Significance
KSHV is the etiological agent of Kaposi sarcoma, primary effusion lymphoma, and the plasma cell variant of multicentric Castleman disease. During latency, this dsDNA tumor virus expresses only a small subset of its more than 90 ORFs. Among these is ORF73/latency-associated nuclear antigen (LANA), which acts as the origin binding protein and chromatin anchor of the extrachromosomal viral genome. This work provides detailed structural insights into the DNA-binding characteristics of LANA. We also report a previously unrecognized, third LANA binding site within the minimal replicator of Kaposi sarcoma herpesvirus. In addition to its mechanistic implications for latent viral persistence, the X-ray crystal structure of LANA bound to LANA binding site 1 DNA may assist in the tailored development of therapeutic LANA inhibitors.
Keywords: X-ray crystallography, gammaherpesvirinae, viral latency, DNA-binding protein, KSHV LANA
Abstract
Kaposi sarcoma herpesvirus (KSHV) persists as a latent nuclear episome in dividing host cells. This episome is tethered to host chromatin to ensure proper segregation during mitosis. For duplication of the latent genome, the cellular replication machinery is recruited. Both of these functions rely on the constitutively expressed latency-associated nuclear antigen (LANA) of the virus. Here, we report the crystal structure of the KSHV LANA DNA-binding domain (DBD) in complex with its high-affinity viral target DNA, LANA binding site 1 (LBS1), at 2.9 Å resolution. In contrast to homologous proteins such as Epstein-Barr virus nuclear antigen 1 (EBNA-1) of the related γ-herpesvirus Epstein-Barr virus, specific DNA recognition by LANA is highly asymmetric. In addition to solving the crystal structure, we found that apart from the two known LANA binding sites, LBS1 and LBS2, LANA also binds to a novel site, denoted LBS3. All three sites are located in a region of the KSHV terminal repeat subunit previously recognized as a minimal replicator. Moreover, we show that the LANA DBD can coat DNA of arbitrary sequence by virtue of a characteristic lysine patch, which is absent in EBNA-1 of the Epstein-Barr virus. Likely, these higher-order assemblies involve the self-association of LANA into supermolecular spirals. One such spiral assembly was solved as a crystal structure of 3.7 Å resolution in the absence of DNA. On the basis of our data, we propose a model for the controlled nucleation of higher-order LANA oligomers that might contribute to the characteristic subnuclear KSHV microdomains (“LANA speckles”), a hallmark of KSHV latency.
Kaposi sarcoma herpesvirus (KSHV) is the only known γ2-herpesvirus of concern to human health. Apart from its involvement in two lymphoproliferative disorders, KSHV plays a vital role in the development of Kaposi sarcoma, the most common form of cancer in patients with AIDS (1, 2). After a primary infection event, the virus establishes lifelong latent persistence in the nuclei of its host cells. A molecular key player in the establishment, maintenance, and regulation of KSHV latency is the latency-associated nuclear antigen (LANA).
KSHV LANA contains 1,162 amino acids in the prototype strain and exerts functions in host cell survival, transcriptional control, latent viral replication, and stable episome segregation during mitosis (3, 4). Its N-terminal domain is separated from its C-terminal domain by a large internal repeat region (5, 6). Although the N-terminal domain and the internal repeat region are predicted to be only poorly structured (7), the C-terminal domain comprises a stable 3D structure with a strong hydrophobic core (8, 9). LANA’s C-terminal domain binds to the LANA binding sites (LBS) within the viral terminal repeats (TRs) in a sequence-specific manner (10, 11). Therefore, this domain is referred to as the DNA-binding domain (DBD). In contrast, the N terminus of LANA binds to nucleosome core particles, and thereby tethers the viral genomes to host chromatin (12). Because LANA itself is not known to possess any enzymatic activity, its functions largely rely on the recruitment of cellular and viral interaction partners (3, 4).
We and others have previously solved the crystal structure of the KSHV LANA DBD in the absence of DNA (8, 9). The homodimeric domain exhibits structural and functional similarities to the DBDs of the Epstein-Barr virus nuclear antigen 1 (EBNA-1) and the papillomavirus E2 protein (8, 9, 13, 14). Unlike these orthologs, the LANA DBD can also form higher-order oligomers via lateral oligomerization sites; however, structural investigation of complexes comprising the LANA DBD and DNA has so far been hampered by their low solubility (15, 16). Here, we used structure-guided mutagenesis to circumvent this obstacle. Specifically, two non-overlapping DNA-binding surfaces on the LANA DBD were independently mutated, which allowed the separate structural investigation of their respective DNA complexes. We present the crystal structure of a LANA DBD multiple-point mutant in complex with LBS1 and use a complementary mutant in low-resolution electron microscopy to show that the LANA DBD can oligomerize on unspecific DNA by using a second mode of DNA interaction.
The X-Ray Structure of the LANA DBD Bound to LBS1 DNA
To understand the sequence-specific interaction between KSHV LANA and its high-affinity target DNA at the 3D level, we used a multiple-point mutant of LANA(1008–1146) for cocrystallization with a 20-bp wild-type LBS1 fragment. A modification of the protein’s surface characteristics became necessary after previous attempts to obtain monodisperse samples of the wild-type LANA DBD in complex with DNA had failed (15, 16). In particular, our mutant contains a total of nine point mutations for charge inversion of the lysine patch (K1055E, K1138S, K1140D, K1141D), partial arginine patch neutralization (R1039Q, R1040Q), oligomerization site perturbation (A1121E), and surface entropy reduction (K1109A, D1110A) (Fig. S1 A and B). The design of this mutant is reported elsewhere in detail (16). Importantly, none of these mutations are located in the proximity of the sequence-specific DNA-binding surface. A 3D superposition of the resulting crystal structure with the DNA-free wild-type LANA DBD dimer structure yields an rmsd of only 0.4 Å (Fig. S3A). This shows that the overall protein geometry is virtually unaffected by our mutagenesis. It also implies that the protein backbone conformation does not significantly change on DNA binding.
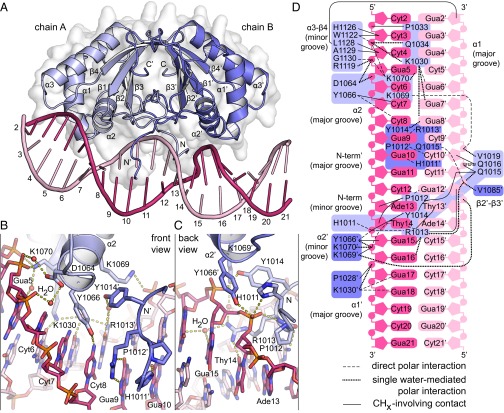
The complex structure was solved at a resolution of 2.9 Å (Fig. 1 and Table S1). In this complex, the homodimeric protein retains almost perfect twofold symmetry. A 3D alignment of the two subunits yields an rmsd of only 0.3 Å. However, the DNA is bound to the LANA DBD dimer with remarkable asymmetry. In this regard, LANA differs from EBNA-1 and E2, both of which bind their palindromic target sequences in a symmetric fashion (13, 14) (Fig. S3 C and D).
Fig. 1.
Structure of KSHV LANA bound to LBS1 DNA. (A) Crystal structure of an oligomerization-deficient LANA(1008–1146) multiple-point mutant bound to 20-bp LBS1 DNA. (B) Close-up of the primary site of DNA recognition. (C) Close-up of the symmetry-related secondary site. (D) Structure-derived interaction chart. LBS1 sequence numbering corresponds to a previous study (17).
The primary site of direct DNA sequence recognition spans positions Gua5 to Gua10 in the major groove of LBS1 (Fig. 1B). This site includes DNA sequence positions Cyt6, Cyt7, and Cyt8, which were previously found to be most crucial for efficient binding (17). The primary site is recognized by helices α1 and α2 of protein chain A and the N′-terminal arm of chain B, as indicated in Fig. 1A. Previous studies pointed toward Y1066 on helix α2 as a key residue in LBS1 recognition (11, 15). Indeed, Y1066 forms a direct hydrogen bond to Cyt8 and also interacts with Cyt7 via pi-stacking. Other direct DNA base interactions at the primary site include K1030:Gua6′ (helix α1), H1011′:Gua10, P1012′:Cyt10′, and R1013′:Gua9 (N′-terminal arm). In addition to these direct interactions, numerous water-mediated hydrogen bonds contribute to indirect sequence recognition, and thus further increase the specificity of the interaction (Fig. 1D).
The observed asymmetry of the DNA implies that the two identical LANA molecules interact differently with the DNA. The symmetry-related equivalents of the above LANA moieties bind the DNA mainly at its backbone (Fig. 1C). Apart from stabilizing the overall complex, such interactions are generally known to support sequence-specific DNA recognition by sensing the sequence-dependent local flexibility of the DNA (18). Y1066′ on helix α2′ of chain B along with H1011, P1012, and R1013 on the N-terminal arm of chain A slightly distort the DNA backbone. This occurs at the only two consecutive Ade:Thy base pairs within LBS1. Moreover, the side chain of R1013 reaches into the minor groove of the DNA, where it promotes narrowing of that groove (19) and thus supports the backbone distortion. Direct DNA base interactions at this secondary site on LBS1 include K1030′:Gua17/Gua18 (helix α1), P1012:Gua12′, and R1013:Thy14 (N-terminal arm). The latter two occur in the minor groove. Consistent with these observations, it was previously found that DNA sequence substitutions within the secondary site on LBS1 affect LANA binding to a lesser extent than those in the primary site (17). Thus, the overall asymmetry of our complex structure is well supported by mutagenesis data.
The overall bend angle of LANA-bound LBS1 DNA was previously reported to be 57° (20). However, the absolute bend angle in our structure is only about 25°. This discrepancy can mainly be explained by crystal packing constraints and is unlikely to be related to the mutations in our protein (Fig. S3B). In fact, our mutant binds to LBS1 with KD = 72 nM (Fig. S3 E and F). This is well consistent with a previous measurement yielding KD = 85 nM for wild-type LANA(1011–1153) (9).
The Arrangement of LANA on the KSHV Minimal Replicator
The minimal replicator is the shortest cis-acting KSHV DNA sequence that confers LANA-dependent replication on plasmids (21). The 78-bp element lies in the TRs of the virus and includes the high-affinity LANA binding site LBS1 at its center. Next to LBS1, a second low-affinity site (LBS2) was previously recognized by an additional DNase I footprint on an ApaI fragment of the TR (10). Moreover, a 32-bp sequence on the other side of LBS1 was also found to be essential for replication (21). In unawareness of its precise function, this site was initially termed the replication element. Notably, the replication element was only partially covered by the DNase I footprinting probe that led to the identification of LBS2. Careful inspection of the DNA sequence revealed that the replication element harbors a third potential LANA binding site of inverse orientation (LBS3), which to our knowledge was previously not described (Fig. 2 A and B).
Fig. 2.
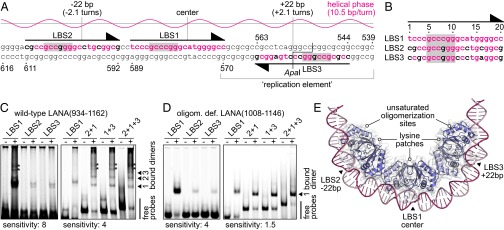
The three LANA binding sites of the KSHV minimal replicator. (A) Sequence of the minimal replicator with the helical phase for standard B-DNA shown above. The primary sites of DNA recognition are shaded in gray. Nucleotide positions identical to those in LBS1 are shown in red letters. Sequence numbering corresponds to a previous study (44) (GenBank: U86666.1). (B) Sequence alignment of the three LANA binding sites. Sequence numbering is as in Fig. 1. (C) EMSA using wild-type LANA(934–1162) (+) or GST as a control (−). An interpretation of the band patterns is provided on the right. Bands of unknown composition are marked with asterisks. These bands may reflect complexes with DNA bound to the lysine patch of LANA (compare Fig. 3). The left gel was exposed with higher sensitivity to visualize the weak bands of the low-affinity core sites. (D) EMSA using the oligomerization-deficient LANA(1008–1146) mutant (+) or GST as a control (−). An interpretation of the bands is provided on the right. The left gel was exposed with higher sensitivity. (E) Model of three LANA dimers bound to the minimal replicator, assuming that binding to LBS2 and LBS3 is structurally similar to LBS1.
We demonstrate binding of wild-type LANA(934–1162) to each of the three LANA binding sites (Fig. 2C, Left). Similarly, our oligomerization-deficient LANA(1008–1146) multiple-point mutant, which was used for cocrystallization with LBS1, also binds to each of the three sites independently (Fig. 2D, Left). Because the latter has a sufficiently high solubility in the presence of DNA, it was applied to affinity measurements using isothermal titration calorimetry (ITC; Fig. S3 E and F). These measurements demonstrate that LBS2 and LBS3 have approximately a hundredfold lower affinity than LBS1. This difference can readily be rationalized by the Cyt8Gua transversion within the primary site of sequence recognition in both LBS2 and LBS3 (17) (compare Fig. 1B). The other substitutions outside the primary sites of LBS2 and LBS3 may also contribute to the decreased affinity for LANA (Fig. 2B).
According to their spacing of 22 bp each, all of the three sites are located on the same face of the DNA (Fig. 2A). It is thus expected that the neighboring LANA dimers directly interact with each other through their hydrophobic lateral oligomerization sites (8, 9). These interactions can be assumed to cooperatively stabilize the whole complex (10) and to impose a strong overall bend on the minimal replicator (9, 15, 20) (Fig. 2E). To confirm cooperativity, electrophoretic mobility shift assays (EMSAs) were also performed with two (LBS2+1 or LBS1+3) and three (LBS2+1+3) LANA binding sites per probe. Using wild-type LANA(934–1162), this resulted in multiple LANA:DNA complexes, indicative of concurrent LANA binding to more than one site (Fig. 2C, Right). In contrast, our oligomerization-deficient LANA(1008–1146) mutant formed mainly single bands in comparable assays (Fig. 2D, Right), suggesting it preferentially binds to only one site per probe molecule. In conclusion, LANA oligomerization supports binding to both of the low-affinity sites. By sterical considerations, an interaction between LBS1-bound and LBS3-bound LANA dimers is only possible because of their asymmetrical DNA binding: if DNA binding was symmetrical, as in the case of EBNA-1, the proteins would bind on different faces of the DNA.
The LANA DBD Exhibits Two DNA-Binding Modes
In solution, the purified LANA DBD mainly exists as a dimer (8). However, previous crystal structures of the wild-type LANA DBD showed either pentameric or tetrameric rings of dimers (8, 9). Here, we present yet another crystal form of wild-type LANA(1013–1149), in which the molecules are arranged as a right-handed, continuous spiral of 4.8 LANA dimers per turn (Fig. 3 A and B and Table S1). All observed LANA oligomers are assembled via the same hydrophobic oligomerization interface that acts as a hinge, and thus allows for the observed intermolecular structural variability (Fig. S4C). Moreover, in all oligomeric LANA assemblies, the sequence-specific DNA-binding sites are located on the outward-directed surface (Fig. 3B, Left). However, the opposite side of the individual dimers is also highly positively charged: On the inside of all assemblies, patches of 14 lysine side chains per LANA dimer are aligned to form a continuous basic surface (Fig. 3B, Right and Fig. S4 A and B). Moreover, in both the rings and the spiral, the inner diameter is sufficiently large to accommodate double-stranded DNA. We thus hypothesized that the LANA DBD also has the capability to assemble into a ring or spiral around DNA, most probably with the positively charged lysine patches contacting the negatively charged DNA in a sequence-independent manner.
Fig. 3.

Structure of the LANA spiral. (A) Top view of the spiral crystal structure of wild-type LANA(1013–1149) shown in cartoon representation (Left) and in an electrostatic surface potential representation (Right). (B) Side view of a full helical repeat of 24 dimers showing the electrostatic surface potential on the outside (Left) and inside (Right) of the spiral. (C and D) Negative-stain EM of a specific DNA-binding-deficient LANA(1013–1162) multiple-point mutant bound to 207 bp/70 nm nonspecific DNA. Kinks in the regular complexes are marked in D. (E) A LANA DBD mutant as in C and D additionally carrying the A1121E mutation loosely bound to 207 bp/70 nm DNA. (F) A LANA DBD mutant as in C and D additionally carrying the mutations K1055E, K1109E, and K1138E in the presence of 207 bp/70 nm DNA. (G) The same mutant as in C and D in the absence of DNA.
To test this hypothesis, we used negative stain electron microscopy. Initial tests were performed on wild-type LANA DBD constructs in the presence of unspecific DNA fragments or viral TR DNA (Fig. S4 E–G). However, we found exclusively dense microaggregates of heterogeneous structure. To simplify the system, we next mutated the sequence-specific DNA-binding surface of the LANA DBD to abolish any DNA binding at this site. For this purpose, mutations R1013G, Y1014N, Q1016D, K1030D, R1032Q, Y1066S, K1069E, and K1070Q were incorporated into the LANA(1013–1162) sequence, which removes the key tyrosines and inverts the overall charge of the site (Fig. S1 A and B). This mutant, along with all other LANA DBD mutants used in this study, possesses a melting point above 68 °C, which indicates its structural integrity at room temperature (Fig. S1D). Similar to the wild-type LANA DBD (8), the mutants also form regular dimers in solution, as determined by small-angle X-ray scattering (Fig. S2).
This specific DNA-binding-deficient mutant was now found to form regular complexes on blunt-ended DNA of 207 bp (Fig. 3 C and D). The length of these spiral-like assemblies exceeds the length of a single DNA fragment, which is about 70 nm. This suggests that the properly ordered LANA oligomers can coat naked DNA via their basic interior surface, thus allowing for noncovalent concatamerization of individual complexes in vitro. Indeed, the complexes occasionally show kinks that likely indicate gaps between individual DNA molecules (arrowheads in Fig. 3D). As a control, we prepared two further LANA DBD mutants. First, an oligomerization-deficient variant of the LANA(1013–1162) mutant also contained the A1121E mutation (8). This variant did not form any regular complexes, but associated only loosely with the DNA molecules (Fig. 3E). Second, another variant of the LANA(1013–1162) mutant had a partial charge inversion at its lysine patch (K1055E, K1109E, K1138E). This variant did not form complexes with DNA at all (Fig. 3F). In addition, in the absence of DNA, the specific DNA-binding-deficient mutant did not form any higher-order assemblies (Fig. 3G).
This analysis shows that the LANA DBD can form ordered assemblies on DNA of arbitrary sequence in vitro. This property is dependent on LANA’s oligomerization site and its lysine patch. However, the arrangement of the DNA-induced LANA oligomers appears to be less regular than the spiral in our crystal structure, which also prevents a reliable deduction of helical parameters. This observation can readily be rationalized by the intrinsic variability of the oligomer interface (Fig. S4C) and the lack of crystal-packing restraints. Notably, both the oligomerization site and the lysine patch of LANA were previously found to be essential for higher oligomerization in pull-down assays, LANA-dependent plasmid replication, and the formation of the characteristic KSHV microdomains (“LANA speckles”) in cells (8, 9).
Discussion
KSHV LANA binds to its target DNA within the viral TRs in a sequence-specific manner. This interaction is considered a key event in KSHV latency, as it promotes latent viral replication, proper episome segregation, and transcriptional repression (22–24). Here, we found that LANA also exhibits a second, sequence-independent DNA-binding mode. For the purpose of structural investigation, we separated the two DNA-binding modes by rational mutagenesis.
The 3D crystal structure of the LANA:LBS1 complex provided the basis for a plausible model to describe the arrangement of three LANA dimers on the KSHV minimal replicator. In contrast to the symmetrical binding of EBV EBNA-1 and HPV E2 to their specific DNA-binding sites, LANA binds to the minimal replicator in an asymmetric manner. Binding of three LANA dimers, whose oligomerization is mediated by an oligomerization interface centered around alanine 1121 (8), is facilitated by the presence of three, rather than the hitherto known two, LANA binding sites (Fig. 2). LBS1 represents a high-affinity binding site for LANA, whereas LBS2 and LBS3 exhibit approximately a hundredfold lower binding affinity (Fig. S3 E and F). In this assembly of three LANA dimers on DNA, the protein complex presumably follows the shape of a semicircle, where the lysine patches of the individual dimers are oriented toward the center. Intriguingly, LANA can use these lysine patches as a secondary DNA-binding site for higher oligomerization into spiral-like complexes around DNA. The semicircular assembly of LANA on the minimal replicator structurally resembles a section of three consecutive LANA dimers from our spiral crystal structure (compare Figs. 2E and 3A).
We propose that the KSHV minimal replicator may act as a structural template for the nucleation of LANA spirals in trans (Fig. 4). Such spirals may assemble on remote sites in the viral genome that are not directly defined by their DNA sequence. Once formed, the nucleator complex may recruit more free and/or LBS-bound LANA dimers to cooperatively assemble a spiral of unknown length. Functionally, this self-assembly of LANA oligomers may be involved in the formation of the latent subnuclear KSHV microdomains (LANA speckles), which can be visualized by fluorescence microscopy (25–27). These microdomains possibly also contain a plethora of cellular proteins (28), many of which are known to directly interact with LANA. In addition, an oligomer might also incorporate LANA binding sites from within the host genome, which would further contribute to the physical link between virus and host chromatin (29, 30). Alternatively, the minimal replicator may induce LANA spiral assembly directly on unspecific host DNA, which would have a similar effect.
Fig. 4.
Hypothetical model for LANA spiral assembly. (A) A nucleator is formed once three LANA dimers bound to the minimal replicator catch a remote segment of free DNA on their lysine patches. (B) Recruitment of more LANA molecules leads to spiral formation in trans. Histones and chromatin structure are omitted for clarity.
In particular, the DNA-bound spiral-like assembly of LANA might represent a scaffold for association with other DNA-binding, spiral-shaped host assemblies such as, for example, the origin recognition complex or the DNA polymerase clamp loader (31, 32). Both of these complexes have been shown to directly interact with LANA and to facilitate latent viral replication (33–35). Hypothesizing that LANA spirals may be induced in trans, the initiation sites for latent KSHV replication would not necessarily have to be restricted to the TR region. Indeed, it was found that the replication initiation sites are distributed throughout the entire viral genome (36).
The actions of LANA on DNA may be controlled by CpG methylation of its specific target sites, posttranslational modification of the protein, and binding of interaction partners. In this context, it may be noted that two host proteins, bromodomain-containing proteins 2 and 4, were previously found to directly interact with both of LANA’s DNA-binding surfaces (8, 9). Future research will shed yet more light on the regulation of this multifunctional viral protein in its physiological environment.
Materials and Methods
Crystallization and X-Ray Structure Determination.
All protein samples for structural studies were prepared as described previously and carry the nonnative sequence Gly-Ser at their N terminus (8). Crystallization, X-ray data acquisition, and phasing of the LANA(1008–1146) mutant bound to LBS1 DNA was described previously (16) (Table S1). The dataset was recorded on beamline P11 of the PETRA III synchrotron, DESY, at T = 293 K. Refinement was done using Coot and Phenix.Refine (37, 38).
The spiral crystal structure of wild-type LANA(1013–1149) was obtained as follows: 0.4 µL of 31.5 mg/mL protein in 5 mM BisTris⋅Cl and 5 mM DTT at pH 6.5 were equilibrated against a reservoir solution of 170 mM lithium acetate and 18% (wt/vol) PEG3350 in a sitting drop setup at 20 °C. After 2 d, crystals were detached from the carrier surface by adding 1 µL of 2 M ammonium formate. The crystals were cryo-cooled in LN2 within 30 s after detachment. A number of more efficient cryoprotectants either failed to detach the crystals or led to their rapid disintegration. A dataset was collected at cryogenic temperature on beamline 14.1 of the BESSY II at the Helmholtz Zentrum Berlin (HZB), Germany (39). The dataset was processed up to the second ice ring at 3.70 Å, using XDS (40). The first ice ring in the resolution range between 3.93 and 3.87 Å was excluded from processing. Molecular replacement was performed with Phaser-MR, using chain A of PDB entry 2YPY (38). Refinement was carried out in Coot and Refmac, using reference model restraints from the MR model (37, 41) (Table S1).
The structural model in Fig. 2E was assembled in Coot, incorporating tailor-made DNA segments from the 3D-DART web server (42).
Interaction Studies.
EMSAs were carried out using untagged wild-type LANA(934–1162) or the oligomerization-deficient LANA(1008–1146) mutant in the presence of 5′ Dy682-labeled DNA probes (IBA Lifesciences) in a sample buffer of 30 mM Tris⋅Cl at pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM EDTA, 10% (vol/vol) glycerol, 0.25% Tween 20, 1 mM DTT, 0.5 mg/mL BSA, and 0.75 mg/mL poly(dI-dC), as described previously (8). The probes cover nucleotide positions 570–589 (LBS1), 592–611 (LBS2), 544–563 (LBS3), 570–611 (LBS2+1), 544–589 (LBS1+3), and 544–611 (LBS2+1+3) with respect to the sequence given in Fig. 2A. ITC is described under SI Experimental Procedures.
Second Mode DNA-Binding Reactions and Electron Microscopy.
The nonspecific DNA fragment was prepared by PCR using PfuTurbo DNA polymerase (Agilent), which creates exclusively blunt ends. The fragment covers positions 52–258 (207 bp) of plasmid pEGFP-N1 (Clontech, GenBank: U55762.1) and does not contain any KSHV-specific sequence. The viral TR DNA fragment was excised from the pGTR4 vector by NotI digest (43). Both DNA fragments were purified by ion exchange chromatography on a MonoQ column (GE Healthcare) and were subsequently desalted in 10-kDa MWCO spin filters. The protein samples were cleared from aggregates by centrifugation for 20 min at 109,000 × g directly before use. Binding reactions contained 5 µg/mL DNA with 25 µg/mL protein in 20 mM BisTris⋅Cl, 10 mM NaCl at pH 6.5. Mixtures were incubated at 4 °C overnight. Quality assessment of our various LANA DBD mutants by differential scanning fluorometry and small-angle X-ray scattering is described under SI Experimental Procedures.
For electron microscopy, the samples were adsorbed to a 40-nm carbon foil after partial withdrawal from the mica support, floating on a drop (40 µL) of reaction mixture for 30 s. After blotting with tipped filter paper, the carbon foil was transferred and detached completely from the mica, face down, onto a drop of 4% (wt/vol) uranyl acetate at pH 4.0. After about 5 s, the carbon foil was picked up with a reticulum foil layered Cu grid (300 mesh), was blotted thoroughly oblique over edge with filter paper, and was then air-dried. Samples were analyzed with an energy-filter transmission electron microscope (Libra 120 plus; Zeiss) in the elastic bright-field mode (energy slit-width, 12 eV; objective aperture, 60 µm; beam current, 1 µA; illumination aperture, 0.5 mrad) within a nominal magnification of ×25,000. Electron micrographs were recorded with a 2,048 × 2,048 16-bit bottom-mount CCD camera (SharpEye) under medium-dose conditions at −1.5 µm defocus.
Supplementary Material
Acknowledgments
We thank Dr. Andrea Scrima (Helmholtz Centre for Infection Research, Braunschweig, Germany) for sharing crystallization equipment. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) Collaborative Research Centre SFB900 “Chronic Infections: Microbial Persistence and Its Control” (to T.F.S.), by Helmholtz-Gemeinschaft Deutscher Forschungszentren Grant VH-GS-202 to the Helmholtz Centre for Infection Research GradSchool, and by DFG Emmy Noether Young Investigator Grant LU1471/3-1 (to T.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4UZB and 4UZC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421804112/-/DCSupplemental.
References
- 1.Schulz TF. The pleiotropic effects of Kaposi’s sarcoma herpesvirus. J Pathol. 2006;208(2):187–198. doi: 10.1002/path.1904. [DOI] [PubMed] [Google Scholar]
- 2.Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas ME, Kaye KM. The latency-associated nuclear antigen, a multifunctional protein central to Kaposi’s sarcoma-associated herpesvirus latency. Future Microbiol. 2011;6(12):1399–1413. doi: 10.2217/fmb.11.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma SC, Lan K, Robertson E. Structure and function of latency-associated nuclear antigen. Curr Top Microbiol Immunol. 2007;312:101–136. doi: 10.1007/978-3-540-34344-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rainbow L, et al. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71(8):5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kedes DH, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Invest. 1997;100(10):2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 8.Hellert J, et al. A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine gammaherpesvirus LANA proteins. PLoS Pathog. 2013;9(10):e1003640. doi: 10.1371/journal.ppat.1003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domsic JF, Chen HS, Lu F, Marmorstein R, Lieberman PM. Molecular basis for oligomeric-DNA binding and episome maintenance by KSHV LANA. PLoS Pathog. 2013;9(10):e1003672. doi: 10.1371/journal.ppat.1003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garber AC, Hu J, Renne R. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J Biol Chem. 2002;277(30):27401–27411. doi: 10.1074/jbc.M203489200. [DOI] [PubMed] [Google Scholar]
- 11.Kelley-Clarke B, et al. Determination of Kaposi’s sarcoma-associated herpesvirus C-terminal latency-associated nuclear antigen residues mediating chromosome association and DNA binding. J Virol. 2007;81(8):4348–4356. doi: 10.1128/JVI.01289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbera AJ, et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science. 2006;311(5762):856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 13.Bochkarev A, et al. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84(5):791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 14.Hegde RS, Grossman SR, Laimins LA, Sigler PB. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359(6395):505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 15.Han SJ, Hu J, Pierce B, Weng Z, Renne R. Mutational analysis of the latency-associated nuclear antigen DNA-binding domain of Kaposi’s sarcoma-associated herpesvirus reveals structural conservation among gammaherpesvirus origin-binding proteins. J Gen Virol. 2010;91(Pt 9):2203–2215. doi: 10.1099/vir.0.020958-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellert J, Krausze J, Schulz TF, Lührs T. Crystallization, room-temperature X-ray diffraction and preliminary analysis of Kaposi’s sarcoma herpesvirus LANA bound to DNA. Acta Crystallogr F Struct Biol Commun. 2014;70(Pt 11):1570–1574. doi: 10.1107/S2053230X14019906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan V, Komatsu T, Ballestas ME, Kaye KM. Definition of sequence requirements for latency-associated nuclear antigen 1 binding to Kaposi’s sarcoma-associated herpesvirus DNA. J Virol. 2004;78(24):14033–14038. doi: 10.1128/JVI.78.24.14033-14038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolstorukov MY, Jernigan RL, Zhurkin VB. Protein-DNA hydrophobic recognition in the minor groove is facilitated by sugar switching. J Mol Biol. 2004;337(1):65–76. doi: 10.1016/j.jmb.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong LY, Wilson AC. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen induces a strong bend on binding to terminal repeat DNA. J Virol. 2005;79(21):13829–13836. doi: 10.1128/JVI.79.21.13829-13836.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, Renne R. Characterization of the minimal replicator of Kaposi’s sarcoma-associated herpesvirus latent origin. J Virol. 2005;79(4):2637–2642. doi: 10.1128/JVI.79.4.2637-2642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J, Garber AC, Renne R. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J Virol. 2002;76(22):11677–11687. doi: 10.1128/JVI.76.22.11677-11687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284(5414):641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 24.Renne R, et al. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2001;75(1):458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao SJ, et al. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2(8):925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 26.Simpson GR, et al. Prevalence of Kaposi’s sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348(9035):1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 27.Kedes DH, et al. The seroepidemiology of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus): Distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2(8):918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 28.Kaul R, Verma SC, Robertson ES. Protein complexes associated with the Kaposi’s sarcoma-associated herpesvirus-encoded LANA. Virology. 2007;364(2):317–329. doi: 10.1016/j.virol.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercier A, Arias C, Madrid AS, Holdorf MM, Ganem D. Site-specific association with host and viral chromatin by Kaposi’s sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation. J Virol. 2014;88(12):6762–6777. doi: 10.1128/JVI.00268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J, et al. LANA binds to multiple active viral and cellular promoters and associates with the H3K4methyltransferase hSET1 complex. PLoS Pathog. 2014;10(7):e1004240. doi: 10.1371/journal.ppat.1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun J, et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20(8):944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429(6993):724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 33.Lim C, Sohn H, Lee D, Gwack Y, Choe J. Functional dissection of latency-associated nuclear antigen 1 of Kaposi’s sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J Virol. 2002;76(20):10320–10331. doi: 10.1128/JVI.76.20.10320-10331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma SC, Choudhuri T, Kaul R, Robertson ES. Latency-associated nuclear antigen (LANA) of Kaposi’s sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J Virol. 2006;80(5):2243–2256. doi: 10.1128/JVI.80.5.2243-2256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Q, et al. Kaposi’s sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence. Proc Natl Acad Sci USA. 2014;111(32):11816–11821. doi: 10.1073/pnas.1404219111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma SC, et al. Single molecule analysis of replicated DNA reveals the usage of multiple KSHV genome regions for latent replication. PLoS Pathog. 2011;7(11):e1002365. doi: 10.1371/journal.ppat.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat. 2012;19(Pt 3):442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.van Dijk M, Bonvin AM. 3D-DART: A DNA structure modelling server. Nucleic Acids Res. 2009;37(web server issue):W235–W239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grundhoff A, Ganem D. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J Virol. 2003;77(4):2779–2783. doi: 10.1128/JVI.77.4.2779-2783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus. Virology. 1997;236(1):147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.