates its UB ligase activity in vitro and promotes MOM ubiquitylation in vivo (11, 13, 14). Additionally, overexpression of UBS65E/D mutants as mimetics of p-S65-UB in cells has been reported to activate PARKIN (13–15). However, these studies have differed as to the relative importance of PARKIN and UB phosphorylation on pathway activation. In an alternative model, PINK1 initially phosphorylates a pool of PARKIN on S65 without a requirement for retention of PARKIN on mitochondria, resulting in PARKIN-dependent ubiquitylation of MOM proteins (12, 13, 15). Newly synthesized UB chains are then phosphorylated by PINK1, facilitating recruitment of PARKIN through its p-S65-UB binding ability, resulting in both further PARKIN recruitment and additional MOM ubiquitylation (12). Although PARKIN S65 phosphorylation increases its affinity for p-S65-UB by 20-fold (12), phospho-UB on the MOM could potentially serve to recruit pools of both phosphorylated and unphosphorylated PARKIN.
Dissecting roles of UB and PARKIN phosphorylation must take into account the complexity inherent in this system, where UB (i) is transferred between multiple active sites involving the UB-conjugating enzyme UBCH7 and PARKIN, (ii) is linked to primary sites on MOM proteins, (iii) is phosphorylated by PINK1 to generate forms recognized by PARKIN itself, and (iv) is also recognized by UB receptors that promote mitophagy. PARKIN itself has a UBL domain with many similarities to UB, including being phosphorylated on the conserved S65. This system is made more complex by the fact that tools for studying phosphorylation of UB are limited largely to overexpression of phosphomimetics in vivo (13–15). However, as demonstrated here, neither UB nor PARKIN phosphomimetics faithfully mimic the biochemical activities of phosphorylated PARKIN and UB in vitro, raising questions as to the in vivo roles of PINK1-dependent phosphorylation on PARKIN and/or UB deduced using these approaches.
Here, we used a UB-replacement strategy (16) to deplete all four mRNAs encoding UB in cultured cells while expressing UBS65A or UB mutants lacking individual Lys residues involved in chain assembly, thereby allowing an assessment of the role of UB phosphorylation and individual chain linkage types in PARKIN activation, MOM protein ubiquitylation, PARKIN retention on mitochondria, and mitophagy. Moreover, we developed in vitro pulse–chase and multiple-turnover assays to examine the role of p-S65-UB in individual steps in UB transfer by PARKIN. These studies rationalize many previous observations (10–15, 17) and provide further insight into how phosphorylation of both UB and PARKIN promotes a “feed-forward” mechanism controlling mitochondrial ubiquitylation and mitophagy.
Results and Discussion
S65D/E Does Not Mimic S65 Phosphorylation in UB or PARKIN.
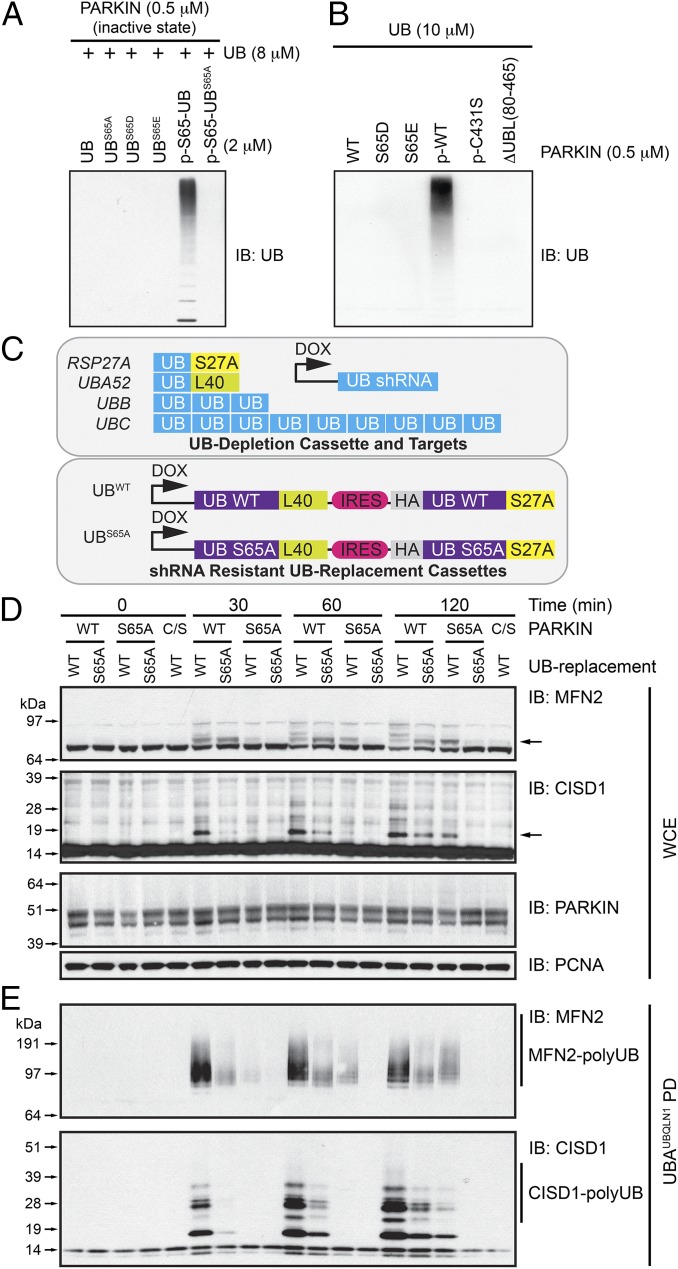
Previous efforts to define roles of phosphorylation of UB S65 in mammalian cells relied on overexpressed UBS65D/E or PARKINS65D/E mutants (13–15). We found that although p-S65-UB robustly activated UB chain synthesis by unphosphorylated PARKIN in the presence of unphosphorylated UB, as reported previously (11–14, 17), UBS65D or UBS65E did not (Fig. 1A and Fig. S1A). Likewise, PARKIN stoichiometrically phosphorylated on S65 (p-WT PARKIN) displayed robust UB chain synthesis activity as described previously (12), whereas PARKINS65D or PARKINS65E did not (Fig. 1B and Fig. S1C). As with p-S65-UB, UBS65D and UBS65E were used poorly for chain synthesis by p-WT PARKIN, as seen previously (12, 17) (Fig. S1B). These data indicate that UB and PARKIN S65D/E mutants do not behave as true phosphomimetics in biochemical assays measuring PARKIN UB chain assembly activity.
Fig. 1.
UB-replacement reveals roles of p-S65 in UB and PARKIN in PINK1-dependent ubiquitylation of outer mitochondrial membrane proteins. (A and B) Effect of phosphomimetics on PARKIN UB chain synthesis. (A) Indicated UB mutants or p-S65-UB was assayed for chain assembly with unphosphorylated PARKIN and UB. (B) Indicated PARKIN proteins, including p-WT PARKIN as a control for activated PARKIN, were assayed for chain synthesis with UB. (C) Schematic of the UB-replacement system used here to replace endogenous UB with UBS65A (16). (D) MFN2 and CISD1 ubiquitylation is reduced in cells wherein UB is replaced by UBS65A. UBWT- and UBS65A-replacement cells expressing WT, S65A, or catalytic Cys-to-Ser PARKIN were depolarized for the indicated times with AO, and whole-cell extracts (WCE) were subjected to immunoblotting (IB) with α-MFN2 and α-CISD1 antibodies. PARKIN and proliferating cell nuclear antigen (PCNA) levels were determined as loading controls. (E) Same as D, except polyubiquitylated proteins were purified using immobilized HALO-UBAUBQNL1. After washing, proteins were subjected to immunoblotting with α-MFN2 and α-CISD1. PD, pull-down.
UB-Replacement Reveals Requirement for PINK1-Dependent UB Phosphorylation in Mitochondrial Polyubiquitylation in Vivo.
To test roles of UB phosphorylation directly in vivo, we used a UB-replacement strategy in U2OS cells (16), wherein all endogenous copies of UB are depleted by doxycycline (DOX)-inducible RNAi while simultaneously expressing shRNA-resistant UBWT and UBS65A fused to the N terminus of ribosomal proteins L40 and S27a from a DOX-responsive promoter (Fig. 1C). Immunoblotting (Fig. S1D) of extracts demonstrated similar levels of UBWT and UBS65A proteins in both monomeric and conjugated forms after DOX induction for 5 d, and UB-absolute quantification (AQUA) proteomics revealed ∼11% endogenous UB remaining in UBS65A-replacement cells (Fig. S1E), comparable to the level of endogenous UB seen previously with other UB mutants (16). UBWT and UBS65A-replacement cells have similar cell proliferation profiles, suggesting that expression of UBS65A in the context of 11% residual WT UB is compatible with many cellular functions (Fig. S1F). Because U2OS cells lack detectable PARKIN and do not support mitochondrial ubiquitylation in response to depolarization, UB-replacement cells were engineered to express low levels of PARKINWT, nonphosphorylatable PARKINS65A, or catalytically defective PARKINC431S. These cells were left untreated or depolarized with antimycin A and oligomycin A (AO) over a 2-h time course and ubiquitylation of two known PARKIN targets, MFN2 and CISD1, examined by immunoblotting of whole-cell extracts (Fig. 1D) or after enrichment of ubiquitylated mitochondrial proteins with immobilized haloalkane dehalogenase (HALO)-tagged–UBAUBQLN1 (Fig. 1E), which preferentially binds all forms of polyubiquitin (poly-UB) chains. In whole-cell extracts, MFN2 monoubiquitylation was prominent at 30 min after depolarization, including in UBS65A-replacement cells. However, over time, this product was converted to more slowly migrating species in UBWT-replacement cells, shown clearly as polyubiquitylated species in the HALO-UBAUBQLN1–enriched samples (Fig. 1 D and E). In contrast, in UBS65A-replacement cells, monoubiquitylated MFN2 is maintained at 2 h, but the extent of MFN2 polyubiquitylation was greatly reduced (Fig. 1 D and E). Similar results were seen with CISD1, although monoubiquitylated forms were kinetically delayed (Fig. 1 D and E). UBWT-replacement cells expressing PARKINS65A or PARKINC431S displayed no or very low activity toward MFN2 and CISD1, as seen previously (12) (Fig. 1 D and E). We infer that UBS65-phosphorylation promotes UB chain synthesis on PARKIN’s substrates.
Correlation Between Defects in PARKIN-Mediated UB Chain Synthesis and Absence of UB Phosphorylation on Depolarized Mitochondria.
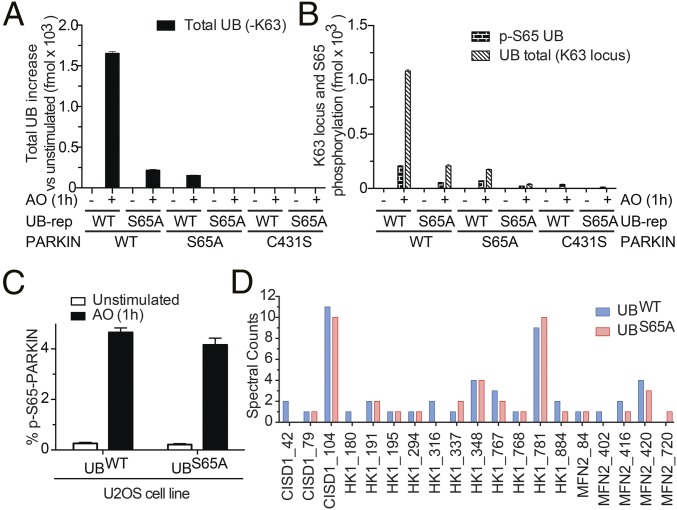
We used UB-AQUA (12) to assess total UB, chain linkage types, and the stoichiometry of UB phosphorylation on UB chains purified from mitochondria (Fig. 2A and Fig. S1G) quantitatively, although we were unable to quantify K63 linkages derived from the inducibly expressed UBS65A mutant in UBS65A-replacement cells due to the replacement of Ser by Ala. As expected (12), depolarization led to an increase in the total abundance of UB associated with depolarized mitochondria in cells expressing PARKINWT (∼1,600 fmol) relative to nondepolarized cells or to cells expressing PARKINC431S (Fig. 2A). K6, K11, K48, and K63 UB chains were found (Fig. S1G), as seen in HeLa cells (12). However, in UBS65A-replacement cells, there was a drastic reduction in the abundance of K6, K11, and K48 chains detected; from 314 fmol with UBWT to 36 fmol with UBS65A, which was comparable to the levels of UB chain formation seen in UBWT-replacement cells expressing PARKINS65A (38 fmol) (Fig. S1G). Thus, UB chain assembly on damaged mitochondria is defective in UBS65A-replacement cells. Consistent with previous results in HeLa cells (12), we found that ∼19% of mitochondrial UB purified using HALO-UBAUBQLN1 from UBWT-replacement cells expressing PARKINWT was phosphorylated on S65 (203 vs. 1,082 fmol), and this species was largely absent in cells expressing PARKINS65A or PARKINC431S (Fig. 2B). In UBS65A-replacement cells, 52 fmol of p-S65-UB was detected, apparently via phosphorylation of residual endogenous UB (226 fmol) (Fig. 2B). Thus, defects in UB chain synthesis in UBS65A-replacement cells correlate with loss of UB phosphorylation on depolarized mitochondria.
Fig. 2.
UB chain assembly on mitochondria, but not primary ubiquitylation or PARKINS65 phosphorylation, is reduced in UBS65A-replacement cells. (A) Cells in which endogenous UB is replaced by phosphorylatable UBS65A display reduced UB ligation to mitochondria in response to depolarization. UB-AQUA–based quantification of total UB on mitochondria from indicated UB-replacement cells 1 h after depolarization. Error bars represent triplicate measurements (±SEM). (B) As in A, but depicting abundance of p-S65-UB and total K63 locus. Error bars represent triplicate measurements (±SEM). (C) Extent of GFP-PARKIN phosphorylation on S65 within its UBL is unchanged in UBS65A-replacement cells. α-GFP immune complexes from the indicated cells were digested with trypsin and subjected to AQUA proteomics using p-S65 PARKIN heavy reference peptides. (D) Mitochondria were isolated from the indicated cells, ubiquitylated proteins were isolated with HALO-UBAUBQLN1 and HALO-UBADSK2, and di-Gly–containing peptides were isolated using α–di-Gly antibodies before analysis by liquid chromatography/tandem MS. The numbers of spectral counts identified for CISD1, HK1, and MFN2 are shown.
Absence of UB Phosphorylation on Mitochondria Without UB Chain Synthesis.
Previous experiments (12) and those experiments presented above used HALO-UBAUBQLN1 for enrichment of poly-UB chains. To rule out a pool of UB (e.g., monomeric UB that preexists on healthy mitochondria) that could serve as a substrate for PINK1 independent of PARKIN-dependent chain synthesis, we used parallel reaction monitoring (PRM) to detect p-S65 within the K63 locus in UB in total purified mitochondria from nondepolarized cells expressing PARKINWT and de-polarized cells expressing either PARKINWT or PARKINC431S. The p-S65-UB was readily detected in depolarized cells expressing PARKINWT, but the abundance of p-S65-UB in cells expressing PARKINC431S was near background levels and unchanged relative to cells without depolarization (Fig. S1I). Thus, detectable mitochondrial UB phosphorylation by PINK1 requires chain synthesis by PARKIN.
Depolarization-Dependent PARKIN S65 Phosphorylation and Activation in UBS65A-Replacement Cells.
PARKIN phosphorylation on S65 and activation as measured by oxy-ester formation with PARKINC431S (3, 12, 18, 19) can occur in the absence of PARKIN-dependent UB-chain assembly on depolarized mitochondria (12). Moreover, S65 phosphorylation in PARKIN activates the ability of C431 to react with UB-vinyl sulfone in vitro (12). We found that GFP-PARKINC431S in either UBWT- or UBS65A-replacement cells formed an oxy-ester with UB with equivalent efficiency upon depolarization (Fig. S1H), and the extent of S65 PARKIN phosphorylation was equivalent with GFP-PARKINWT as measured by AQUA proteomics (Fig. 2C). Thus, PINK1-dependent PARKIN phosphorylation and subsequent charging to form an oxy-ester occur efficiently in UBS65A-replacement cells.
Efficient Primary Ubiquitylation of MOM Proteins in UBS65A-Replacement Cells.
We next tested whether “primary” MOM ubiquitylation (i.e., UB-modified Lys on proteins other than UB itself) occurs in UBS65-replacement cells by purifying di-Gly–containing peptides (20) from mitochondrial UB enriched using HALO-UBAUBQNL1 and HALO-UBADSK2. In response to depolarization, we identified 101 primary di-Gly sites in 40 MOM proteins in UBWT-replacement cells (Fig. S1J and Dataset S1), in agreement with prior reports (20–22). Only 10 di-Gly sites were identified without depolarization, and only five were found in control UBWT-replacement cells expressing PARKINC431S (Dataset S1). At least one primary ubiquitylation site was detected in all but 12 substrates in UBS65A-replacement cells, and those ubiquitylation sites not detected in UBS65A-replacement cells were present in UBWT-replacement cells at only one spectral count (Dataset S1). Moreover, the number of spectral counts for well-characterized substrates was generally comparable (Fig. 2D). Thus, despite a defect in global chain synthesis on mitochondria, PARKIN-dependent primary ubiquitylation of substrates can occur with high efficiency at least on a subset of what may be preferred sites in UBS65A-replacement cells.
Defective PARKIN Mitochondrial Recruitment and Mitophagy in UBS65A-Replacement Cells.
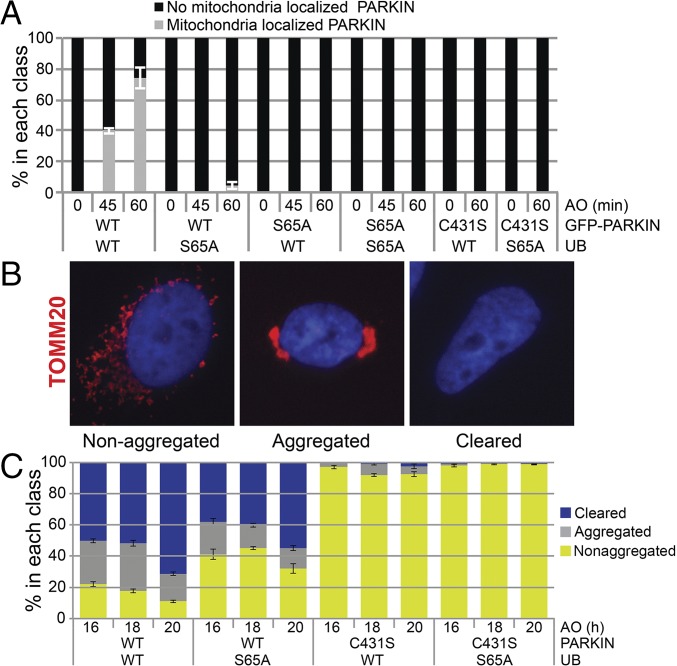
We next examined recruitment of GFP-PARKINWT to damaged mitochondria in UB-replacement cells. Forty percent and 73% of UBWT-replacement cells displayed GFP-PARKINWT localization on mitochondrial 20 kDa outer membrane protein (TOMM20)-positive mitochondria at 45 and 60 min after depolarization, respectively, whereas this figure was reduced to <2% in UBS65A-replacement cells (Fig. 3A and Fig. S2 A and B). As seen previously (12), GFP-PARKINS65A and GFP-PARKINC431S displayed very low to no mitochondrial recruitment (Fig. 3A and Fig. S2B).
Fig. 3.
Impaired PARKIN recruitment and mitophagy in UBS65A-replacement cells. (A) UBS65A-replacement cells display defects in translocation of GFP-PARKIN to mitochondria in response to depolarization. The indicated UB-replacement cells were depolarized with AO, stained for mitochondria with α-TOMM20 antibodies, and imaged for mitochondrial GFP-PARKIN colocalization. The percentage of cells displaying mitochondrial GFP-PARKIN localization was quantified (n > 100 per condition). Error bars represent triplicate measurements (±SD). (B) Classes of depolarization-dependent mitophagy scored after imaging of cells stained with α-TOMM20 20 h after treatment with AO. “Non-aggregated” refers to distributed mitochondria, “Aggregated” refers to perinuclear aggregation of mitochondria, and “Cleared” refers to cells that are devoid of mitochondria. (C) UBS65A-replacement cells display a delay in mitophagy. Quantification of α-TOMM20 staining in UBWT- and UBS65A-replacement cells (n > 100). Error bars represent triplicate measurements (±SEM).
We next examined mitophagy (as measured by immunofluorescence with α-TOMM20) in UB-replacement cells (Fig. 3B) by quantifying the percentages of cells with nonaggregated mitochondria, aggregated mitochondria, and cleared mitochondria (Fig. 3B). UBWT-replacement cells expressing PARKINWT displayed aggregated or cleared mitochondria in 79%, 82%, and 90% of cells at 16 h, 18 h, and 20 h, respectively, whereas the corresponding UBS65A-replacement cells displayed 59%, 57%, and 70% aggregated or cleared mitochondria, respectively (Fig. 3 B and C). As expected, PARKINC431S-expressing cells were defective in mitophagy and displayed >95% nonaggregated mitochondria irrespective of the form of UB replacement. Thus, UBS65 phosphorylation contributes to PARKIN retention on mitochondria, and the efficiency of mitophagy. This mitophagy defect could be greater in the context of a true UBS65A genetic model.
UB Chain Linkage Types Influence Mitophagy.
PARKIN assembles canonical and noncanonical UB chains in vitro and in vivo in response to mitochondrial depolarization (12) (Fig. S1G). We used the UB-replacement system to examine the roles of UB chain linkage types in mitochondrial ubiquitylation and mitophagy (Fig. S3A). Immunoblotting of whole-cell extracts (Fig. S3B) or HALO-UBAUBQLN1-captured UB chains from mitochondria (Fig. S3C) derived from cells individually expressing K6R, K11R, K48R, or K63R (Fig. S3A) showed that the abundance of CISD1 and MFN2 conjugates was largely insensitive to linkage type. Thus, at least for these substrates, no single linkage type is absolutely required for chain building. In contrast, mitophagy is significantly delayed in UBK6R- and UBK63R-replacement cells, with 68% of UBWT cells displaying cleared or aggregated mitochondria but only 46% and 43% of UBK6R- and UBK63R-replacement cells, respectively, displaying cleared or aggregated mitochondria at 20 h after depolarization (Fig. S3D). Involvement of K6 and K63 chains in mitophagy was also reported recently based on overexpression of UB mutants (22).
For K63 UB chains, one obvious potential role would be recognition by autophagy receptors, including optineurin (OPTN) and sequestosome-1 (p62), which have been implicated in promoting mitochondrial clustering and/or mitophagy and shown to bind K63 chains (23–25). Given the proximity of S65 and K63 in UB (17), we asked whether UBS65 phosphorylation in K63 UB chains (with a stoichiometry of 0.52) affects receptor interactions. GST-OPTN associated efficiently with K63 chains but not with K48 or monoubiquitin (Fig. S3E), and this association was reduced upon chain phosphorylation (Fig. S3 E, H, and I). Analogous experiments with nuclear dot protein 52 (NDP52) and p62 gave similar results (Fig. S3 F and G). These data suggest that mechanisms other than direct recognition of phosphorylated K63 UB chains by these receptors may promote mitophagy.
p-S65-UB Promotes UB Transfer in Single-Turnover Assays.
p-S65-UB binds to unphosphorylated PARKIN and promotes UB chain synthesis in vitro (12, 13, 17). To test whether other known phosphorylated forms of UB (p-S20 and p-S57) are also capable of activating UB chain synthesis by PARKIN, we synthesized these three forms of p-UB using an Escherichia coli phosphoserine expression system (26). Only p-S65-UB activated unphosphorylated PARKIN (Fig. S4 A and B), and this activation required the classical UB “hydrophobic patch,” because p-S65-UBI44A failed to activate (Fig. S4A). In contrast, p-S65-UB inhibits the ability of stoichiometrically phosphorylated p-WT PARKIN to build chains with unphosphorylated UB, potentially due to competition with UB in the conjugation cascade. Indeed, p-S65-UB is used very poorly by p-WT PARKIN to build chains when present as the only source of conjugatable UB (12, 17) (Fig. S1B).
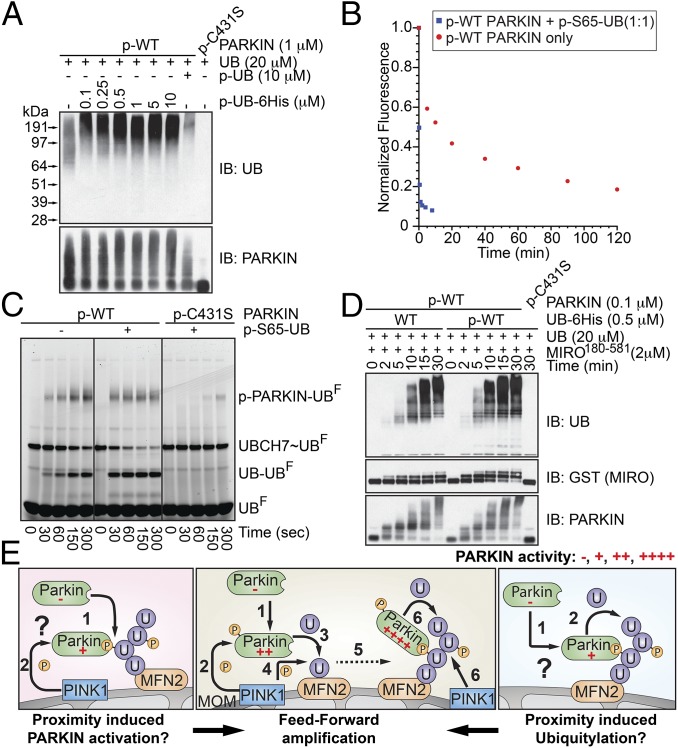
To rationalize the various findings with p-S65-UB (10–14, 17) (Figs. S1 A and B and S4 A and B), we considered that C-terminally blocked, nonconjugatable p-S65-UB could mimic forms of p-S65-UB linked to mitochondrial proteins, whereas p-S65-UB with a free C terminus could engage in the catalytic cycle with inhibitory consequences. Indeed, we found that nonconjugatable p-S65-UB-6His not only fails to inhibit, but actually further stimulates, UB chain synthesis by p-WT PARKIN in vitro (Fig. 4A). Time-course analysis of p-WT PARKIN activation in vitro using UB-AQUA demonstrated a 2,473-fold increase in chain assembly rate relative to unphosphorylated PARKIN, as seen previously (12), and adding p-S65-UB-6His increased this increase further to 4,398-fold (Fig. S4C). Thus, binding of p-WT PARKIN to nonconjugatable p-S65-UB may serve to enhance catalysis further, in addition to having a potential tethering function.
Fig. 4.
Additional element in PINK1-dependent feed-forward signaling: p-S65-UB activation of UB transfer from PARKIN to substrates. (A) Nonconjugatable form of p-S65-UB activates UB chain synthesis by p-WT PARKIN and is not inhibitory in vitro. p-WT or p-C431S PARKIN was incubated with either p-S65-UB or the indicated amount of p-S65-UB-6His, and mixtures were supplemented with ATP, E1, UBCH7, and UB to allow for chain assembly. After 30 min, reaction mixtures were subjected to immunoblotting with α-UB and α-PARKIN antibodies. (B) p-S65-UB greatly stimulates transfer of Lys-free UB from UBCH7 to PARKIN in single-turnover assays. UBCH7 was charged with E1, UBF, and ATP, and purified by gel filtration. UBCH7∼UBF was mixed with p-WT PARKIN in the presence or absence of p-S65-UB, and reactions were quenched at the indicated times postmixing. Reaction mixtures were separated by SDS/PAGE, and UBF was detected (Fig. S4D). Loss of UBCH7∼UBF was plotted. (C) As in Fig. S4E, but chased in the presence of UB (300 μM) to allow di-UB synthesis to be monitored. (D) Nonconjugatable p-S65-UB promotes PARKIN-dependent transfer of UB to MIRO in multiple-turnover assays. p-WT or p-C431S PARKIN was incubated with UB and GST-MIRO180–581 in the presence of ATP, E1, and UBCH7, and reactions were quenched and subjected to SDS/PAGE and immunoblotting with α-UB, α-GST, or α-PARKIN at the indicated times. (E) Schematic of a unifying feed-forward model for the PINK1-PARKIN pathway. (Middle) Mitochondrial damage leads to PARKIN recruitment to mitochondria (1) and its PINK1-dependent phosphorylation (2), resulting in the activation of its UB ligase activity. Because activated PARKIN promotes primary substrate ubiquitylation (3), this process increases the density of UB on mitochondria and provides a substrate for phosphorylation by PINK1 (4). We hypothesize that maximal UB amplification involves recruitment of phospho-PARKIN to phospho-UB chains through a direct interaction, and maximal chain synthesis reflects the fact that phospho-PARKIN has significantly higher specific activity than unphosphorylated PARKIN bound to phospho-UB. (Right) Newly modified mitochondrial proteins with phospho-UB may possibly also recruit nonphosphorylated PARKIN (1) to promote chain synthesis or ubiquitylation of MOM proteins (2), albeit with rates lower than the rates seen with phospho-PARKIN. (Left) Moreover, binding of unphosphorylated PARKIN to phospho-UB (1) may facilitate PARKIN access to PINK1, providing further amplification. A combination of these mechanisms may occur. P, phosphorylation; U, ubiquitin.
To define functions of p-S65-UB further, we developed pulse–chase assays that monitor transfer of fluorescently labeled Lys-free UB (UBF) from UBCH7∼UBF to p-WT PARKIN itself (auto-ubiquitylation) or to either free unlabeled UB to form di-UB or the known mitochondrial PARKIN substrate mitochondrial Rho GTPase (MIRO) tagged with GST. Adding stoichiometric p-S65-UB to p-WT PARKIN greatly stimulated transfer from UBCH7∼UBF to PARKIN (Fig. 4B and Fig. S4D). Fifty percent of UBCH7∼UBF is discharged within 10 s in the presence of p-WT PARKIN and p-S65-UB, whereas this process takes 50 min in the absence of p-S65-UB (Fig. 4B and Fig. S4D), suggesting that p-S65-UB could stimulate this step potentially by more than two orders of magnitude. This stimulation required PARKIN’s active site Cys (Fig. S4E) and the canonical RING binding residue (F63) in UBCH7 (Fig. S4F). Very high concentrations of p-WT PARKIN were required to stimulate UBCH7∼UB discharge at a rate similar to the stoichiometric pWT-PARKIN⋅p-S65-UB complex (Fig. S4G). Rate enhancements for transfer from UBCH7∼UBF to UB (di-UB synthesis) and GST-MIRO were also seen in pulse–chase and multiple-turnover assays (Fig. 4 C and D and Fig. S4H). Probing the mechanism further, we found that unphosphorylated PARKINC431S failed to assemble a stable complex with UBCH7C86K-UBiso as assessed by gel filtration (Fig. S4I). In contrast, addition of p-S65-UB led to loss of free UBCH7C86K-UBiso consistent with the formation of a stable PARKINC431S⋅UBCH7C86K-UBiso⋅p-S65-UB complex (Fig. S4I). Taken together, these data suggest that forms of p-S65-UB that cannot be used in chain assembly reactions, such as poly-p-S65-UB chains on mitochondria, are capable of further activating p-WT PARKIN beyond the activity it possesses by direct PINK1 phosphorylation, and that this process may occur, in part, by facilitating association with charged UBCH7.
Implications of UBS65 Phosphorylation for PARKIN Function.
These studies provide further insight into a feed-forward mechanism underlying PARKIN activation by PINK1 (12) (Fig. 4E). Replacement of UB with UBS65A has major effects on the assembly of poly-UB chains on damaged mitochondria, recruitment of PARKIN to mitochondria, and the efficiency of mitophagy. In contrast, PARKIN phosphorylation on S65 by PINK1, PARKIN activation, and primary ubiquitylation of MOM proteins all occur largely on schedule in UBS65A-replacement cells, which are defective in accumulation of p-S65-UB on the MOM. Although we cannot rule out a role for phosphorylation of residual UB in UBS65A-replacement cells, these results indicate a very profound role for UB phosphorylation in late steps of the pathway and, in particular, in the synthesis and subsequent phosphorylation of UB chains on mitochondria that appear to be central to the amplification mechanism (Fig. 4E).
UB phosphomimetics have been used to address the mechanism of PARKIN activation, but we find that S65D/E mutants in UB and PARKIN poorly recapitulate PARKIN activation in vitro, and thus may be more appropriate for modeling loss of function upon phosphorylation rather than activation. Using PRM proteomics of total mitochondria, we confirmed our previous finding (12) that phosphorylation of UB on mitochondria requires depolarization-dependent chain synthesis by PARKIN (Fig. S1I). This finding, together with the fact that PARKIN recruitment is defective in UBS65A-replacement cells, indicates that bulk recruitment and chain elongation occur downstream of initial PARKIN activation and primary ubiquitylation events (Fig. 4E, Middle). Although our data suggest that initial PARKINS65 phosphorylation occurs independent of MOM UBS65 phosphorylation, it is possible that an important aspect of the feed-forward mechanism is recruitment of a pool of unphosphorylated PARKIN to p-S65-UB on the MOM. This recruitment could serve two purposes (Fig. 4E, Left and Right). First, such recruitment could enhance MOM ubiquitylation through direct activation of PARKIN UB ligase activity, albeit with a lower specific activity than phosphorylated PARKIN (12). Second, such recruitment could enhance phosphorylation of PARKINS65 by increasing the proximity of PARKIN and PINK1.
An alternative model involves phosphorylation of free UB by PINK1, which could then bind and activate PARKIN (11, 13, 14). We examined the levels of monomeric p-S65-UB in PINK1+/+ and PINK1−/− HeLa cells expressing PARKINWT or PARKINC431S with or without depolarization. We found a 0.163% increase in p-S65-UB in PARKINWT cells after depolarization from its resting state of 0.189% relative to the total monomeric pool of UB, although we cannot rule out the possibility that a portion of this increase derives from damaged MOM UB chains, because a smaller increase (0.053%) was observed in PARKINC431S (Fig. S4J). However, the finding that PARKINS65A is not efficiently recruited to the MOM and cannot mount an efficient feed-forward response (12) (Figs. 1 D and E and 3A) indicates the absence of a prominent role for monomeric p-S65-UB in PARKIN activation, given that unphosphorylated PARKINS65A is still activated by p-S65-UB in vitro (12).
Signal amplification also may be manifested by binding of p-S65-UB chains to phosphorylated PARKIN, as indicated by increased rates of UB transfer from p-WT PARKIN to substrates and itself in single-turnover and multiple-turnover assays upon binding to nonconjugatable forms of p-S65-UB (Fig. 4E). We speculate that the density of primary substrates, and conjugated UB molecules used as acceptors for chain synthesis, may have distinct properties that are reflected in the single- and multiple-turnover assays reported here. Phosphorylation of PARKIN alone promotes opening of its active site for reaction with UB-vinyl sulfone (12), but both PARKIN phosphorylation and binding to p-S65-UB greatly enhance association with charged UBCH7 (Fig. S4I). The biochemical basis for these activation steps remains to be determined.
These studies extend our understanding of a feed-forward mechanism (12) wherein PINK1-dependent phosphorylation of PARKIN promotes initial isopeptide bond formation between UB and Lys residues on MOM proteins and newly conjugated UB is then phosphorylated by PINK1 (Fig. 4E). These events are hypothesized to promote PARKIN retention on damaged mitochondria as well as additional chain synthesis through both PARKIN localization and increased ubiquitylation activity. Amplification of UB density on mitochondria may promote mitophagy. Further work is necessary to determine whether primary substrate specificity is a component of the feed-forward mechanism. A key aspect of this model is supported by the finding that optimal binding of PARKIN to UB chains occurs when both PARKIN and UB are phosphorylated on S65 (Kd of ∼20 nM) (12), although the affinity of unphosphorylated PARKIN for p-S65-UB (∼370 nM) may also be sufficient to support recruitment as described above (12). Additional studies are required to understand mechanisms by which UB chains and phosphorylation ultimately mediate mitophagy. Although the autophagy receptors OPTN and p62 bind K63-linked UB chains and are thought to direct mitochondrial aggregation and clearance (23, 25), we find that substoichiometric phosphorylation of K63 chains reduces their in vitro binding to OPTN, p62, and NDP52 (Fig. S3 E–G). We speculate that bias against phosphorylation of K63 chain linkages relative to other linkages in vivo (12) may ensure available unphosphorylated K63 linked chains as effective binding sites for autophagy receptors for recognition of damaged mitochondria. Taken together, our data provide a framework for understanding how positive effects of UB and PARKIN phosphorylation manifested through activation of ubiquitylation, combined with negative effects that either prevent unwanted interactions or indirectly promote specificity by masking alternative interaction surfaces, may provide multiple mechanisms that amplify feed-forward signaling of mitophagy.
Materials and Methods
Full methods are provided in SI Materials and Methods. UB-replacement cells were produced as described (16). Mitochondrial purification and UB-AQUA proteomics were performed as described (12). Mitophagy in U2OS cells was performed 16–20 h after depolarization with antimycin A and OA by following α-TOMM20 staining by immunofluorescence. GFP-PARKIN translocation to mitochondria was determined by immunofluorescence after staining of depolarized cells with α-TOMM20 antibodies. Ubiquitylation assays were performed using multiple-turnover assays with the indicated concentrations of PARKIN, UB, p-S65-UB, and UBCH7. Single-turnover assays were performed with UBCH7∼UBF, wherein UBF was tagged with fluorescein and all Lys residues were preplaced with Arg. Reactions were monitored by gel electrophoresis followed by fluorescence detection.
Supplementary Material
Acknowledgments
We thank James Chen (University of Texas Southwestern Medical Center) for the UB-replacement system, Sean Beausoleil (Cell Signaling Technologies) for AQUA peptides, Craig Braun and Steve Gygi (Harvard Medical School) for assistance with MS, and the Nikon Imaging Center at Harvard Medical School for assistance with microscopy. This work was supported by NIH Grant R37NS083524 (to J.W.H.); the American Lebanese Syrian Associated Charities and NIH Grants R37GM069530, R01GM077053, and P30CA021765 (to B.A.S.); and NIH Grant K01 DK098285 (to J.A.P.). B.A.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506593112/-/DCSupplemental.
References
- 1.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85(2):257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32(15):2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riley BE, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340(6139):1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 5.Chaugule VK, et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30(14):2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spratt DE, et al. A molecular explanation for the recessive nature of parkin-linked Parkinson’s disease. Nat Commun. 2013;4:1983. doi: 10.1038/ncomms2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondapalli C, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2(5):120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiba-Fukushima K, et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazlauskaite A, et al. Phosphorylation of Parkin at Serine65 is essential for activation: Elaboration of a Miro1 substrate-based assay of Parkin E3 ligase activity. Open Biol. 2014;4:130213. doi: 10.1098/rsob.130213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazlauskaite A, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460(1):127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ordureau A, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56(3):360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205(2):143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 15.Shiba-Fukushima K, et al. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet. 2014;10(12):e1004861. doi: 10.1371/journal.pgen.1004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36(2):302–314. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wauer T, et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34(3):307–325. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarou M, et al. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200(2):163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng X, Hunter T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 2013;23(7):886–897. doi: 10.1038/cr.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarraf SA, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496(7445):372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bingol B, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510(7505):370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham CN, et al. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat Cell Biol. 2015;17(2):160–169. doi: 10.1038/ncb3097. [DOI] [PubMed] [Google Scholar]
- 23.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6(8):1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Wijk SJ, et al. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol Cell. 2012;47(5):797–809. doi: 10.1016/j.molcel.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci USA. 2014;111(42):E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aerni HR, Shifman MA, Rogulina S, O’Donoghue P, Rinehart J. Revealing the amino acid composition of proteins within an expanded genetic code. Nucleic Acids Res. 2015;43(2):e8. doi: 10.1093/nar/gku1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.