Significance
A comprehensive view of protein folding is crucial for understanding how misfolding can cause neurodegenerative diseases and cancer. When using physical or chemical perturbations, NMR spectroscopy is a powerful tool to reveal a shift in the native conformation toward local intermediates that act as seeds for misfolding. Using NMR, we show that the ensemble of dry and wet molten-globule intermediates populated by high pressure is different from that found when urea is used. The dissimilar actions of urea and pressure can be summarized by their “pull” and “push” effects, respectively. By combining NMR and small-angle X-ray scattering (SAXS), we demonstrate the action of urea at the initial stages of unfolding and the dominance of a direct interaction mechanism for urea-induced protein denaturation.
Keywords: protein folding, hydrostatic pressure, urea, NMR, SAXS
Abstract
High pressure (HP) or urea is commonly used to disturb folding species. Pressure favors the reversible unfolding of proteins by causing changes in the volumetric properties of the protein–solvent system. However, no mechanistic model has fully elucidated the effects of urea on structure unfolding, even though protein–urea interactions are considered to be crucial. Here, we provide NMR spectroscopy and 3D reconstructions from X-ray scattering to develop the “push-and-pull” hypothesis, which helps to explain the initial mechanism of chemical unfolding in light of the physical events triggered by HP. In studying MpNep2 from Moniliophthora perniciosa, we tracked two cooperative units using HP-NMR as MpNep2 moved uphill in the energy landscape; this process contrasts with the overall structural unfolding that occurs upon reaching a threshold concentration of urea. At subdenaturing concentrations of urea, we were able to trap a state in which urea is preferentially bound to the protein (as determined by NMR intensities and chemical shifts); this state is still folded and not additionally exposed to solvent [fluorescence and small-angle X-ray scattering (SAXS)]. This state has a higher susceptibility to pressure denaturation (lower p1/2 and larger ΔVu); thus, urea and HP share concomitant effects of urea binding and pulling and water-inducing pushing, respectively. These observations explain the differences between the molecular mechanisms that control the physical and chemical unfolding of proteins, thus opening up new possibilities for the study of protein folding and providing an interpretation of the nature of cooperativity in the folding and unfolding processes.
It is known that proteins are far from equilibrium during folding reactions, and they undergo a wide range of conformational states to reach the global folding minimum. Various physical and chemical strategies, such as the use of high temperature, high pressure, protonation, altered ionic strength, and harsh denaturants, are commonly used to disturb folding species to promote the formation of rarely observed folding intermediates. From the thermodynamic point of view, any perturbing agent affecting protein folding is controlled by Le Chatelier’s principle. For instance, increasing the concentration of urea shifts the folding equilibrium toward the unfolded state because of the increased preferential binding of urea to this state. In the case of pressure, the smaller volume of the unfolded state is favored at high pressure because it only affects the volumetric properties of the molecule.
The energy landscape theory of folding assumes that protein folding is the progressive organization of an ensemble of partially folded structures through which the protein passes on its way to a native conformation (1–3). Accordingly, proteins have a rugged funnel-like landscape that is biased toward their native structure due to evolution (3). Obtaining structural and dynamic information on multiple-stage protein intermediates that follow the folding trail may increase our knowledge of protein misfolding, which is associated with amyloidosis, prion formation, and the occurrence of several diseases, including Parkinson’s disease, Huntington’s disease, spongiform encephalopathy, and, more recently, cancer (4–6). In these diseases, proteins tend to traverse the “wrong side of the funnel” (7, 8).
The effects of pressure on proteins were discovered in early experiments with egg albumin (9). Currently, pressure is extensively used in various biological and biotechnological applications (10–13) and is one of the most promising variables enabling the structural analysis of these protein substates. The application of pressure to a protein forces the water shell into the protein, thus shifting the equilibrium of the system from the native state to an intermediate or to the unfolded state. Pressure appears to favor water infiltration into the protein and the disassembly of protein cavities (14), leading to increased hydration and decreased partial molar volume. Unlike high temperatures, which cause systematic changes in the total energy and volume of the molecule, high pressure affects only the volumetric properties of the molecule. However, under physiological conditions, water solvation dictates protein folding (15).
High pressure can be coupled with various spectroscopic techniques, including methods based on the intrinsic fluorescence of Trp residues (16), Fourier-transform infrared spectroscopy (FTIR) (17), NMR (13, 18, 19), small-angle X-ray scattering (SAXS) (20, 21), microsecond pressure-jump coupled to fluorescence lifetime (22), and, more recently, circular dichroism (CD) (23). To better understand local changes, high-pressure NMR (HP-NMR) is adopted as the most informative approach (18, 19). SAXS is another useful tool for assessing changes in protein folding and the size and shape of macromolecules in solution (24–26).
Most denaturants affect protein folding through their binding properties. Urea is one of the most commonly used chemical denaturants for the study of protein folding and thermodynamics. Two main theories exist to explain why urea induces protein denaturation (27, 28). The first theory hypothesizes an indirect mechanism by which urea alters the water structure and leads to hydrophobic group solvation. The second view is based on the direct interaction of urea with the protein. Most studies are based on model compounds and theoretical and modeling approaches, such as molecular dynamics (29–35).
The binding of urea to proteins is based on the ability of urea to displace water molecules from the first solvation shell (29), thus increasing the system entropy and weakening the hydrophobic effect (36). Pioneer calorimetric studies on protein–urea interactions were developed by Makhatadze and Privalov (37). Urea is also believed to form hydrogen bonds with the amide unit of peptide bonds, as shown by calorimetric measurements of cyclic dipeptides (36) and H/D exchange by 1D NMR of an alanine-based model compound (38). A two-stage kinetic mechanism for the action of urea has been proposed based on extensive (microsecond) molecular-dynamics simulations (29). However, at equilibrium conditions, experimental insights into the action of urea at the initial stages of denaturation are lacking. A long-standing controversy in the literature concerns the contribution of nonpolar groups, peptide backbone interactions, or both as the driving force behind urea-induced protein denaturation (32, 33, 39, 40). Experimental efforts were mostly based on measurements of transfer free energies (41, 42) of amino acid side chains, peptide backbones, or random-coil polypeptide chains, in which packing defects and the overall 3D architecture of a native polypeptide chain were neglected.
Here, we studied Moniliophthora perniciosa necrosis- and ethylene-inducing protein 2 (MpNep2). We systematically studied protein folding in response to chemical (urea), physical (pressure), or both perturbations using HP-NMR spectroscopy and 3D low-resolution shape reconstructions from SAXS data to better understand how each of the two disturbing agents (pressure and urea) affect protein folding and to examine how urea denatures proteins. The use of these synergistic techniques (HP-NMR and SAXS) provides reliable structural information on local/global intermediates within protein energy landscapes. We provide evidence of a conformational state in which urea binds to the protein without promoting full denaturation by populating a dry molten globule (DMG). This state is more sensitive to pressure (lower p1/2) due to the presence of a pulling force and an increased volume change.
Results
Assessing the Effects of Pressure and Temperature on Protein Unfolding Based on Trp Fluorescence and NMR Spectroscopy.
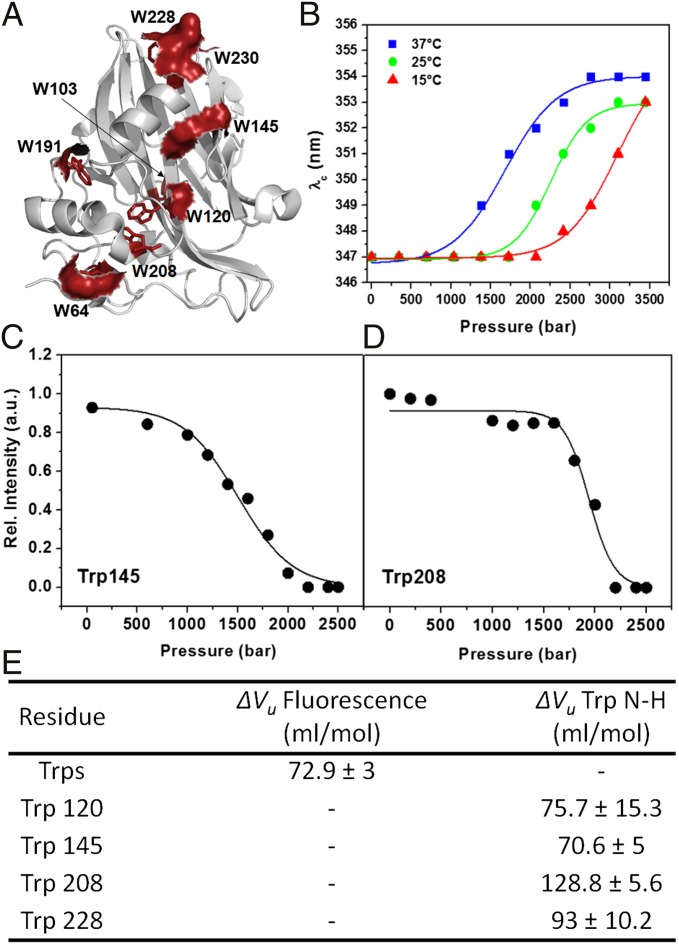
Steady-state fluorescence spectroscopy is commonly used to probe conformational transitions in proteins as a function of the perturbing agent. MpNep2 contains multiple Trp residues (Fig. 1A), thus enabling the observation of its global conformational changes. A surface representation of the eight Trp residues in the MpNep2 crystal structure reveals that only two residues (Trp103 and Trp208) are completely buried in the structure, which is consistent with its center of spectral mass at ∼28,850 cm−1. We used various temperatures and increasing pressures to observe complete unfolding of this protein, which occurred at 37 °C and 2.5 kbar, yielding a center of spectral mass at ∼28,300 cm−1, which is close to that of free tryptophan in solution. Titrations at 15 and 25 °C were not sufficient to shift the equilibrium to the unfolded state, indicating that transient high-energy substates were formed (Fig. 1B).
Fig. 1.
(A) A schematic representation of the secondary structural elements of MpNep2 (gray). Trp side chains are highlighted as red sticks, and the corresponding solvent-accessible surfaces of these residues are shown. (B) Plots of the emission wavelength (λc) obtained from Trp spectra for pressures ranging from 0.001 to 3.5 kbar at 15, 25, and 37 °C. (C and D) Cross-peak relative intensity profile vs. pressure obtained from 1H–15N HSQC experiments at 37 °C for H–N Trp145 and 208 resonances. (E) ΔVu values obtained by fluorescence spectroscopy and NMR.
ΔVu values for H–N of Trp residues were obtained from fits of the 1H–15N heteronuclear single-quantum coherence (HSQC) peak intensity vs. pressure increase (Fig. 1 C and D) and revealed different contributions among them. The transitions were analyzed as two-state processes. Trp208 showed the biggest ΔVu contribution (128.8 ± 5.6 mL/mol) compared with the other Trp residues (Fig. 1E) and to the overall ΔVu obtained from Trp fluorescence (72.9 ± 3 mL/mol), as expected for a buried residue upon unfolding.
NMR Spectroscopy Revealing Site-Specific Changes Caused by High Pressure and the Addition of Urea.
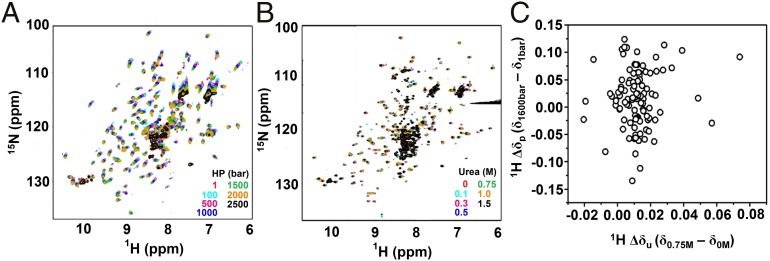
Fig. 2 A and B show a set of merged 1H–15N HSQC spectra obtained at 25 °C for pressures ranging from 0.001 to 2.5 kbar and urea concentrations ranging from 0 to 1.5 M, respectively. Based on a qualitative evaluation of the HSQC NMR spectra, we were able to study local conformational diversity (43) based on two straightforward parameters: (i) the measurement of systematic chemical shifts and (ii) line broadening of the corresponding 1H–15N cross-peaks. We observed systematic changes in chemical shifts for all 1H–15N cross-peaks with increasing pressure up to 2 kbar, without losing spectral dispersion (Fig. 2A). In contrast to this observation, less chemical shift contribution was observed upon urea titration (Fig. 2B). At 25 °C and 2.5 kbar, most cross-peaks disappeared, strongly suggesting the occurrence of a greater conformational change consistent with unfolding. However, a residual population of low-intensity peaks [37 out of 144 analyzed peaks (25.7%)] remained visible in this condition. At 15 °C and 2.5 kbar, these residual peaks represented 45 of the 112 analyzed peaks (40.1%), but at 37 °C and 2 kbar, these cross-peaks were no longer observed, a condition under which the unfolded species was obtained. As shown in Fig. 2A, a cluster of peaks is commonly observed in the central part of the spectrum under these conditions. At 1.5 M urea (Fig. 2B), we observed the following: (i) some cross-peaks were no longer visible, (ii) most of the cross-peaks were drastically reduced in intensity, and (iii) some new cross-peaks were present in the spectrum. At higher urea concentrations (>2 M), the protein was totally unfolded, as reflected by intermediate exchanging rate acquisition (i.e., NMR peak disappearance).
Fig. 2.
Pressure and urea perturb MpNep2 through different mechanisms. Representative 1H–15N HSQC spectra of MpNep2 for various pressures (A) and urea concentrations (B) at 25 °C. (C) Correlation plot of 1H Δδ obtained by high pressure (δ1,600bar − δ1bar) and urea (δ0.75M − δ0M).
We also recorded 1H–15N HSQC spectra at 5 and 10 °C with increasing pressure up to 2.5 kbar to trap and stabilize an increased residual population of the low-intensity cross-peaks previously identified at higher temperatures (15 and 25 °C at 2.5 kbar). Under these conditions, we observed a dispersion profile for the HSQC spectrum at 1 bar that differed from the results observed at higher temperatures (SI Appendix, Fig. S1). Under pressures higher than 1 kbar, we observed protein unfolding at both temperatures. These observations indicate that highly energetic intermediates of MpNep2 may be trapped and stabilized at temperatures lower than 15 °C. The observed spectral changes were totally reversible under all conditions, as shown by decompression spectra (SI Appendix, Fig. S1). In particular, at 5 °C, one can clearly deduce that cold denaturation assisted by pressure is present (SI Appendix, Fig. S1).
The systematic changes in chemical shift resulting from high-pressure treatment are shown as 1H and 15N chemical shifts (ΔδH and ΔδN) from the values observed at 1 bar for each temperature and are plotted as a function of the residue number (SI Appendix, Fig. S2). Because the changes in peak position were systematic and almost linear for pressure increases up to 2 kbar, sequential assignments were straightforward, made by following and extrapolating the earliest assignments (at room temperature and 1 bar) (44). We obtained an average 1H chemical shift of 0.03 ± 0.06 ppm over all residues by comparing the values obtained at 2 kbar to those obtained at 1 bar for 15 and 25 °C. For the average 15N value under the same conditions, we obtained 0.47 ± 0.33 ppm at 15 °C and 0.46 ± 0.36 ppm at 25 °C. The higher sensitivity of the ΔδH values to pressure is directly associated with the shortening of hydrogen bonds between 1H nuclei and the solvent shell and, to a lesser extent, with the shortening of those hydrogen bonds participating in secondary and tertiary contacts. Based on our Δδ analysis at 25 °C, we detected 24 1H nuclei that underwent chemical shifts at least 100% greater than the average value for all residues (SI Appendix, Fig. S2A). Mapping these residues in the MpNep2 structure (SI Appendix, Fig. S2B) revealed that pressure globally shortens hydrogen bonds, similar to observations made in other proteins, including hen lysozyme (45) and basic pancreatic trypsin inhibitor (46). We observed that significant changes were found in or near loop regions (residues V22, D24, A33, S34, G35, Y55, S65, A77, A91, T92, I110, D157, S173, T179, T187, and N212) and within regions of protein secondary structure (residues T40, T81, V100, A123, Y165, A198, N203, and R205). In addition to aiding in the identification of hydrogen bond length changes, 15N shifts also provided insights regarding torsion angle changes in the backbone. We detected 18 15N nuclei that underwent significant changes (SI Appendix, Fig. S2C); of these, 10 out of 18 were common to those changes observed for the 1H nucleus (A33, S34, G35, T81, Y165, S173, T179, A198, N203, and R205).
Because chemical shifts are very sensitive to changes in molecular structure, the chemical shift dependence on pressure was examined more closely. Of the residual 1H–15N cross-peaks observed at 25 °C and 2.5 kbar, most exhibited a linear dependence on pressure for the Δδ 1H and 15N shifts (SI Appendix, Fig. S3 A and B), indicative of a linear response of hydrogen bond distance and torsion angles to pressure. Although most 1H–15N cross-peaks changed linearly with pressure, we detected several ΔδH and ΔδN that changed nonlinearly, as shown in SI Appendix, Fig. S3 C–E. For these nonlinear changes, we used sigmoidal, hyperbolical, and polynomial equations to fit the data, revealing that the behavior of some residues in response to pressure was complex. Although affected to a lesser extent compared with pressure, the chemical-shift dependence on urea might provide clues about the binding sites of urea to the protein surface.
Chemical-shift perturbation (CSP) analysis is extremely powerful and can help map changes in the local environment of a nuclei due to aromatic ring current effects, peptide bond anisotropy, electrostatic interactions, and hydrogen bonding (47). Using CSP, we mapped the primary (C52, D80, H118, D135, G143, W145, E192, and I220) and secondary (G35, T40, A91, G115, D119, W191, and A211) preferential binding sites of urea to MpNep2 (SI Appendix, Fig. S4 A–C). Notably, excluding C52, all mapped residues were on the protein surface and presented labile oxygen and nitrogen atoms from the backbone and side chains (SI Appendix, Fig. S4 B and C). Finally, the lack of correlation between 1H Δδ obtained with high pressure (δ1,600bar − δ1bar) and with urea (δ0.75M − δ0M) (Fig. 2C) indicates that physical and chemical agents affect protein structure through different mechanisms.
Measuring Pressure Effects Based on Peak Intensities.
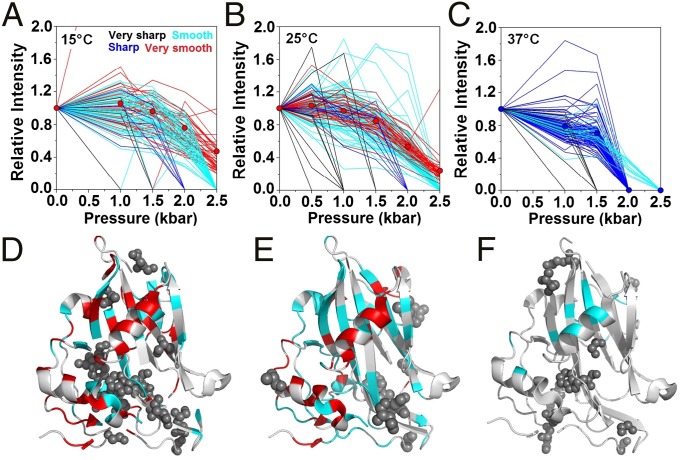
Another important parameter that is commonly used to measure the degree of local changes is the line broadening of 1H–15N cross-peaks as a function of increasing pressure. Line broadening reflects dynamic effects occurring on different timescales, ranging from thermal motions to slow-intermediate conformational exchange (43). Fast-intermediate conformational exchange results in NMR spectra with broader lines (i.e., peak intensity reduction) may account for protein–solvent interactions or protein unfolding events. Using pressure as the perturbing agent, we observed various intensity changes among the 1H–15N correlations; these intensity variations were classified as very sharp, sharp, smooth, and very smooth, and are represented in Fig. 3 A–C in black, blue, cyan, and red, respectively. Increasing the temperature from 15 to 25 °C resulted in an inverse relationship between the number of residues participating in the smooth and very smooth decaying groups (from 47 residues at 15 °C to 79 residues at 25 °C for the smooth decaying group and from 45 residues at 15 °C to 37 residues at 25 °C for the very smooth decaying group). Several residues participating in the smooth group at 25 °C presented a mountain-like intensity distribution, which was not observed at 15 °C. In the very smooth decaying group, we observed that, at 25 °C, the average intensity decay among all residues was more pronounced than that observed at 15 °C, reaching 0.47 at 15 °C/2.5 kbar compared with 0.24 at 25 °C/2.5 kbar (Fig. 3 A and B). At 37 °C, most peaks shifted from the very smooth and smooth groups to the sharp decaying group (Fig. 3C), and only 14 residual peaks were observed in the smooth decaying group (Q43, A84, V88, L99, V124, V125, F126, N155, I156, G183, G193, R199, G221, and S225).
Fig. 3.
Line-broadening analysis of the response to increasing pressure at various temperatures. (A–C) Changes in resonance intensities to increasing pressure are classified as very sharp (black), sharp (blue), smooth (cyan), and very smooth (red) decaying groups at 15, 25, and 37 °C. Average results for the very smooth decaying group are shown for 15 and 25 °C, and average results for the sharp decaying group for 37 °C are shown as solid circles. (D–F) Mapping of very sharp (spheres), smooth and very smooth decaying groups onto the MpNep2 crystal structure (PDB ID code 3ST1) for the results obtained at 15, 25, and 37 °C. The color scheme used is the same as that used in A, B, and C.
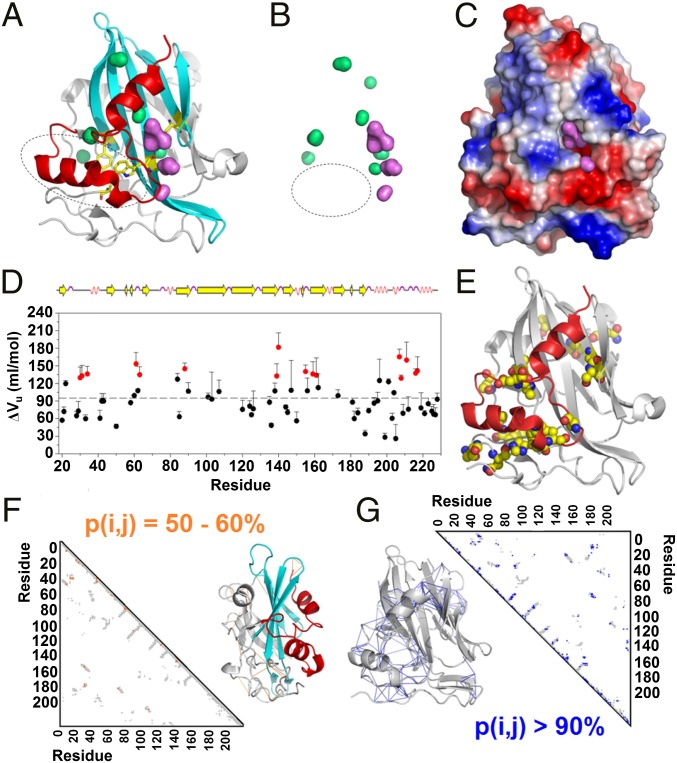
Qualitatively, the sharper the transition, the greater the sensitivity to pressure associated with larger volume changes. Observation of these groups in the MpNep2 structure yielded insights about the mechanism by which pressure affects MpNep2 protein folding. We observed that 6 out of 14 side chains (Y55, A57, Y87, A98, W120, and S140) belonging to the very sharp decaying group at 15 °C (represented as black spheres in Fig. 3 D–F) were located internally within the protein. A comparison of the intensity distributions between 15 and 25 °C revealed that residues participating in the smooth decaying group were primarily located in the main β-sheet region of the MpNep2 structure and that residues in the very smooth decaying group were mostly located in helical regions. At the structural level, two cooperative units are revealed by the mapped residues upon the pressure increase: the two most C-terminal α-helices of the protein and the main β-sheet in the protein core. The two most C-terminal α-helices form a hydrophobic bulk sandwiched with the protein manifold that is not associated with the presence of solvent-excluded cavities, as theoretically calculated (48) based on the MpNep2 crystal structure (49) (Fig. 4 A and B). It is possible that this bulk (containing the side chains of A57, Y87, and W120) is the region that is most sensitive to compression effects and might be affected by the penetration of water into a large cavity that is lined with acidic residues and exposed to the solvent (Fig. 4C). Of note, the N–H bond between W120 and S140 is located deep within this exposed cavity. Through this mechanism, the protein may lose hydrophobic interactions within the bulk, while maintaining the helical segments within the cooperative unit. In line with this, several residues located in this C-terminal cooperative unit, including His207, Trp208, Ala211, Ala216, and Asp217, shared the highest ΔVu contribution compared with the other residues (Fig. 4 D and E and SI Appendix, Fig. S5).
Fig. 4.
The proposed mechanism for pressure unfolding mediated by cooperative units. (A) The crystal structure of MpNep2 highlights the two cooperative units detected by pressure titration (red and cyan). (B) Surface distribution of MpNep2 solvent-exposed (violet) and solvent-excluded cavities (green). The circles shown in A and B represent the hydrophobic cavities. (C) Electrostatic surface of MpNep2, highlighting the acidic cavern that mediates water penetration and the initiation of protein unfolding. (D) ΔVu values among residues for MpNep2 calculated from the loss of 1H–15N HSQC cross-peak intensities at 37 °C. The highlighted values (red) correspond to those H–N resonances that are higher than the average ΔVu value plus one time SD (94.6 ± 33.9 mL/mol). (E) A crystal structure highlighting the residues that had higher contributions (red labeled in D) to volume change during pressure-dependent protein unfolding. (F and G) MpNep2 contact maps (gray) representing the probability of contacts between 50–60% (orange) and >90% (blue). Contact formation probabilities between 50–60% and >90% for each pair of residue mapped at 1,000 bar and 25 °C are highlighted in the crystal structures as orange and blue solid lines, respectively.
Finally, fractional contact maps at 1 kbar and 25 °C displayed a higher probability of contact formation [p(i, j) > 90%] among residues located internal to these cooperative units compared with those located in the N-terminal loop region [50% < p(i, j) < 60%] (Fig. 4 F and G and SI Appendix, Fig. S6). Altogether, we conclude that the initial steps for pressure-dependent protein unfolding occur at the most extreme N- and C-terminal regions of the protein, a process that would convert the protein to a molten-globule state (50, 51).
Measuring Urea Effects Based on NMR Peak Intensities and SAXS.
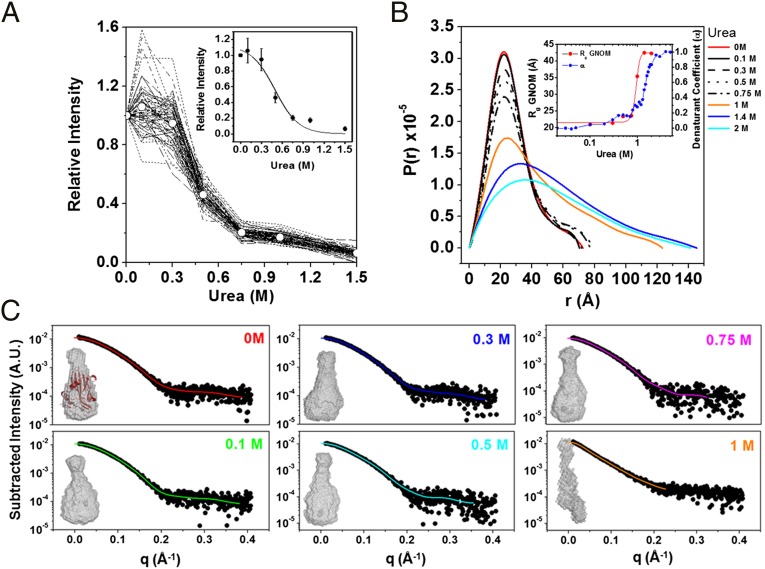
When urea was the perturbing agent, a similar decay transition was detected among all analyzed resonances; however, no values reached zero, even in concentrations of urea as high as 1.5 M (Fig. 5A) at 25 °C (unlike the results obtained by varying the pressure). Because NMR data indicate the presence of a unique cooperative decay upon treatment with urea, we designed single-point SAXS experiments under equilibrium using subdenaturing concentrations of urea, which were determined based on Trp fluorescence spectroscopy experiments (44), to probe conformational states that occur immediately before protein denaturation. The distance distribution function P(r) revealed an overall similar shape in the range of 0–0.75 M urea (Fig. 5B), with discrete increases in Dmax from 72.6 Å at 0 M to 77 Å at 0.75 M urea, and with a range of Rg from 21 Å at 0 M to 23.5 Å at 0.75 M. These observations were confirmed by ab initio low-resolution shape reconstructions using the X-ray scattering profile as input (Fig. 5C). As revealed by SAXS, the protein architecture remained firmly packed at urea concentrations of up to 0.75 M (Fig. 5 B and C); however, a homogenous decrease in peak intensity was observed by NMR at this urea concentration.
Fig. 5.
Mechanism for chemical unfolding. (A) Line-broadening analysis of the response to increasing urea concentrations at 25 °C. The changes in resonance intensities in response to increasing urea concentrations are uniformly distributed. Average results (solid circles) were used to fit the sigmoidal transition (Inset). (B) Distance distribution function of MpNep2 for various subdenaturing urea concentrations obtained from X-ray scattering. The Inset shows Rg values in response to increasing concentrations of urea for MpNep2 calculated using GNOM software (red circles) and superimposed with the denaturation fraction (blue circles) obtained from fluorescence data (44). (C) Three-dimensional molecular shape reconstructions for various subdenaturing urea concentrations. Subtracted scattering plots were fitted using the GNOM suite to generate shape restorations.
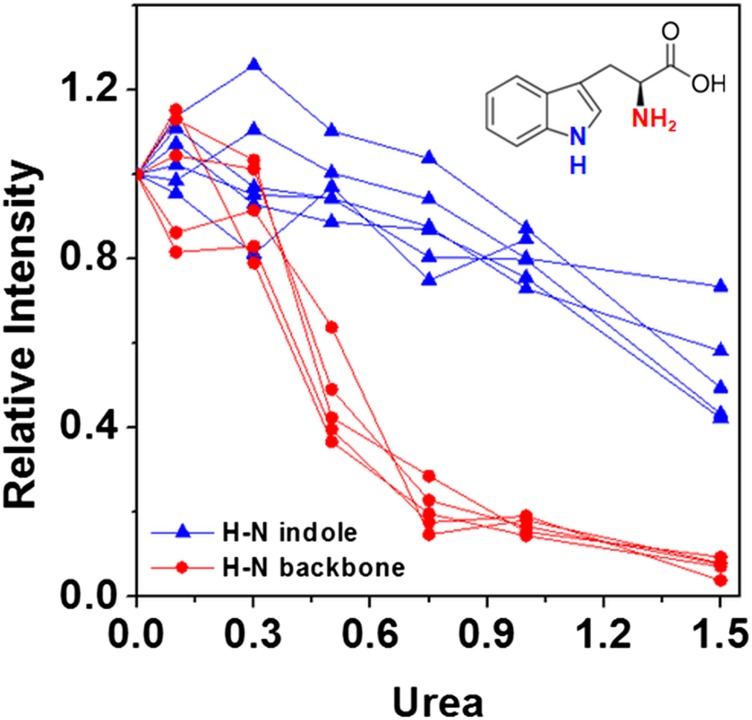
Because SAXS data revealed a packed/folded structure at 0.75 M urea and because NMR line-broadening effects are commonly associated with changes in protein dynamics due to a mixture of protein–solvent interactions or unfolding effects, for MpNep2, the effects observed by NMR urea titrations on peak intensities exhibit major urea-binding contributions rather than protein unfolding. Kratky plots obtained from the SAXS data confirm that MpNep2 is folded at 0.75 M urea (SI Appendix, Fig. S7). Increasing the urea concentration from 0.75 to 1 M led to an abrupt increase in the dimensional values, as observed for Dmax and Rg, and a loss of protein structure; this finding reveals that urea, unlike pressure, appears to form a global cooperative ensemble of structures. We previously obtained a U1/2 value of 1.41 M using Trp fluorescence spectroscopy (44). Using NMR spectroscopy, based on the average intensity of the cross-peaks in Fig. 5A, we calculated a U1/2 value of 0.5 M, which is quite different from that calculated by fluorescence. However, the SAXS data are consistent with our previous fluorescence data (Rg as a function of urea concentration; Fig. 5B, Inset). Unlike the SAXS and fluorescence data, the NMR data provide evidence for local specific changes at each N–H bond. This apparent discrepancy between U1/2 values is explained by the major contribution of preferential binding of urea to the protein before unfolding (likely due to N–H bonds with the protein backbone and side chains) as reported by NMR. As discussed below, the binding of urea is likely to replace water molecules and result in the formation of a dry globule, which remains compact (SAXS data), with the Trp protected from solvent relaxation (fluorescence data) and with urea predominantly bound to the protein surface at concentrations below 0.75 M (CSP analysis; SI Appendix, Fig. S4). The H–N from the Trp indole groups were not affected by urea up to 0.75 M and began a smooth decrease at higher concentrations that was accompanied by structure disruption (Fig. 6). The homogenous effect on the H–N peptide bond (reported by peak intensities) is probably due to urea covering the protein surface and slowing the backbone dynamics and not to the fact that it is the site of preferential binding (better reported by CSP analysis).
Fig. 6.
Trp line-broadening analysis of the response to increasing urea for backbone and indole H–N resonances.
Experimental Evidence for a Push-and-Pull Model.
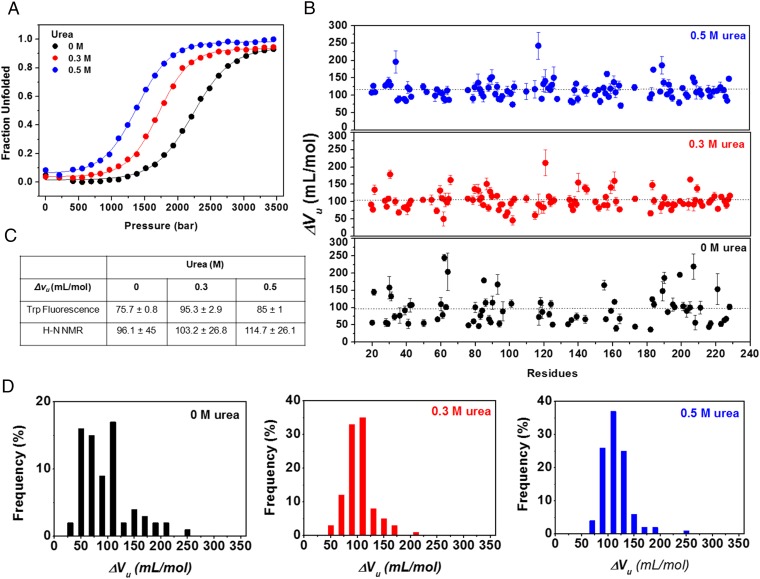
The overall results presented above indicate that urea preferentially binds to protein (“pull” effect), whereas pressure favors hydration (“push” effects). If true, pressure and urea have opposite effects at the initial stages under equilibrium, i.e., preferential hydration and preferential urea binding, and the combination of the two methods should lead to a mixture of these two effects. Fig. 7 shows the effects of pressure on MpNep2 at urea concentrations of 0.3 and 0.5 M, and SI Appendix, Fig. S8, shows the effects on the SH2 domain from c-Abl tyrosine kinase at concentrations of 4–4.8 M. These urea concentrations were sufficient to lead to preferential binding, but not unfolding, at atmospheric pressures as determined by urea titrations (44) (SI Appendix, Fig. S8A). Increasing the urea concentration shifted the pressure denaturation curves to lower pressure, as determined by fluorescence spectroscopy (Fig. 7A and SI Appendix, Fig. S8B) and NMR (Fig. 7B). ΔVu increased significantly, and a decrease in the heterogeneity of the pressure effects on ΔVu was observed, as determined by the frequency distribution of the averaged residues (Fig. 7 C and D). SI Appendix, Table S1, summarizes the unanimous ΔVu behavior upon increasing urea concentrations for three different protein models, including skeletal troponin C in the apo and holo states, MpNep2, and an SH2 domain. Additionally, similar behavior has been observed for different mutants of staphylococcal nuclease (SNAse) with increasing concentrations of guanidinium chloride (Gdm-Cl) (52, 53).
Fig. 7.
The push-and-pull model revealed by high-pressure titrations at different subdenaturing concentrations of urea for MpNep2. (A) Pressure-induced unfolding at 0, 0.3, and 0.5 M urea monitored by Trp fluorescence. (B) Volume changes upon unfolding (ΔVu) as a function of the protein sequence at 0, 0.3, and 0.5 M urea. (C) ΔVu values obtained by Trp fluorescence and from 1H–15N HSQC NMR spectra fits. (D) Distribution of ΔVu values obtained for MpNep2 at 0, 0.3, and 0.5 M urea.
Discussion
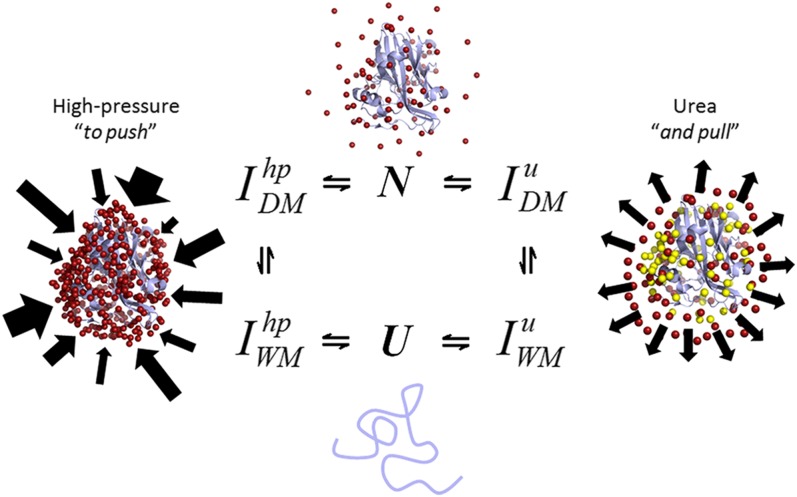
Here, we present the push-and-pull hypothesis to account for the effects of chemical and physical (pressure) denaturation of the folded state of a protein. Although both chemical and pressure denaturation processes are controlled by Le Chatelier's principle, the molecular mechanisms are different. We know that pressure shifts the equilibrium toward the denatured state due to the negative volume change, which is generally explained by a combination of several effects: the disruption of water-excluded cavities in the protein interior, electrostriction of broken salt bridges, and general water solvation (10–14). Monitoring each amino acid residue by NMR has permitted us to map the progressive effects of pressure. At low pressures, linear effects are observed as pressure is increased due to the shortening of hydrogen bonds. The population of these low-lying excited states is a springboard to partially unfolded states. Thus, pressure leads to a homogeneous shrinking of the protein, followed by an unfolding intermediate with part of the hydrophobic core exposed and, ultimately, full denaturation (Fig. 8).
Fig. 8.
Urea and high pressure results in different local unfolding states. The ensemble of dry (DM) and wet molten globule (WM) intermediates (I) in response to high pressure and urea are different according to NMR spectroscopy. A schematic of the push-and-pull hypothesis used to explain the mechanisms of the physical and chemical unfolding of proteins. Water molecules are indicated by red spheres, and urea molecules are indicated by yellow spheres. Arrows represent the inhomogeneous effect of pressure in contrast to that of urea.
In contrast, urea denaturation does not affect protein conformation until cooperative unfolding to the denatured state occurs. The “push-and-pull” mechanism has a dual character, explaining the general differences expected after denaturation with pressure vs. chemical agents. The use of a two-state assumption is the simplest thermodynamic analysis for both urea and pressure effects, and the analysis would be little affected if more complex assumptions were used.
Much debate regarding the contributions of cavities and hydration to the negative volume change upon unfolding, as well as compressibility effects, exists (10–14). Using NMR, the “squeezing” of the hydrogen bonds in parallel with the heterogeneous release of the water-excluded cavities by hydration was explored. Because we obtained high-precision data, we now understand these molecular events in detail.
In contrast to the pushing effects of pressure, we show for the first time (to our knowledge) how urea acts along a protein, pulling its structure as it preferentially binds to the protein (Fig. 8). Our data clearly demonstrate that the most significant contribution is the “direct” urea–protein interaction rather than an “indirect” mechanism caused by a change in the water structure. The pressure data at 0.3 and 0.5 M urea (Fig. 7 A–D) are crucial pieces of evidence supporting the action of the push-and-pull mechanism in MpNep2 denaturation and other protein models (SI Appendix, Table S1). At concentrations of 0.3 and 0.5 M, urea covers the protein surface and is preferentially bound to generate a hidden intermediate that has increased cavities (larger ΔVu) in addition to the expected destabilization to pressure (lower p1/2). In parallel, a “homogenization” of the pressure effect occurs. Weber and coworkers (54, 55) postulated a similar mechanism to explain the increased pressure sensitivity and homogenization caused by subdenaturing concentrations of urea.
How much of the results obtained for MpNep2 can be generalized to other proteins? In SI Appendix, Table S1, we show data that are available for pressure and urea denaturation of different proteins. In the case of apomyoglobin, although separate elegant works on pressure denaturation (56) and urea denaturation (57) exist, NMR data have been extensively obtained at 8 M urea and not at intermediate urea concentrations. Considerable fluorescence and NMR data are available for the urea and pressure denaturation of the N and C domains of troponin C (24, 58–60). Clearly, MpNep2 is highly sensitive to urea and pressure. We believe that the separation of the effects of urea covering and binding to the protein surface (decrease in NMR signal before unfolding with modest changes in chemical shifts) and cooperative urea denaturation might vary from protein to protein. For the studied SH2 domain, we observed that urea binding most significantly affects chemical shifts rather than line broadening (SI Appendix, Fig. S8 C and D). Thus, different protein models may have different susceptibilities to urea-binding effects, particularly considering the difference in folding stability investigated herein (MpNep2: ΔGu = 2.41 kcal/mol; SH2: ΔGu = 7.36 kcal/mol). The NMR spectra might respond differently depending on the exchange rate of the system upon binding (fast exchange, implying systematic chemical shifts, and intermediate exchange, indicating line broadening). The opposing NMR effects observed during urea binding to MpNep2 (major contribution to peak intensities and less to chemical shifts) and SH2 (less contribution to peak intensities and more to chemical shifts) confirm that the effects of urea binding are protein sensitive. In the fast-exchange regime, CSP analysis is the most common parameter to define binding sites on proteins (47) and is most often influenced by through-space and through-bond interactions of 1H and 15N nuclei. The summation of the interacting effects results in the observed shifts and the primary and secondary binding sites of urea to MpNep2 and to the SH2 domain (SI Appendix, Figs. S4 A–C and S8 E and F).
We cannot distinguish whether urea is preferentially bound to the protein backbone or to its side chains. Although we clearly show that the backbone N–H NMR intensity of MpNep2 decreases more than the imine N–H Trp (Fig. 6) and that all primary and secondary binding sites possess exposed side chains at the protein surface (SI Appendix, Figs. S4 and S8), the binding location cannot be determined with certainty. Based on a “universal-backbone” transfer model and molecular-dynamic simulations, the backbone and side chains equally contribute to urea-induced protein denaturation (61). In the slow-intermediate analysis performed for MpNep2, we believe that the dramatic decrease in resonance intensity is caused by urea covering the protein surface and slowing its backbone dynamics before unfolding. During this water exclusion, urea preferentially binds to specific residues on the protein surface (monitored by CSP). Our data agree with data from Guinn et al. (31), who showed that urea accumulates moderately at amide O and weakly at aliphatic C using model compounds. The pressure denaturation data at 0.5 M urea that shows that the volume change has increased and that p1/2 has decreased add evidence to this theory. An analogy of blowing up a bag with air (urea) in contrast to a packed bag (without urea) can be generated.
Site-Specific Effects of Pressure.
Concerning the effects of pressure below 1.5 kbar, our data clearly show the effects of pressure on hydrogen bonds, which are likely due to compressibility effects. However, the changes in water density around the more hydrophobic groups lead to the observed volume changes and pressure effect heterogeneity. The charged and polar groups on the protein surface are already electrostricted and suffer smaller pressure effects than the hydrophobic and less polar groups. A new methodology, pressure perturbation calorimetry (PPC), has made it possible to evaluate the changes in hydration of the side chains as a function of temperature (62–64). In PPC, the thermal expansion coefficient of a peptide or a protein is obtained directly from the heat absorbed or released after small pressure jumps. The apparent opposing effects of pushing water into the protein upon increasing pressure and the thermal expansion coefficients (α) as a function of temperature, α(T), may be reconciled when the terms participating in the volume change of a protein are considered separately [i.e., υint, sum of van der Waals volumes of all atoms and internal cavities; υhyd, volume associated with the hydration of the solvent-accessible surface of the protein (relative to the bulk); and υt, the thermal volume related to the molecular vibrations of the protein and the solvent] (63). This approach emphasizes the specific contribution of each of these terms to the overall volume change of a protein, which may vary depending on whether one uses one physical perturbation (e.g., high pressure up to 2,500 bar in case of HP-NMR) or two (e.g., low pressure around 5.5 bar and high temperature in case of PPC, where the density of the systems is hardly affected, but the thermal energy changes). Increasing the temperature of a system at low pressure has a greater contribution to υt than to υint or υhyd, and consequently leads to the expansion of the solvent away from the protein surface. The result differs from that obtained by applying high pressure at room temperature, where υhyd and υint have a greater contribution than υt, thereby rendering the push effect of the solvent predominant.
Using a line-broadening analysis, we observed that pressure affects MpNep2 folding heterogeneously, leading to the local unfolding of specific regions of the protein architecture (U`, U``, …, U`n states), as shown in Fig. 3 D–F. Based on this evaluation, we identified the two most C-terminal α-helices and the core β-sheet as cooperative folding units. Taking these cooperative units together with the residues clustered in the very sharp decaying group and the fractional contact map analysis, we were able to probe the segments of the protein in which the tertiary architecture was affected by pressure first. We noted that, in the initial stage of unfolding, pressure effects are governed by volume decreases due to hydrophobic solvation. Three residues, A57, Y87, and W120, located in a hydrophobic pocket, exhibited very sharp transitions at pressures up to 1.5 kbar, suggesting that hydrophobic solvation in this compartment might be caused by water penetration into the nearby acidic exposed cleft. In line with this, several residues located in this C-terminal cooperative unit made greater contributions to ΔVu. Cooperative folding units have been explored in several proteins, including cytochrome c (65) and apocytochrome b562 (66), using different perturbing agents.
Despite recent evidence that pressure unfolds proteins due to the disruption of solvent-excluded microcavities in the unfolded state (14), we did not observe cavities located near this hydrophobic bulk suffering from pressure effects, as theoretically calculated from the crystal structure of the studied protein.
Long-lived NOE contacts between water and interior amide hydrogen contacts in NOESY spectra were not detected at 2.5 kbar for ubiquitin (67), even though simulations at higher pressures (3 kbar) were able to detect water penetration (68, 69). This could be explained by equilibrium shifts from a DMG (at 2.5 kbar) to a wet molten globule (>3 kbar) for ubiquitin (67–70). For the MpNep2 structure, we believe that protein unfolding mediated by pressure occurs as the result of mixing events. At pressures of up to 1.5 kbar, we detected H–N bonds located within a hydrophobic bulk, suggesting that the volume is reduced due to hydrophobic solvation in the initial steps. This state is likely a DMG conformation similar to that found in ubiquitin (67). However, at higher pressures, we cannot exclude the possibility that water penetration into solvent-excluded cavities contributes to this effect because the cooperative β-sheet unit participates in the formation of these excluded voids together with other segments of the protein.
The Direct Interaction Mechanism Is Dominant in Chemical Unfolding and Leads to the Pull Effect.
Using NMR spectroscopy, we calculated a U1/2 value of 0.5 M based on the average intensity of the residues in Fig. 5A, which is significantly different from that calculated by fluorescence and SAXS (U1/2 values around 1.4 M). The apparent discrepancy between the U1/2 values can be explained by the preferential binding and covering of the protein by urea at a slow-intermediate exchange rate without a drastic effect on the structure. Urea binding was also mapped for the SH2 domain, but in a fast-exchange regime using CSP analysis (SI Appendix, Fig. S8 E and F). SAXS reconstructions revealed that the MpNep2 architecture was maintained up to 0.75 M urea, which is largely consistent with the NMR observations (relative intensity did not reach zero during urea titration).
The formation of a state with attenuated NMR intensities at low urea concentration is strong evidence supporting the formation of a DMG at equilibrium conditions. This hypothesis was further verified by treating this state (at 0.5 M urea) with pressure and observing the dry-globule behavior. As predicted by our push-and-pull model, the protein is destabilized, exhibits larger volume changes, and is less heterogeneous. The DMG state has been postulated previously and observed in some experiments (71). An early experiment on guanidinium-induced unfolding of RNase revealed a DMG state as determined by NMR (72). Stopped-flow NMR spectroscopy also detected a DMG state for dihydrofolate reductase (73). In these two cases, there was fast formation of an intermediate that had an expanded form but no increase in solvent exposure. In a recent kinetic study of monellin unfolding, FRET revealed a fast expansion of the native state before unfolding (74). More recently, in the case of villin headpiece (HP35), the DMG intermediate was characterized by conformational fluctuations at equilibrium (75). In silico kinetic studies of lysozyme denaturation by urea (29) have revealed that urea accumulates around the lysozyme, expelling water molecules from the first hydration shell of the protein. These authors also found that urea penetrated the hydrophobic core before water, forming a “dry globule.”
Detection of DMG species is elusive because the tryptophan residues are still buried in the interior of the protein and there is not enough exposed hydrophobic surface to be detected by other fluorescence probes (71). In the case of MpNep2, we detected the DMG intermediate at low urea concentrations when the tryptophan environment was not affected (no significant hydration) and no significant changes in the overall packing (SAXS) occurred (Fig. 5). Only changes in NMR intensity were detected as well as some CSPs due to urea binding. Unfortunately, the long acquisition times required for NMR make it difficult to detect this intermediate in the unfolding kinetics for MpNep2. It was peculiar that the DMG state of MpNep2 was populated over a short range of urea concentrations. In the case of villin headpiece (HP35), the DMG intermediate could also be observed by monitoring conformational fluctuations at equilibrium (75).
Taken together, these observations provide us with mechanistic explanations of how urea controls protein unfolding. Our results clearly establish the direct interaction mechanism as the dominant mechanism of urea-induced protein denaturation as shown by the peak intensities and systematic chemical shifts. The ensemble of conformations populated at low urea concentrations is consistent with a DMG in which the protein surface is dominated by urea instead of water molecules.
Tracing some parallels with high pressure, we can state that pressure affects protein structure due to compressibility effects leading to a shortening of protein–protein and protein–solvent H bonds; this phenomenon contrasts with the “H-bond shift” that occurs during urea binding (Fig. 8). Consequently, pressure ultimately pushes water molecules against protein atoms, heterogeneously affecting the structure, unlike urea, which homogenously pulls water molecules away from the solvation shell due to its binding properties. Water pull would drive the conformation into an unstable DMG state, thus explaining why small additional increases in urea concentration result in cooperative unfolding (Fig. 8). Similarly, pulling the structure would make it more sensitive to pressure (lower p1/2), with a tendency to have larger volume changes, as verified in Fig. 7 and SI Appendix, Table S1. The separate and combined thermodynamic effects of urea and pressure have allowed us to provide a push-and-pull molecular interpretation for these effects, taking advantage of site-specific (NMR) and global (fluorescence and SAXS) methods. These interpretations provide insights that illuminate the cumulative effects of using subdenaturing concentrations of urea and pressure to measure protein folding and stability. In conclusion, we present experimental data introducing the push-and-pull hypothesis, which may explain differences between the molecular mechanisms involved during the physical and chemical unfolding of proteins. These observations open up possibilities for the study of protein folding and provide a new interpretation to explain the nature of the cooperative behavior of proteins during folding reactions.
Materials and Methods
Protein Preparation and Assignment.
The SH2 domain from c-Abl protein and MpNep2 were purified and assigned as previously described (25, 44). For the high-pressure fluorescence and NMR experiments, size exclusion chromatography in Superdex 75 10/300 (GE Life Science) was performed immediately before each experiment using 20 mM Tris⋅HCl (pH 7.4) containing 80 mM NaCl.
Fluorescence Spectroscopy.
High-pressure fluorescence emission measurements were recorded using an ISSK2 spectrofluorometer (ISS, Inc.) equipped with a high-pressure cell (ISS, Inc.). Trp emission spectra were collected from 0.001 to 3.5 kbar in 0.345-kbar increments at 15, 25, and 37 °C. Samples (5 µM) were left for 10 min at each pressure before excitation. Trp residues were excited at 280 nm using a slit width of 1 mm, and the emission was recorded from 300 to 400 nm using a slit width of 1 mm. Experiments were performed three times with different preparations, and the results are expressed as the mean. Changes in the fluorescence spectra recorded at each pressure were quantified as the center of spectral mass (per centimeter) as previously described (44). Stability measurements were performed using urea, as previously reported for the MpNep2 protein (44). All of the transitions were analyzed as two-state processes.
HP-NMR Spectroscopy.
High-pressure heteronuclear 1H–15N HSQC NMR spectra were acquired using a zirconia NMR tube with an internal diameter of 3 mm and an outer diameter of 5 mm (Daedalus Innovations) at 5, 10, 15, 25, and 37 °C and a Bruker Avance III 800-MHz spectrometer. An Xtreme-60 Syringe pump system (Daedalus Innovations) was used to generate pressures from 1 to 20, 50, 200, 400, 600, 800, 1,000, 1,200, 1,400, 1,600, 1,800, 2,000, 2,200, 2,400, and 2,500 bar at each of the temperatures listed above.
For thermodynamic analysis, a first-order sigmoidal equation was used to fit 1H–15N HSQC cross-peak intensity vs. pressure for those resonances that could be followed throughout the unfolding reaction (more than 70 H–N fits). Volume changes (ΔVu) were calculated as described previously (59) assuming two-state transitions.
For the chemical titration, 1H–15N HSQC spectra were recorded at different urea concentrations in a BMS-003 Shigemi NMR tube at 25 °C. The 1H dimension was acquired in 1,024 increments, and for the 15N dimension, 128 complex points were collected; eight scans were collected at each increment. CSP analysis was performed at 1, 3, and 5 M urea against 1H–15N HSQC NMR spectra at 0 M urea for the SH2 domain and at 0.3, 0.5, and 0.75 M urea for MpNep2 using the following equation:
where ΔδH and ΔδN represent the chemical shift variations of 1H and 15N, respectively, between different concentrations of urea to 0 M. At all pressures, chemical shifts were referenced to the methyl signal of DSS. All spectra were processed using Topspin 3.11. Chemical shift values were measured using the CARA 1.8.4 suite or CCPN Analysis 2.4.1.
Computational Analysis.
We used the Monte Carlo method included in the McVol (48) program to detect internal cavities and surface clefts in the MpNep2 crystal structure [Protein Data Bank (PDB) ID code 3ST1]. The algorithm was applied using a probe sphere of 1.1 Å, 250 Monte Carlo steps per Å3 of the molecule, and 2,500 dots per atom on the dotted surface. Contact maps were generated using the CMView 1.1 suite implemented in PyMOL. Pairs of residues (i, j) separated by a distance of 8 Å were considered among Cα. The MpNep2 crystal structure (3ST1) was used as the template. The probability of contact formation ρ(i, j) at 1 kbar and 25 °C was calculated as the product of the relative intensity for the amide resonances of two residues in contact and classified as shown in SI Appendix, Fig. S6.
Scattering Data Collection and Analysis.
SAXS data were collected using the SAXS1 small-angle scattering beamline of the National Synchrotron Light Laboratory (Campinas, Brazil), using a 300K Pilatus detector, 84 mm × 107 mm (Dectris); a mica sample cell for liquids and a wavelength of 0.155 nm were used at room temperature. The position of the direct beam in the detector and the detector inclination were calibrated using silver behenate (76). X-ray photons elastically scattered from the samples were taken from a single 10-s frame for each condition. The modulus of the scattering vector q was calculated according to the equation q = (4π/λ) sin 2θ, where λ is the wavelength used and 2θ is the scattering angle. Scattering from each sample was corrected by subtracting the contribution from the buffer (20 mM phosphate containing 80 mM NaCl at pH 7.4) and the corresponding urea concentration, and the resulting data were fitted using autoGNOM (77) implemented in the PrimusQt suite. The value of Rg was determined using GNOM; a range of input distances was tested to best fit the experimental data. The value of Rg was also determined from the angular coefficient (α) of the low q region (q < 1.3/Rg) in Guinier plots. Fourier transformation of the scattered particles shows the distribution of the interatomic distances P(r) in the molecules, enabling a prediction of particle shape. All samples were measured at 2.5 mg/mL. The sample-detector distance was set to 941.7 mm, enabling detection over a q range of 0.107–4.07 nm−1. The samples were centrifuged at 10,000 × g for 10 min at room temperature immediately before data acquisition. Low-resolution shape restorations were performed using 10 independent calculation trials, as previously described (44).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Council of Technological and Scientific Development and Carlos Chagas Filho Foundation for Research Support in the State of Rio de Janeiro (to J.L.S.). G.A.P.d.O. is a recipient of a postdoctoral fellowship from the Brazilian government agency National Postdoctoral Program/Coordination for the Improvement of Higher Education Personnel. This work was aided by the Brazilian Synchrotron Light Laboratory under Proposals SAXS1-15168 and SAXS1-16149.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500352112/-/DCSupplemental.
References
- 1.Levinthal C. Are there pathways for protein folding? J Chim Phys. 1968;65(1):44–45. [Google Scholar]
- 2.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: The energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 3.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14(1):70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Ishimaru D, et al. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry. 2003;42(30):9022–9027. doi: 10.1021/bi034218k. [DOI] [PubMed] [Google Scholar]
- 5.Ano Bom APD, et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J Biol Chem. 2012;287(33):28152–28162. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva JL, De Moura Gallo CV, Costa DC, Rangel LP. Prion-like aggregation of mutant p53 in cancer. Trends Biochem Sci. 2014;39(6):260–267. doi: 10.1016/j.tibs.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 8.Silva JL, et al. Ligand binding and hydration in protein misfolding: Insights from studies of prion and p53 tumor suppressor proteins. Acc Chem Res. 2010;43(2):271–279. doi: 10.1021/ar900179t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridgman PW. The coagulation of albumen by pressure. J Biol Chem. 1914;19:511–512. [Google Scholar]
- 10.Silva JL, Weber G. Pressure stability of proteins. Annu Rev Phys Chem. 1993;44:89–113. doi: 10.1146/annurev.pc.44.100193.000513. [DOI] [PubMed] [Google Scholar]
- 11.Silva JL, Foguel D, Royer CA. Pressure provides new insights into protein folding, dynamics and structure. Trends Biochem Sci. 2001;26(10):612–618. doi: 10.1016/s0968-0004(01)01949-1. [DOI] [PubMed] [Google Scholar]
- 12.Silva JL, et al. High-pressure chemical biology and biotechnology. Chem Rev. 2014;114(14):7239–7267. doi: 10.1021/cr400204z. [DOI] [PubMed] [Google Scholar]
- 13.Akasaka K, Kitahara R, Kamatari YO. Exploring the folding energy landscape with pressure. Arch Biochem Biophys. 2013;531(1-2):110–115. doi: 10.1016/j.abb.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Roche J, et al. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci USA. 2012;109(18):6945–6950. doi: 10.1073/pnas.1200915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung MS, García AE, Onuchic JN. Protein folding mediated by solvation: Water expulsion and formation of the hydrophobic core occur after the structural collapse. Proc Natl Acad Sci USA. 2002;99(2):685–690. doi: 10.1073/pnas.022387699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Royer CA. Probing protein folding and conformational transitions with fluorescence. Chem Rev. 2006;106(5):1769–1784. doi: 10.1021/cr0404390. [DOI] [PubMed] [Google Scholar]
- 17.Dzwolak W, Kato M, Taniguchi Y. Fourier transform infrared spectroscopy in high-pressure studies on proteins. Biochim Biophys Acta. 2002;1595(1-2):131–144. doi: 10.1016/s0167-4838(01)00340-5. [DOI] [PubMed] [Google Scholar]
- 18.Akasaka K. Probing conformational fluctuation of proteins by pressure perturbation. Chem Rev. 2006;106(5):1814–1835. doi: 10.1021/cr040440z. [DOI] [PubMed] [Google Scholar]
- 19.Dyson HJ, Wright PE. Unfolded proteins and protein folding studied by NMR. Chem Rev. 2004;104(8):3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 20.Winter R. Synchrotron X-ray and neutron small-angle scattering of lyotropic lipid mesophases, model biomembranes and proteins in solution at high pressure. Biochim Biophys Acta. 2002;1595(1-2):160–184. doi: 10.1016/s0167-4838(01)00342-9. [DOI] [PubMed] [Google Scholar]
- 21.Schroer MA, et al. High-pressure SAXS study of folded and unfolded ensembles of proteins. Biophys J. 2010;99(10):3430–3437. doi: 10.1016/j.bpj.2010.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Prigozhin MB, Schulten K, Gruebele M. Observation of complete pressure-jump protein refolding in molecular dynamics simulation and experiment. J Am Chem Soc. 2014;136(11):4265–4272. doi: 10.1021/ja412639u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerch MT, Horwitz J, McCoy J, Hubbell WL. Circular dichroism and site-directed spin labeling reveal structural and dynamical features of high-pressure states of myoglobin. Proc Natl Acad Sci USA. 2013;110(49):E4714–E4722. doi: 10.1073/pnas.1320124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Oliveira GAP, et al. Insights into the intramolecular coupling between the N- and C-domains of troponin C derived from high-pressure, fluorescence, nuclear magnetic resonance, and small-angle X-ray scattering studies. Biochemistry. 2013;52(1):28–40. doi: 10.1021/bi301139d. [DOI] [PubMed] [Google Scholar]
- 25.de Oliveira GAP, et al. Intramolecular dynamics within the N-Cap-SH3-SH2 regulatory unit of the c-Abl tyrosine kinase reveal targeting to the cellular membrane. J Biol Chem. 2013;288(39):28331–28345. doi: 10.1074/jbc.M113.500926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mertens HDT, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J Struct Biol. 2010;172(1):128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Das A, Mukhopadhyay C. Urea-mediated protein denaturation: A consensus view. J Phys Chem B. 2009;113(38):12816–12824. doi: 10.1021/jp906350s. [DOI] [PubMed] [Google Scholar]
- 28.Zhou R, Li J, Hua L, Yang Z, Berne BJ. Comment on “urea-mediated protein denaturation: A consensus view.”. J Phys Chem B. 2011;115(5):1323–1326, discussion 1327–1328. doi: 10.1021/jp105160a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua L, Zhou R, Thirumalai D, Berne BJ. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc Natl Acad Sci USA. 2008;105(44):16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canchi DR, Paschek D, García AE. Equilibrium study of protein denaturation by urea. J Am Chem Soc. 2010;132(7):2338–2344. doi: 10.1021/ja909348c. [DOI] [PubMed] [Google Scholar]
- 31.Guinn EJ, Pegram LM, Capp MW, Pollock MN, Record MT., Jr Quantifying why urea is a protein denaturant, whereas glycine betaine is a protein stabilizer. Proc Natl Acad Sci USA. 2011;108(41):16932–16937. doi: 10.1073/pnas.1109372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auton M, Bolen DW. Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc Natl Acad Sci USA. 2005;102(42):15065–15068. doi: 10.1073/pnas.0507053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auton M, Holthauzen LMF, Bolen DW. Anatomy of energetic changes accompanying urea-induced protein denaturation. Proc Natl Acad Sci USA. 2007;104(39):15317–15322. doi: 10.1073/pnas.0706251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennion BJ, Daggett V. The molecular basis for the chemical denaturation of proteins by urea. Proc Natl Acad Sci USA. 2003;100(9):5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zangi R, Zhou R, Berne BJ. Urea’s action on hydrophobic interactions. J Am Chem Soc. 2009;131(4):1535–1541. doi: 10.1021/ja807887g. [DOI] [PubMed] [Google Scholar]
- 36.Zou Q, Habermann-Rottinghaus SM, Murphy KP. Urea effects on protein stability: Hydrogen bonding and the hydrophobic effect. Proteins. 1998;31(2):107–115. [PubMed] [Google Scholar]
- 37.Makhatadze GI, Privalov PL. Protein interactions with urea and guanidinium chloride. A calorimetric study. J Mol Biol. 1992;226(2):491–505. doi: 10.1016/0022-2836(92)90963-k. [DOI] [PubMed] [Google Scholar]
- 38.Lim WK, Rösgen J, Englander SW. Urea, but not guanidinium, destabilizes proteins by forming hydrogen bonds to the peptide group. Proc Natl Acad Sci USA. 2009;106(8):2595–2600. doi: 10.1073/pnas.0812588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nozaki Y, Tanford C. The solubility of amino acids and related compounds in aqueous urea solutions. J Biol Chem. 1963;238(12):4074–4081. [PubMed] [Google Scholar]
- 40.Wang A, Bolen DW. A naturally occurring protective system in urea-rich cells: Mechanism of osmolyte protection of proteins against urea denaturation. Biochemistry. 1997;36(30):9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 41.Tanford C. Isothermal unfolding of globular proteins in aqueous urea solutions. J Am Chem Soc. 1964;86(10):2050–2059. [Google Scholar]
- 42.Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 43.Valente AP, Miyamoto CA, Almeida FCL. Implications of protein conformational diversity for binding and development of new biological active compounds. Curr Med Chem. 2006;13(30):3697–3703. doi: 10.2174/092986706779026147. [DOI] [PubMed] [Google Scholar]
- 44.de Oliveira GA, et al. Moniliophthora perniciosa necrosis- and ethylene-inducing protein 2 (MpNep2) as a metastable dimer in solution: Structural and functional implications. PLoS One. 2012;7(9):e45620. doi: 10.1371/journal.pone.0045620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamatari YO, et al. Response of native and denatured hen lysozyme to high pressure studied by 15N/1H NMR spectroscopy. Eur J Biochem. 2001;268(6):1782–1793. [PubMed] [Google Scholar]
- 46.Li H, Yamada H, Akasaka K. Effect of pressure on individual hydrogen bonds in proteins. Basic pancreatic trypsin inhibitor. Biochemistry. 1998;37(5):1167–1173. doi: 10.1021/bi972288j. [DOI] [PubMed] [Google Scholar]
- 47.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Till MS, Ullmann GM. McVol—a program for calculating protein volumes and identifying cavities by a Monte Carlo algorithm. J Mol Model. 2010;16(3):419–429. doi: 10.1007/s00894-009-0541-y. [DOI] [PubMed] [Google Scholar]
- 49.Zaparoli G, et al. The crystal structure of necrosis- and ethylene-inducing protein 2 from the causal agent of cacao’s Witches’ Broom disease reveals key elements for its activity. Biochemistry. 2011;50(45):9901–9910. doi: 10.1021/bi201253b. [DOI] [PubMed] [Google Scholar]
- 50.Silva JL, Silveira CF, Correia Júnior A, Pontes L. Dissociation of a native dimer to a molten globule monomer. Effects of pressure and dilution on the association equilibrium of arc repressor. J Mol Biol. 1992;223(2):545–555. doi: 10.1016/0022-2836(92)90669-b. [DOI] [PubMed] [Google Scholar]
- 51.Peng X, Jonas J, Silva JL. Molten-globule conformation of Arc repressor monomers determined by high-pressure 1H NMR spectroscopy. Proc Natl Acad Sci USA. 1993;90(5):1776–1780. doi: 10.1073/pnas.90.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roche J, et al. Remodeling of the folding free energy landscape of staphylococcal nuclease by cavity-creating mutations. Biochemistry. 2012;51(47):9535–9546. doi: 10.1021/bi301071z. [DOI] [PubMed] [Google Scholar]
- 53.Roche J, et al. Effect of internal cavities on folding rates and routes revealed by real-time pressure-jump NMR spectroscopy. J Am Chem Soc. 2013;135(39):14610–14618. doi: 10.1021/ja406682e. [DOI] [PubMed] [Google Scholar]
- 54.Da Poian AT, Oliveira AC, Gaspar LP, Silva JL, Weber G. Reversible pressure dissociation of R17 bacteriophage. The physical individuality of virus particles. J Mol Biol. 1993;231(4):999–1008. doi: 10.1006/jmbi.1993.1347. [DOI] [PubMed] [Google Scholar]
- 55.Weber G, Da Poian AT, Silva JL. Concentration dependence of the subunit association of oligomers and viruses and the modification of the latter by urea binding. Biophys J. 1996;70(1):167–173. doi: 10.1016/S0006-3495(96)79557-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kitahara R, Yamada H, Akasaka K, Wright PE. High pressure NMR reveals that apomyoglobin is an equilibrium mixture from the native to the unfolded. J Mol Biol. 2002;320(2):311–319. doi: 10.1016/S0022-2836(02)00449-7. [DOI] [PubMed] [Google Scholar]
- 57.Mohana-Borges R, Goto NK, Kroon GJ, Dyson HJ, Wright PE. Structural characterization of unfolded states of apomyoglobin using residual dipolar couplings. J Mol Biol. 2004;340(5):1131–1142. doi: 10.1016/j.jmb.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 58.Suarez MC, et al. Role of hydration in the closed-to-open transition involved in Ca2+ binding by troponin C. Biochemistry. 2003;42(18):5522–5530. doi: 10.1021/bi027102h. [DOI] [PubMed] [Google Scholar]
- 59.Suarez MC, Rocha CB, Sorenson MM, Silva JL, Foguel D. Free-energy linkage between folding and calcium binding in EF-hand proteins. Biophys J. 2008;95(10):4820–4828. doi: 10.1529/biophysj.108.135715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rocha CB, et al. Volume and free energy of folding for troponin C C-domain: Linkage to ion binding and N-domain interaction. Biochemistry. 2008;47(17):5047–5058. doi: 10.1021/bi702058t. [DOI] [PubMed] [Google Scholar]
- 61.Moeser B, Horinek D. Unified description of urea denaturation: Backbone and side chains contribute equally in the transfer model. J Phys Chem B. 2014;118(1):107–114. doi: 10.1021/jp409934q. [DOI] [PubMed] [Google Scholar]
- 62.Mitra L, Smolin N, Ravindra R, Royer C, Winter R. Pressure perturbation calorimetric studies of the solvation properties and the thermal unfolding of proteins in solution—experiments and theoretical interpretation. Phys Chem Chem Phys. 2006;8(11):1249–1265. doi: 10.1039/b516608j. [DOI] [PubMed] [Google Scholar]
- 63.Schweiker KL, Makhatadze GI. Use of pressure perturbation calorimetry to characterize the volumetric properties of proteins. Methods Enzymol. 2009;466:527–547. doi: 10.1016/S0076-6879(09)66022-5. [DOI] [PubMed] [Google Scholar]
- 64.Pandharipande PP, Makhatadze GI. Thermal expansivities of peptides, polypeptides and proteins as measured by pressure perturbation calorimetry. Methods. 2015;76:61–66. doi: 10.1016/j.ymeth.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269(5221):192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuentes EJ, Wand AJ. Local stability and dynamics of apocytochrome b562 examined by the dependence of hydrogen exchange on hydrostatic pressure. Biochemistry. 1998;37(28):9877–9883. doi: 10.1021/bi980894o. [DOI] [PubMed] [Google Scholar]
- 67.Fu Y, et al. Coupled motion in proteins revealed by pressure perturbation. J Am Chem Soc. 2012;134(20):8543–8550. doi: 10.1021/ja3004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Day R, García AE. Water penetration in the low and high pressure native states of ubiquitin. Proteins. 2008;70(4):1175–1184. doi: 10.1002/prot.21562. [DOI] [PubMed] [Google Scholar]
- 69.Imai T, Sugita Y. Dynamic correlation between pressure-induced protein structural transition and water penetration. J Phys Chem B. 2010;114(6):2281–2286. doi: 10.1021/jp909701j. [DOI] [PubMed] [Google Scholar]
- 70.Kitahara R, Yokoyama S, Akasaka K. NMR snapshots of a fluctuating protein structure: Ubiquitin at 30 bar-3 kbar. J Mol Biol. 2005;347(2):277–285. doi: 10.1016/j.jmb.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 71.Baldwin RL, Frieden C, Rose GD. Dry molten globule intermediates and the mechanism of protein unfolding. Proteins. 2010;78(13):2725–2737. doi: 10.1002/prot.22803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kiefhaber T, Labhardt AM, Baldwin RL. Direct NMR evidence for an intermediate preceding the rate-limiting step in the unfolding of ribonuclease A. Nature. 1995;375(6531):513–515. doi: 10.1038/375513a0. [DOI] [PubMed] [Google Scholar]
- 73.Hoeltzli SD, Frieden C. Stopped-flow NMR spectroscopy: Real-time unfolding studies of 6-19F-tryptophan-labeled Escherichia coli dihydrofolate reductase. Proc Natl Acad Sci USA. 1995;92(20):9318–9322. doi: 10.1073/pnas.92.20.9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jha SK, Udgaonkar JB. Direct evidence for a dry molten globule intermediate during the unfolding of a small protein. Proc Natl Acad Sci USA. 2009;106(30):12289–12294. doi: 10.1073/pnas.0905744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reiner A, Henklein P, Kiefhaber T. An unlocking/relocking barrier in conformational fluctuations of villin headpiece subdomain. Proc Natl Acad Sci USA. 2010;107(11):4955–4960. doi: 10.1073/pnas.0910001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang TC, Toraya H, Blanton TN, Wu Y. X-ray powder diffraction analysis of silver behenate, a possible low-angle diffraction standard. J Appl Cryst. 1993;26:180–184. [Google Scholar]
- 77.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.