Significance
Plant roots function as an interface between plants and the complex soil environment. Root systems of higher plants consist of different root types (RTs) that maximize their adaptive potential in heterogenous soil for nutrient uptake and anchorage. This study pioneers the molecular examination of individual RTs of adult rice root systems. The global signature of the transcriptional activity of each RT reveals significant quantitative and qualitative differences that predict functional diversity and specialization. Interaction with naturally prevalent beneficial mycorrhizal fungi profoundly modulated the relationship across the RTs such that the crown root transcriptome resembled that of lateral roots. The alteration of secondary cell wall synthesis in colonized roots is consistent with previously reported changes in root system architecture of mycorrhizal plants.
Keywords: rice, root system, arbuscular mycorrhizal symbiosis, transcriptome, secondary cell wall
Abstract
Root systems consist of different root types (RTs) with distinct developmental and functional characteristics. RTs may be individually reprogrammed in response to their microenvironment to maximize adaptive plasticity. Molecular understanding of such specific remodeling—although crucial for crop improvement—is limited. Here, RT-specific transcriptomes of adult rice crown, large and fine lateral roots were assessed, revealing molecular evidence for functional diversity among individual RTs. Of the three rice RTs, crown roots displayed a significant enrichment of transcripts associated with phytohormones and secondary cell wall (SCW) metabolism, whereas lateral RTs showed a greater accumulation of transcripts related to mineral transport. In nature, arbuscular mycorrhizal (AM) symbiosis represents the default state of most root systems and is known to modify root system architecture. Rice RTs become heterogeneously colonized by AM fungi, with large laterals preferentially entering into the association. However, RT-specific transcriptional responses to AM symbiosis were quantitatively most pronounced for crown roots despite their modest physical engagement in the interaction. Furthermore, colonized crown roots adopted an expression profile more related to mycorrhizal large lateral than to noncolonized crown roots, suggesting a fundamental reprogramming of crown root character. Among these changes, a significant reduction in SCW transcripts was observed that was correlated with an alteration of SCW composition as determined by mass spectrometry. The combined change in SCW, hormone- and transport-related transcript profiles across the RTs indicates a previously overlooked switch of functional relationships among RTs during AM symbiosis, with a potential impact on root system architecture and functioning.
Root systems of higher plants take up nutrients and water and provide anchorage to the ground. These functions are shared among distinct root types (RTs) of which the sturdier tap or crown roots are implicated in anchorage, and the finer, soil-exploring lateral roots in foraging for nutrients and water (1). However, although RT-specific growth responses to abiotic stimuli have been described (2), the molecular basis of this functional diversification within the root system remains poorly characterized. In recent years, significant efforts have been devoted to the examination of transcriptomes of individual root cell types of Arabidopsis thaliana and rice (3–6) and have provided important insight into root cell patterning and functional specialization. Such experiments have focused typically on young, developing tissue in the primary root. In contrast, the transcriptional differences among RTs from a mature root stock remain unknown. In addition to innate, within-root system diversity, root systems are asymmetrically influenced by the abiotic and biotic environment. For example, due to heterogeneities in the soil, each individual root might encounter different nutrient concentrations or soil moisture (7, 8). In addition, soil microorganisms release signaling molecules and induce changes in the microenvironment of the root through their physiological activities (9). The response of individual RTs to these alterations will have an important impact on plant survival. Understanding root system behavior at the RT level is therefore of great importance in rational breeding approaches for greater plant stress tolerance and crop productivity.
Most plants, including major agricultural crops, engage in evolutionarily ancient arbuscular mycorrhizal (AM) symbioses with glomeromycotan fungi. These fungi colonize the root intracellularly and increase plant mineral nutrition through uptake of nutrients via an extended extraradical hyphal network and subsequent release by highly branched hyphal structures, the arbuscules that colonize root cortex cells (10, 11). Typically, AM colonization is unevenly distributed within the root system (12). This is particularly evident in rice root systems that consist of three major RTs: weakly colonized crown roots (CRs), strongly colonized large lateral roots (LLRs), and fine lateral roots (FLRs), which are not colonized (13, 14). Differential colonization suggests that reciprocal root-fungal signaling events prior and during colonization, and plant physiological changes in response to colonization, are RT specific and likely impact nutrition and root system development. Indeed, AM colonization induces developmental responses that lead to changes in root system architecture (12). In rice, these responses are most pronounced in CRs, which grow longer and increase the production of lateral roots (13). Such proliferative root system changes potentially impact plant survival under stress conditions such as drought or nutrient scarcity.
The architectural changes of the rice root system during colonization and the difference in colonization pattern among the RTs prompted the hypothesis that the transcriptome of each rice RT must be differentially reprogrammed during AM colonization, resulting in distinct developmental and physiological responses. To address this hypothesis, we generated whole genome transcriptomic data for the three rice RTs, individually collected from root systems that were either noncolonized (NC) or colonized by the AM fungus Rhizophagus irregularis (M). We report substantial differences among the transcriptomes of rice CRs and lateral roots. CRs display high expression levels of large numbers of genes involved in hormonal activity and secondary cell wall (SCW) biosynthesis, whereas lateral roots show a stronger expression of certain nutrient transporters. Importantly, we observed RT-specific responses to AM colonization that included a switch in the class of hormone-related genes and a large and systemic decrease in SCW gene expression in CRs, accompanied by changes in cell wall phenolics.
Results
Global Patterns in the RT Transcriptome.
To monitor the transcriptomes of the RTs, CRs, LLRs, and FLRs were collected from mock inoculated and R. irregularis colonized mature root stocks of rice plants at V8, corresponding to late tillering stage (15). Significant variation was expected between biological replicates (SI Appendix, Fig. S1 A and B) because of the limited synchrony of (i) the developmental stages of each RT and of (ii) fungal colonization dynamics. Before collecting RTs, the root samples were examined microscopically to confirm efficient colonization of mycorrhizal roots and absence of colonization in control samples. In inoculated root systems, the LLRs were the most strongly colonized with a total root length colonization of 69.5% (±3.5%), followed by CRs with 48.2% (±7.2%), and FLRs lacking fungal colonization, confirming earlier observations (13).
Following an exploratory analysis (SI Appendix, Fig. S1 A and B), the number of genes being expressed in each RT was recorded (SI Appendix, Fig. S1C). About one-half to two-thirds of the genes were called “present” in the rice root systems of both NC and M conditions. The three RTs showed a large overlap of transcriptionally active genes across the two treatments. Interestingly, a significant number of genes were expressed in only one or two of the RTs, suggesting RT specificity in gene expression (SI Appendix, Fig. S1C).
Gene expression data (Dataset S1) were analyzed to identify significant changes (FDR 10%) among RTs and mycorrhizal condition using a linear model including RT, inoculation status, and their interaction. This analysis identified 8,980 distinct genes that accumulated differentially between at least one pair of treatments (Dataset S2).
Distinct RT Transcriptomes.
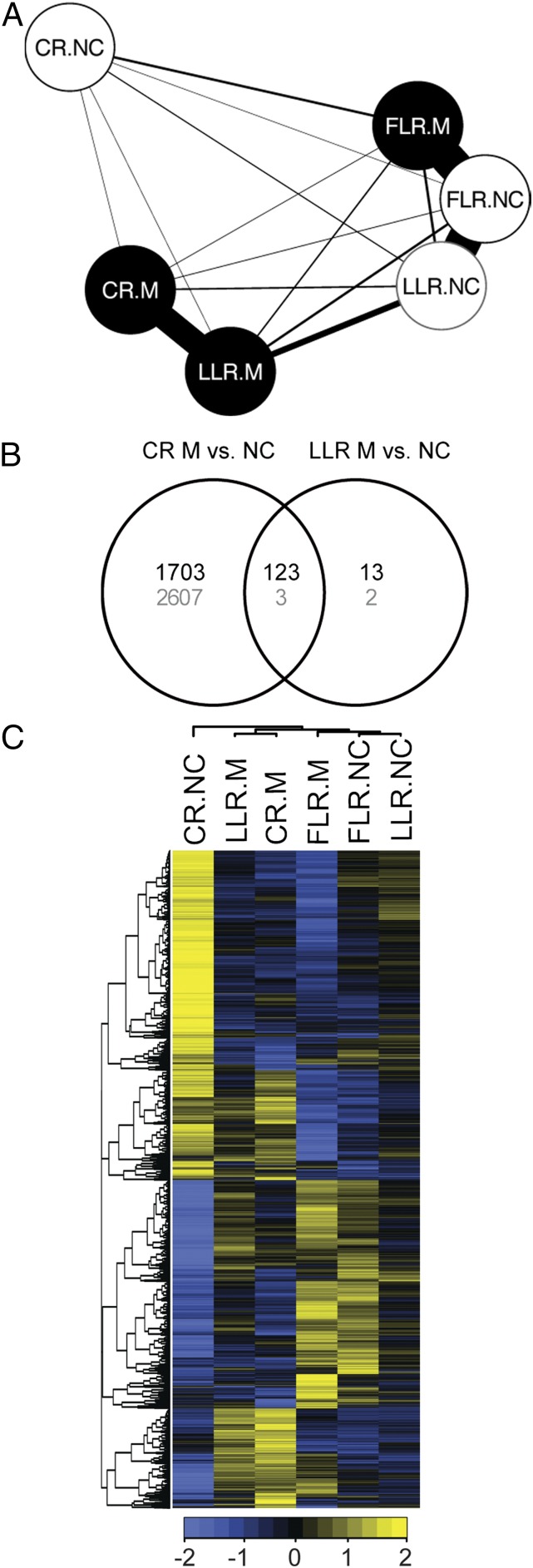
Examination of the transcriptional profiles of noncolonized RTs revealed that the expression profiles of LLRs and FLRs were highly similar (Fig. 1) and no gene was detected by our model to be significantly differently expressed between them (Dataset S2). This is surprising as FLRs differ from LLRs in their reduced number of cell layers and in their tissue composition. However, the transcriptional profiles of LLRs and FLRs were clearly distinct from that of CRs, with 5,591 transcripts accumulating to significantly different levels for the CR–FLR comparison and 1,179 transcripts for the CR–LLR comparison (Dataset S2 and Fig. 1), supporting the notion that CRs and LRs are functionally different (1).
Fig. 1.

Differentially expressed genes among rice root types. Relative expression (scale bar, Z-scale of SDs from the cross-sample mean; blue, below the mean; yellow, above the mean) of 5,630 genes that are differentially expressed (FDR 10%) in crown roots (CRs), large lateral roots (LLRs), and fine lateral roots (FLRs) of rice. Both genes and samples are clustered hierarchically. A large group of genes is distinguished by high relative expression in CRs.
Hormonal and transporter action underpin root system development and functioning (3). We therefore investigated expression patterns of genes associated with these activities. Mapping relative transcript abundance of differentially called genes across the RTs revealed a distinct profile for hormone (SI Appendix, Fig. S2A)- and transport (SI Appendix, Fig. S2B)-associated genes. For either group, relative transcript abundance was highest in CRs for the majority of genes. Among the hormone-related genes, those associated with the hormone auxin were overrepresented (39 of 68, SI Appendix, Fig. S2A), indicative of the main role CRs play during root system architectural changes (13, 16). In addition, the high abundance of transcripts encoding amino acid and peptide transporters in the CRs (23 of 30 significantly differentially expressed) points toward their pronounced contribution to organic as opposed to inorganic nitrogen transport (two of four ammonium transporters). LRs, on the contrary, showed an overall lower number of highly expressed genes. However, transcript levels of inorganic ion transporter-encoding genes were elevated for ammonium, chloride, and magnesium transporter genes among others (SI Appendix, Fig. S2B), thereby illustrating the importance of LRs for the uptake of these essential elements from the soil. LLRs produced an intermediate transcriptional signature, which may be explained by the tissue heterogeneity of this RT, with older aerenchymatic and younger, cortex-containing parts being similar to CRs and FLRs, respectively (15).
RT-Specific Transcriptome Changes in Response to AM.
Examining the impact of colonization by the AM fungus R. irregularis on the transcriptome of individual RTs revealed a total of 8,980 transcripts that accumulated differently in at least one pairwise comparison (Dataset S2). Of these, 76 genes were significant for the interaction between RT and mycorrhizal status, indicating root-type–specific responses to AM.
To reveal relationships among the transcriptional signatures of the three rice RTs, a network representation was produced, with connections inversely proportional to the number of differentially expressed transcripts among each pair of conditions (Fig. 2A). The relationship within the network representation helps illustrate the strong similarity between expression profiles of noncolonized LLRs and FLRs and their distinction from noncolonized CRs. In response to AM symbiosis, both colonized CRs and LLRs showed a significant shift in transcriptional profile (Fig. 2 A and C). Surprisingly, the number of transcripts responding to AM was dramatically higher in CRs (4,436 genes) than in LLRs (141 genes, Fig. 2B), although CRs were colonized to a significantly lower extent than LLRs. This large response brought the transcriptional signature of colonized CRs closer to that of mycorrhizal LLRs than to noncolonized CRs (Fig. 2 A and C and SI Appendix, Fig. S1B). This is a remarkable observation as it suggests that upon AM colonization CRs adopt a transcriptional profile with increased similarity to LLRs. In contrast, the expression signature of the FLRs from colonized root systems remained highly similar to that of noncolonized FLRs and LLRs (Fig. 2 A and C), as no significant transcriptional changes in response to AM colonization were detected in FLRs (10% FDR), consistent with the absence of colonization from this RT (Dataset S2).
Fig. 2.
Similarity among gene expression profiles of colonized and noncolonized rice root types. (A) Network representation of the number of differentially expressed genes (FDR 10%) between each type of sample (CR, crown roots; LLR, large lateral roots; FLR, fine lateral root; open circles, noncolonized; filled circles, mycorrhizal). Line width is inversely proportional to the number of significantly changing genes. (B) Number of genes that are significantly (FDR 10%) up- (black) or down-regulated (gray) in CRs or LLRs upon AM colonization. At FDR 10%, no gene was significantly regulated in FLRs. (C) Relative expression (scale bar, Z-scale of SDs from the cross-sample mean; blue, below the mean; yellow, above the mean) of the 8,980 differentially expressed genes (FDR 10%) in each sample. NC, noncolonized; M, mycorrhizal; root types as in A. Both genes and samples are clustered hierarchically. A large group of genes was distinguished by high relative expression in noncolonized crown roots (CR.NC).
The quantitatively strong response of CRs to AM colonization included a switch in the activities of genes belonging to different hormonal pathways, foremost auxin. The predominantly auxin-associated gene activity of noncolonized CR was suppressed in mycorrhizal CRs (SI Appendix, Fig. S3A), thereby suggesting a profound remodelling of root developmental processes upon AM colonization that may be linked with the well-documented changes in root system architecture of mycorrhizal plants (12, 13).
Transporter-encoding representatives of the same gene family frequently displayed opposing induction patterns between CRs and LLRs and the two inoculation conditions (SI Appendix, Fig. S3B). These families included, ammonium, sulfate, phosphate, and also amino acid and peptide transporters. Such complementary gene induction of different gene family members illustrates a broad shift of the physiologically alternative and specific uptake machineries across RTs and conditions and resembles the well-known expression pattern of phosphate transporter genes that constitutes the molecular basis for the switch from direct to mycorrhizal phosphate uptake upon root colonization with AM fungi (10).
Gene set enrichment analysis across all contrasts detected an overrepresentation of cell wall organization and biosynthesis, with higher gene expression in noncolonized CRs relative to LRs (Fig. 3A and Dataset S3). Closer examination revealed that most genes falling into this functional category were related to biosynthesis of SCW and the regulation thereof, hereafter referred to as “SCW genes” (Fig. 3 B and C, SI Appendix, Fig. S4A and Table S1, and Dataset S3). They included genes encoding the cellulose synthase subunits 4, 7, and 9, that have been linked to secondary cell wall synthesis (17), COBRAs involved in cellulose deposition at the secondary cell wall (18), and also genes participating in lignin biosynthesis, including phenylalanine ammonia lyase and a large number of peroxidase- and dirigent-encoding genes (19, 20) (SI Appendix, Fig. S4 A–C and Table S1, and Datasets S2 and S3). This strong expression of SCW genes lends molecular support to the importance of CRs in anchorage and stabilization of the plant (1).
Fig. 3.
Gene set enrichment analysis identifies cell-wall-biosynthesis–related genes. (A) Enrichment of gene ontology biological process for all contrasts. Relative gene set expression differences (scale bar, fold-change of gene sets; blue, down-regulated; yellow, up-regulated) are shown for all contrasts among noncolonized (NC) and mycorrhizal (M) crown roots (CRs), large lateral roots (LLRs), and fine lateral roots (FLRs). **FDR 10%, *FDR 25%. Microarray-based expression of (B) genes involved in cellulose biosynthesis or SCW modification and (C) genes involved in monolignol biosynthesis in NC and M CRs, large lateral roots (LLRs), and fine lateral roots (FLRs).
A cluster containing 2,607 transcripts was found to be present at high levels specifically in CRs of noncolonized plants and accumulated at reduced levels, comparable to the other root types, following AM colonization (Fig. 2B). This cluster largely drove the shifting relationship between colonized CRs and LLRs observed in the network matrix (Fig. 2A). Gene set enrichment analysis revealed that this cluster included the SCW genes found to be most highly expressed in CRs (Fig. 3 A–C, SI Appendix, Fig. S4 A–C, and Dataset S3). This large-scale RT-specific expression signature may have escaped notice in previous transcriptome analyses as a result of the limited spatial resolution afforded by sampling whole root systems.
Systemic Suppression of SCW Genes by AM.
Decreased expression of SCW genes in a specific RT is a previously unknown feature of root system responses to AM colonization. Therefore, we studied this phenomenon in more detail and addressed whether down-regulation of these genes in mycorrhizal CRs was restricted to colonized cells, to entire CRs containing colonized areas, or whether it occurred systemically. We focused on a set of five representative SCW genes and examined their expression in “split root” systems—a physical separation of the root system into two compartments, of which only one is inoculated with AM fungal spores. Following colonization [at 7 wk postinoculation (wpi), V8 stage of rice development] (15) of one side of the split root, transcripts of all five genes were down-regulated in CRs on both sides of the split root, indicating that AM repression of these genes was systemic and not spatially limited to colonization units (SI Appendix, Fig. S5A). In one of the biological replicates, only LLRs but not CRs established functional symbiosis, as indicated by transcript accumulation of the phosphate transporter 11 (PT11), a mycorrhiza-specific gene exclusively expressed in cells containing arbuscules and required for mycorrhizal phosphate uptake (21). Nonetheless, SCW genes were down-regulated in the CRs of this replicate, indicating that colonization of LLRs alone is sufficient to cause reduced expression of SCW genes in CRs.
To define the tissue domain in CRs in which the SCW gene expression was affected by AM inoculation, we analyzed the promoter activity of one of these genes, CesA4, which was reproducibly and strongly suppressed in mycorrhizal CRs (Fig. 4A), using a GUS reporter line (22). A growing rice plant has CRs of different ages and developmental stages. We categorized these into newly emerged (ECRs), mature (MCRs) and old crown roots (OCRs). In both NC and M plants, ECR that had not yet developed lateral roots, expressed pCesA4-GUS along the whole root length except the tip, whereas OCRs did not show any expression (Fig. 4B). In MCRs, GUS expression was observed in an extended zone behind the root tip. In mycorrhizal root systems, the area and strength of the GUS stain was strongly reduced (Fig. 4B), confirming our microarray and real-time RT-PCR expression analysis and indicating that CesA4 (and possibly other SCW genes) is down-regulated mainly in the newly growing tissue above the root tip, a tissue that later becomes colonized as this part of the root matures.
Fig. 4.
AM colonization reduces SCW marker gene activity and alters the composition of cell wall phenolics in distinct zones of CRs. (A) Real time RT-PCR–based expression pattern of the SCW marker gene CesA4 in noncolonized (NC) and mycorrhizal (M) crown roots (CRs), large lateral roots (LLRs), and fine lateral roots (FLRs). Means of two technical replicates are shown. (B) pCesA4-GUS expression in old (OCRs) and early emerging (ECRs) noncolonized (NC), and mycorrhizal (M) crown roots and in NC vs. M mature crown roots (MCRs). (Scale bars, 1 cm.) (C) Cell wall phenolic acid (lignin precursor) composition of tips (one-third of root length from apex of CRs) of NC and M root systems as determined by HPLC-multiple reaction monitoring (MRM) after alkaline hydrolysis followed by an acid hydrolysis. Data for each hydrolysis subfraction are displayed. Means ± SE of eight biological replicates consisting each of a pool of five root systems are shown in micrograms per gram of cell wall residues (CWRs). S, H, and G refer to the phenolic acid precursors of S, H, and G lignin. S refers to the sum of syringic and sinapic acid; H refers to the sum of 4-OH-benzaldehyde, 4-OH-benzoic acid, p-coumaric acid, and caffeic acid; and G refers to the sum of vanillic acid, ferulic acid, and coniferaldehyde. (D) Details of the measured extracts shown in C for all conditions where changes between NC and M were significant.
Suppression of SCW Genes Is Associated with an Established Symbiosis.
The reduced expression of SCW genes upon inoculation with R. irregularis could occur in response to early presymbiotic or to symbiotic signals (12). To distinguish between these possibilities we quantified transcript accumulation of the five SCW marker genes at an earlier and a later time point, at 3 and 7 wpi, respectively. Down-regulation of SCW genes was observed at 7 wpi, when symbiosis was fully functional, as reflected by the expression of PT11 in LLRs and CRs (SI Appendix, Fig. S5B). At 3 wpi, the five SCW genes were expressed at similar levels in CRs of mock- and AM fungi-inoculated root systems. A generally higher expression of the genes at 3 wpi compared with 7 wpi was also consistent with the strong pCesA4-GUS expression in ECRs (Fig. 4B). This indicated that down-regulation of SCW genes occurred in developmentally older MCRs, and/or required an established symbiosis.
In maize roots, phosphate deficiency is correlated with the induction of genes related to the biosynthesis of cell wall phenolics (23). Thus, improved phosphate nutrition through AM symbiosis (10) could be responsible for decreased SCW gene expression. Rice lines perturbed in PT11 function are unable to take up phosphate via AM (21). However, all representative SCW genes were down- regulated in colonized roots of pt11 RNAi plants (SI Appendix, Fig. S5C). These results indicate that the decreased SCW gene expression during AM is independent of mycorrhizal phosphate uptake.
AM Causes Changes in the Cell Wall Phenolic Acid Composition of CR Tips.
Down-regulation of SCW might presage differences in cell wall composition or levels of SCW metabolites in CRs after AM colonization. We quantified cell wall phenolic acids that are precursors of lignin (24), an important structural component of SCW. According to the pCesA4-GUS expression pattern under our assay conditions, we expected most changes to occur in the part of the CR proximal to the root tip. Therefore, we separated the lower third (“tip”) from the upper part (“top”) before chemical analysis. AM colonization was accompanied by a reduction of syringic acid, 4-OH-benzoic acid, p-coumaric acid, and ferulic acid in root system tips (Fig. 4 C and D), whereas no difference was found for tops (SI Appendix, Table S2). This finding was consistent with the reduced transcript accumulation of genes putatively involved in the biosynthesis of cell wall phenolics, such as genes encoding for instance phenylalanine ammonia lyase (Fig. 3C) and leads to the conclusion that the reduced accumulation of SCW transcripts in CRs during AM was indeed translated into a metabolic-developmental output. Transcription of SCW genes and the resulting SCW composition were predominantly affected in newly growing tissue (tip) following establishment of AM symbiosis.
Discussion
To gain a better understanding of functional differences among rice root types we performed root-type–specific transcriptional profiling of nonstimulated root systems and of root systems colonized by the AM fungus R. irregularis. In noncolonized root systems a strong divergence of the transcriptional profile of CRs from that of both lateral root types was detected, including a high accumulation of transcripts related to SCW biosynthesis, which is in line with the role of CRs in plant stabilization.
Whereas LRs exhibited a transcriptional activity consistent with a role in the uptake of mineral ions, the presence of significantly higher expressed genes encoding transport proteins for organic nitrogen (amino acid and peptide transporters) in CRs was unexpected and contrasted with the small number of inorganic nitrogen (ammonium) transporters. Plants take up soil nitrogen either in the inorganic form as ammonium and nitrate or in the organic form as amino acids, peptides, or proteins (25, 26). In natural ecosystems, the availability of organic nitrogen sources depends on the soil type and may exceed levels of ammonium and nitrate (25). Of the different RTs, rice LRs therefore mostly contribute to uptake of inorganic minerals, whereas CRs appear to predominantly participate in acquisition and/or partitioning of organic nitrogen within the plant (27).
Interestingly, CRs showed a highly divergent pattern of hormone-associated gene activities from lateral RTs. Notably, transcripts encoding auxin-signaling related genes were present at increased levels, suggesting a specific difference in the hormone status of CRs, which at the same time represent the RT with central adaptive significance for root system architecture. Indeed, mimicking a common ecosystem scenario by exposing rice roots to the beneficial AM fungus R. irregularis resulted in the strongest transcriptional response in CRs, altering accumulation of the large group of genes associated with auxin transport and signaling. This result is in line with earlier analyses of AM-induced root system architecture changes that showed CRs to undergo the most apparent developmental modifications, including an increased elongation and production of lateral roots (13). For example, a gain-of-function mutation in OsIAA11 blocks LR development in rice (28) and the reduced expression of OsIAA11 (and possibly other AUX/IAA genes) observed in colonized CRs might be associated with increased LR formation during AM symbiosis (13). Beneficial root-infecting ectomycorrhizal fungi (29–32) and the endophytic fungus Piriformospora indica (33) are known to produce auxin (IAA). However, the absence of genes related to auxin biosynthesis in the genome of R. irregularis (34) argues against AM fungi directly contributing to root auxin distribution. Instead, other factors likely cause changes in auxin signaling in CRs upon AM colonization.
In A. thaliana, a stimulation of LR development was attributed to cell wall softening (35). Consistent with these data and the observed changes in auxin-related genes, the specific down-regulation of SCW synthesis in CRs of mycorrhizal rice root systems might be associated with the observed increase in LR formation during AM symbiosis (12). Interestingly, in both systems cell wall modulation occurred in the tissue zone right behind the root tip, possibly leading to a relaxation of the physical wall constraints, thereby promoting lateral root formation. Colonization by R. irregularis exerted a systemic effect on SCW gene expression predominantly in mature CRs, indicating that an unknown systemic signal is triggered by AM formation. Remarkably, in the basal liverworts Conocephalum conicum, mycorrhizal colonization also leads to cell wall modifications as monitored by diminished autofluorescence, indicative of reduced cell wall phenolics (36). Modification of cell wall properties upon establishment of AM symbioses may therefore correspond to an evolutionarily conserved phenomenon. Because AM fungal genomes do not contain plant cell wall hydrolytic enzymes (34), the down-regulation of plant secondary cell wall genes might be key to AM colonization of CRs as cell wall loosening might facilitate intercellular fungal proliferation. Simultaneously or alternatively, it might be a consequence of carbon partitioning toward the microsymbiont, independent of fungal phosphate delivery (37, 38).
Our work reveals distinct root-organ transcriptional profiles in nonstimulated root systems and a combination of root-organ–specific and overlapping responses to AM colonization, demonstrating that a thorough understanding of root system remodelling in response to external cues will require separate examination of the constituent root types.
Materials and Methods
All rice lines arose in Oryza sativa ssp. japonica cv. Nipponbare. Root systems were dissected with forceps under a stereomicroscope. As CRs emerge directly from the shoot they are readily distinguished and sampled. To minimize developmental variability we sampled only MCRs and OCRs that had already developed lateral roots, whereas LR-free, newly emerging CRs (ECR) were avoided. LLRs and FLRs span diverse developmental stages and both emerge from a CR or LLR. Therefore, it is technically challenging to separate them. Four biological replicates, corresponding each to five pooled plants, were collected for individual RTs and treatments. Plant and fungal growth conditions, and also basic molecular biology methods followed standard protocols described in SI Appendix. Affymetrix Genechip hybridization and processing of the data followed routine manuals recommended by the manufacturers. A detailed description of the bioinformatics approach to both defining differentially expressed genes and gene set enrichment test is given in SI Appendix. Cell-wall–bound phenolic compounds were extracted according to the method reported in ref. 39 with minor modifications, explained in SI Appendix. Phenolic acids were quantified by LC-MS/MS following a protocol as detailed in SI Appendix.
Supplementary Material
Acknowledgments
We thank Yihua Zhou (Chinese Academy of Sciences) for providing seeds from the pCESA4-GUS rice line and members of the U.P. laboratory for help with root-type collection. Real-time RT-PCR analyses were conducted at the DNA Array Facility of Lausanne University. C.G. held fellowships from the Roche Research Foundation (Switzerland) and the Societé Academique Vaudoise (Switzerland). Research in the U.P. laboratory was supported by an Swiss National Science Foundation “Professeur Boursier” Grant PP00A-110874 and the Gatsby Charitable Foundation RG60824.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504142112/-/DCSupplemental.
References
- 1.McCully M, Canny M. Pathways and processes of water and nutrient uptake in roots. Plant Soil. 1988;111:159–170. [Google Scholar]
- 2.Tian H, De Smet I, Ding Z. Shaping a root system: Regulating lateral versus primary root growth. Trends Plant Sci. 2014;19(7):426–431. doi: 10.1016/j.tplants.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Takehisa H, et al. Genome-wide transcriptome dissection of the rice root system: Implications for developmental and physiological functions. Plant J. 2012;69(1):126–140. doi: 10.1111/j.1365-313X.2011.04777.x. [DOI] [PubMed] [Google Scholar]
- 4.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302(5652):1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 5.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 6.Dinneny JR, et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320(5878):942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- 7.Giehl RFH, Gruber BD, von Wirén N. It’s time to make changes: Modulation of root system architecture by nutrient signals. J Exp Bot. 2014;65(3):769–778. doi: 10.1093/jxb/ert421. [DOI] [PubMed] [Google Scholar]
- 8.Hodge A. Root decisions. Plant Cell Environ. 2009;32(6):628–640. doi: 10.1111/j.1365-3040.2008.01891.x. [DOI] [PubMed] [Google Scholar]
- 9.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 10.Smith SE, Smith FA. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu Rev Plant Biol. 2011;62:227–250. doi: 10.1146/annurev-arplant-042110-103846. [DOI] [PubMed] [Google Scholar]
- 11.Gutjahr C, Parniske M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Ann Rev Cell Dev Biol. 2013;29:593–617. doi: 10.1146/annurev-cellbio-101512-122413. [DOI] [PubMed] [Google Scholar]
- 12.Gutjahr C, Paszkowski U. Multiple control levels of root system remodeling in arbuscular mycorrhizal symbiosis. Front Plant Sci. 2013;4:204. doi: 10.3389/fpls.2013.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutjahr C, Casieri L, Paszkowski U. Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling. New Phytol. 2009;182(4):829–837. doi: 10.1111/j.1469-8137.2009.02839.x. [DOI] [PubMed] [Google Scholar]
- 14.Rebouillat J, et al. Molecular genetics of rice root development. Rice. 2009;2:15–34. [Google Scholar]
- 15.Counce P, Keisling T, Mitchell A. A uniform, objective and adaptive system for expressing rice development. Crop Sci. 2000;40:436–443. [Google Scholar]
- 16.Lavenus J, et al. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013;18(8):450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka K, et al. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 2003;133(1):73–83. doi: 10.1104/pp.103.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, et al. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell. 2003;15(9):2020–2031. doi: 10.1105/tpc.011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153(2):402–412. doi: 10.1016/j.cell.2013.02.045. [DOI] [PubMed] [Google Scholar]
- 20.Hosmani PS, et al. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc Natl Acad Sci USA. 2013;110(35):14498–14503. doi: 10.1073/pnas.1308412110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S-Y, et al. Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell. 2012;24(10):4236–4251. doi: 10.1105/tpc.112.104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, et al. A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol Biol. 2009;71(4-5):509–524. doi: 10.1007/s11103-009-9536-4. [DOI] [PubMed] [Google Scholar]
- 23.Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L. Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot. 2008;59(9):2479–2497. doi: 10.1093/jxb/ern115. [DOI] [PubMed] [Google Scholar]
- 24.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 25.Näsholm T, Kielland K, Ganeteg U. Uptake of organic nitrogen by plants. New Phytol. 2009;182(1):31–48. doi: 10.1111/j.1469-8137.2008.02751.x. [DOI] [PubMed] [Google Scholar]
- 26.Tegeder M, Rentsch D. Uptake and partitioning of amino acids and peptides. Mol Plant. 2010;3(6):997–1011. doi: 10.1093/mp/ssq047. [DOI] [PubMed] [Google Scholar]
- 27.Okumoto S, Pilot G. Amino acid export in plants: A missing link in nitrogen cycling. Mol Plant. 2011;4(3):453–463. doi: 10.1093/mp/ssr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z-X, et al. A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol Plant. 2012;5(1):154–161. doi: 10.1093/mp/ssr074. [DOI] [PubMed] [Google Scholar]
- 29.Gay G, Normand L, Marmeisse R, Sotta B, Debaud JC. Auxin overproducer mutants of Hebeloma cylindrosporum Romagnesi have increased mycorrhizal activity. New Phytol. 1994;128:645–657. [Google Scholar]
- 30.Splivallo R, Fischer U, Göbel C, Feussner I, Karlovsky P. Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol. 2009;150(4):2018–2029. doi: 10.1104/pp.109.141325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frankenberger WT, Poth M. Biosynthesis of indole-3-acetic acid by the pine ectomycorrhizal fungus Pisolithus tinctorius. Appl Environ Microbiol. 1987;53(12):2908–2913. doi: 10.1128/aem.53.12.2908-2913.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ek M, Ljungquist PO, Stenstr ÖME. Indole-3-acetic acid production by mycorrhizal fungi determined by gas chromatography-mass spectrometry. New Phytol. 1983;94:401–407. [Google Scholar]
- 33.Sirrenberg A, et al. Piriformospora indica affects plant growth by auxin production. Physiol Plant. 2007;131(4):581–589. doi: 10.1111/j.1399-3054.2007.00983.x. [DOI] [PubMed] [Google Scholar]
- 34.Tisserant E, et al. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci USA. 2013;110(50):20117–20122. doi: 10.1073/pnas.1313452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roycewicz PS, Malamy JE. Cell wall properties play an important role in the emergence of lateral root primordia from the parent root. J Exp Bot. 2014;65(8):2057–2069. doi: 10.1093/jxb/eru056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ligrone R, et al. Glomeromycotean associations in liverworts: A molecular, cellular, and taxonomic analysis. Am J Bot. 2007;94(11):1756–1777. doi: 10.3732/ajb.94.11.1756. [DOI] [PubMed] [Google Scholar]
- 37.Kiers ET, et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science. 2011;333(6044):880–882. doi: 10.1126/science.1208473. [DOI] [PubMed] [Google Scholar]
- 38.Walder F, et al. Mycorrhizal networks: common goods of plants shared under unequal terms of trade. Plant Physiol. 2012;159(2):789–797. doi: 10.1104/pp.112.195727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattila P, Kumpulainen J. Determination of free and total phenolic acids in plant-derived foods by HPLC with diode-array detection. J Agric Food Chem. 2002;50(13):3660–3667. doi: 10.1021/jf020028p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.