Significance
Spatiotemporal characterization of molecular expression during embryonic development is critical for understanding how cells become different and give rise to distinct tissues and organs. Technological advances enabled the measurement of RNAs and proteins in single cells of embryos, but there is very little information on small molecules, metabolites, that are the ultimate indication of the physiological state. To fill this knowledge gap, we developed and used a single-cell technology to find that embryonic cells that give rise to different tissues have characteristically different metabolic signatures that are not simply a reflection of cell pigmentation, yolk content, size, or position in the embryo, but also affect cell fate. This approach is likely to provide new mechanistic insights into early embryo development.
Keywords: single cell, mass spectrometry, metabolomics, embryo development, Xenopus
Abstract
Spatial and temporal changes in molecular expression are essential to embryonic development, and their characterization is critical to understand mechanisms by which cells acquire different phenotypes. Although technological advances have made it possible to quantify expression of large molecules during embryogenesis, little information is available on metabolites, the ultimate indicator of physiological activity of the cell. Here, we demonstrate that single-cell capillary electrophoresis-electrospray ionization mass spectrometry is able to test whether differential expression of the genome translates to the domain of metabolites between single embryonic cells. Dissection of three different cell types with distinct tissue fates from 16-cell embryos of the South African clawed frog (Xenopus laevis) and microextraction of their metabolomes enabled the identification of 40 metabolites that anchored interconnected central metabolic networks. Relative quantitation revealed that several metabolites were differentially active between the cell types in the wild-type, unperturbed embryos. Altering postfertilization cytoplasmic movements that perturb dorsal development confirmed that these three cells have characteristic small-molecular activity already at cleavage stages as a result of cell type and not differences in pigmentation, yolk content, cell size, or position in the embryo. Changing the metabolite concentration caused changes in cell movements at gastrulation that also altered the tissue fates of these cells, demonstrating that the metabolome affects cell phenotypes in the embryo.
The ability to understand the basic mechanisms that regulate embryonic development requires knowledge of the complete suite of biomolecules expressed by each cell in the organism. Although technological advances in single-cell isolation, genome sequencing, and transcriptome analyses have made it possible to determine spatial and temporal changes for large molecules (1, 2), little is known about small-molecular events that unfold in the individual cells (blastomeres) of the embryo. In many animals, mRNAs and proteins synthesized during oogenesis are sequestered to different cytoplasmic domains, which after fertilization then specify the germ-cell lineage and determine the anterior–posterior and dorsal–ventral axes of the embryo. For example, in the South African clawed frog (Xenopus laevis), several mRNAs are localized to the animal pole region (Fig. 1), which later gives rise to the embryonic ectoderm and the nervous system (3), whereas VegT mRNA localization to the vegetal pole specifies endoderm formation (4), and region-specific relocalization of the Wnt and Dsh maternal proteins govern the dorsal–ventral patterning of the embryo (5). However, there is abundant evidence that in developing systems not all transcripts are translated into proteins, and not all proteins are active; therefore, analyses of the mRNAs and proteins within single cells may not reveal the activity state of the cell. In fact, different animal blastomeres of the 16-cell Xenopus embryo that are transcriptionally silent can have very different potentials to give rise to neural tissues (6, 7), even though they seem to express common mRNAs (3). For a deeper understanding of the developmental processes that govern cell-type specification, it would be transformative to assay the activity state of embryonic cells downstream of transcription and translation, at the level of the metabolome, the complete suite of small molecules produced by the cell.
Fig. 1.
Our experimental workflow to uncover small-molecular activity during early-stage embryo development. Single blastomeres were identified and dissected from 16-cell frog (X. laevis) embryos and their metabolomes extracted and measured by a custom-designed single-cell CE-µESI-MS. (Scale bars, 250 µm.)
How transcriptional information translates into small-molecular activity, particularly metabolites, while the different blastomeres commit to specific fates is completely unknown because of a lack of technology capable of measuring such small molecules [molecular weight (MW) <1,500 Da] in single blastomeres. Recent high-resolution transcriptional profiling found multiple waves of activity during early embryogenesis (8), and whole-embryo analyses revealed that transcriptomic events are accompanied by gross proteomic and metabolic changes during the development of Xenopus (9–13) and zebrafish (Danio rerio) (14, 15), raising the question whether these chemical changes are heterogeneous also between individual cells of the embryo at the different developmental stages. However, the challenge has been to collect high-quality signal from the miniscule amounts of small molecules contained within single blastomeres for analysis by MS, the analytical technique of choice for these small molecules. Because different blastomeres in Xenopus give rise to different tissues (16), elucidating the metabolome in individual cells of the embryo holds a great potential to elevate our understanding of the cellular physiology that regulates embryogenesis. The metabolome is particularly informative of a cell’s state because it is highly dynamic, diverse, and sensitive to intrinsic and extrinsic factors. However, to enable the measurement of the metabolome in individual blastomeres, new MS technologies and protocols are needed with the capability to address single cells.
Technological innovations have only recently made it possible to use small-molecular MS for the measurement of single cells, opening new research possibilities in biology. Unlike conventional MS that seeks high coverage of the metabolome by averaging together a large number, often millions, of cells, single-cell MS technologies are purposed to characterize biomolecular events in a cell-specific manner (17–21). For example, targeted experiments by microarrays for MS recently probed the metabolic mechanism of perturbation in yeast cells that were masked by traditional population-averaging approaches (22), and atmospheric-pressure laser desorption/ablation (13, 23) and direct microsampling electrospray ionization (ESI) (24, 25) have also found differences between single cells. Encouraged by the earlier success of capillary electrophoresis (CE) MS in the proteomic analysis of single erythrocytes (26–28), we have recently extended single-cell microdissection and CE-microflow ESI (µESI) MS to small molecules to broaden the coverage of the metabolome via chemical separation (29). By removing detection interferences, single-cell CE-µESI-MS was able to detect various small molecules including neurotransmitters in single neurons of the Aplysia californica. The success of these experiments indicates that single-cell MS serves as a powerful analytical tool for studying the biochemical machinery in the basic functioning building block: the cell.
Here, we demonstrate using single-cell CE-µESI-MS that differential expression of the genome translates to the domain of small metabolites in embryonic cells at the earliest stages of development. Our study uses Xenopus, a favored model for molecular, cell, and developmental biological inquiries, to measure the production of small molecules in central metabolic networks that are essential precursors to the formation of other metabolites as well as peptides and proteins. We selected three different types of blastomeres from the 16-cell embryo (Fig. 1) because they are still relatively large (∼250 µm in spherical diameter with ∼90-nL cell volume) at this developmental stage, are readily identifiable by position and pigmentation, which enables consistent isolation of the same cell from different embryos, and they have distinct developmental fates: The midline dorsal-animal cell (named D11) gives rise primarily to the retina and brain, the midline ventral-animal cell (named V11) gives rise primarily to the head and the trunk epidermis, and the midline ventral-vegetal cell (named V21) gives rise primarily to the hindgut (16) (Fig. 1). Using single-cell CE-µESI-MS and functional experiments including cell lineate tracking, we find that different blastomeres foster cell type-specific metabolites that are able to define cell fate. The data provided herein can be used to give a functional interpretation to existing information on the transcriptome and proteome. Besides providing previously inaccessible information about the activity state of these cells, the results presented here demonstrate that single-cell MS, for example, using CE-µESI-MS, makes it now possible to ask new types of questions for obtaining a broader understanding of cellular processes during development.
Results
Central Metabolic Networks in Single Blastomeres.
Our overall approach to determine small-molecular activity in three different blastomere types is presented in Fig. 1. We isolated the D11, V11, and V21 blastomeres randomly from the left or right sides of embryos produced from two mothers and two fathers from an outbred Xenopus colony, even though this experimental design was likely to increase the biological variability in the single-cell MS data. For each cell type, n = 5 blastomeres (biological replicates) were manually dissected from different embryos (totaling 15 single blastomeres from 15 different embryos) to ensure statistical confidence and also to avoid interblastomere biases based on a common embryo origin. Each blastomere was assigned a unique identifier to help interpret measurement results based on cell type and identity, although these identifiers were not directly used during multivariate data analysis. This experimental design allowed us to ask whether different blastomere types foster characteristic metabolomes.
Small-molecular activity of the blastomeres was characterized by measuring metabolites that formed known central metabolic pathways. Following isolation, small metabolites (MW <500 Da) were extracted from each blastomere in 10 µL of 50% (vol/vol) methanol containing 0.5% (vol/vol) acetic acid. A ∼10-nL volume of the extracts (samples), equivalent to <0.1% of the total volume of the extract or ∼10% of the single blastomere volume, were measured in technical duplicate–quadruplet for four of the n = 5 biological replicates using a single-cell CE-µESI-MS that was built based on our prototype (29). A detailed account of the technology and validation of its analytical performance is provided in SI Appendix. The lower limit of detection of the system was below 10 nM, or 60 amol, for various metabolites, and the measurement reproducibility was ∼5% relative SD (RSD) for separation time with <25% RSD (SI Appendix, Fig. S1) for quantitation (SI Appendix). Because several metabolites accumulate at micromolar–millimolar concentration in whole Xenopus embryos (12), these analytical metrics were sufficient to quantify the production of endogenous small molecules at their native concentrations between single Xenopus blastomeres.
A large number of small molecules were detected in the single blastomeres despite the limited amount of material contained by them. More than 80 different molecular features corresponding to different small molecules were detected in all of the blastomeres (SI Appendix, Table S1). SI Appendix, Fig. S2 exemplifies the separation of more than 15 different small molecules in one measurement. These molecular features were assigned to a total of 40 distinct endogenous small molecules (SI Appendix, Table S2) with exceptional confidence via a multipronged approach, as detailed in SI Appendix and presented in SI Appendix, Fig. S3, encompassing nearly all essential and nonessential amino acids, osmolites, and classical neurotransmitters. This level of coverage of the metabolome from a single blastomere in the 16-cell embryo compares favorably with ∼50 metabolites that were recently identified in a whole Xenopus egg and embryo using liquid chromatography MS (12).
The identified metabolites underpinned central metabolic networks. A comparison of the data (SI Appendix, Table S2) with the Kyoto Encyclopedia of Genes and Genomes (30) found ∼90% of the identified small molecules to be interconnected members of major metabolic pathways in mammalian systems. One such network is reconstructed for Xenopus in SI Appendix, Fig. S2 based on the measured data. Several of the small molecules were shared among the pathways, creating nodes in the network. In agreement, recent experiments on whole Xenopus embryos eloquently demonstrated alanine to be the energy source that rapidly converts to aspartate and glutamate, which then fluxes to the synthesis of multiple amino acids including valine, proline, and glutamine (12). Thus, our measurements captured an information-rich snapshot of small-molecule activity in the blastomeres, providing data to complement already available transcriptomic and some proteomic information in single blastomeres and metabolic and proteomic expression in the whole embryo of Xenopus (11, 12).
Blastomere Type-Specific Small-Molecular Activity.
We performed multivariate analysis to compare small-molecular production between the individual blastomeres. Fig. 2 shows the unsupervised hierarchical cluster analysis (HCA) and heat map of D11, V11, and V21 blastomere extracts, computed based on the relative abundance of 40 of the statistically most significant metabolites. In the resulting dendrogram, the samples formed three main groups, this grouping corresponding to differences in cell type (top axis) and small-molecular composition (left axis), essentially highlighting separate islands in the heat map. Finer features are also apparent within each group of the dendrogram, in cell type and molecular composition. For example, the D111, V111, and V212, and V215 clustered separately from the other blastomeres in the same type with further subclustering evident at lower levels. Elimination of the V111 or D111 or both blastomeres left the clustering of the blastomere extracts unaffected based on cell type (SI Appendix, Fig. S4), demonstrating that all of the measured cells had cell type-characteristic metabolic activity. It is possible that these finer chemical differences are linked to embryo origin from different parents and/or differences between blastomeres collected from the left or right side of the embryo. Supporting this notion, different female frogs have been found to lay eggs with detectable differences in their total metabolome (12), and previous studies have noted left–right asymmetry in serotonin composition and ion flow in the 16-cell embryo (31, 32). Despite these small differences, the data presented herein establish, for the first time to our knowledge, that the small-molecular cell composition of individual cells is heterogeneous in the early-stage embryo; the D11, V11, and V21 blastomeres foster distinct metabolomes, and these differences are reproducible for multiple embryos of different parental origins (n = 5).
Fig. 2.
False-color heat map and hierarchical clustering of small molecules detected in single D11, V11, and V21 blastomeres of 16-cell Xenopus embryos. Metabolite abundances are normalized relative to the mean and shown in false color for the 40 statistically most significant features (P < 0.05). Each blastomere has a unique identifier (bottom axis) with solid lines connecting the technical replicates for four of the n = 5 biological replicates that were measured for each cell type. The data reveal that D11, V11, and V21 cells have distinct small-molecular activities during early embryogenesis. Abbreviations of small molecules are given in SI Appendix, Table S2.
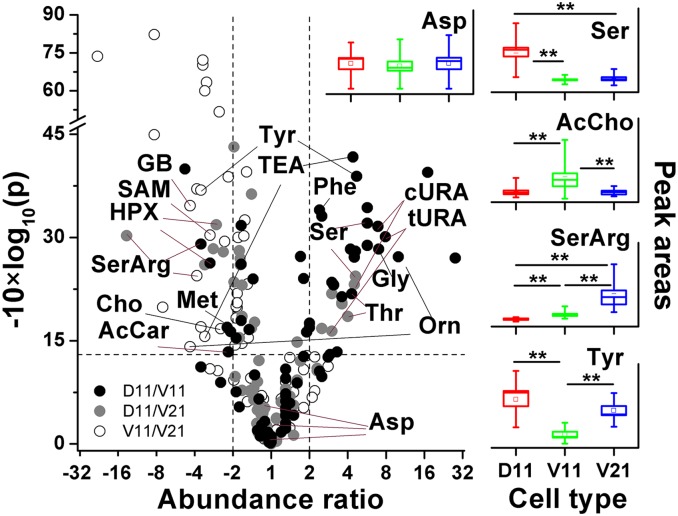
The observed differences in small-molecular activity were further evaluated in the context of the dorsal–ventral–animal–vegetal domains of the embryo. Fig. 3 shows the correlation between statistical significance (P value) and biological significance (fold change) for all detected molecular features (identified or unidentified) on the basis of selected ion electropherographic peak areas measuring relative concentrations. Small-molecular activity was compared between the animal dorsal-to-animal ventral region by D11-to-V11 ratios (D11/V11), the animal dorsal-to-vegetal ventral region by D11-to-V21 (D11/V21) ratios, and the animal ventral-to-vegetal ventral region by V11-to-V21 (V11/V21) ratios. A number of metabolites were produced in indistinguishable abundance between these regions (P > 0.05 or fold change <2.0). For example, aspartate (Fig. 3, Left), glutamate, and glutamine were present in comparable amounts in the extracts, indicating that part of the Ala–Asp–Glu and connected pathways (SI Appendix, Fig. S2) were independent of the type of cell, its physical location, or its activity.
Fig. 3.
Significant differences in the metabolic activity of blastomeres in the animal–vegetal–dorsal–ventral domains of the embryo. (Left) Each data point plots the metabolite signal abundance ratio vs. statistical significance for different small molecules between the D11/V11 (black filled circle), D11/V21 (gray filled circle), and V11/V21 (open circle) cell types. (Right) Box-whisker plots compare cases of select metabolites that have similar and biologically statistically significantly different concentrations in the D11, V11, and V21 blastomeres (square is mean, box is 1 × SE, and whisker is 1.5 × SE.) Statistical significance is marked at *P < 0.05 and **P < 0.005. Small molecules are listed in SI Appendix, Table S2.
In contrast, there were complex patterns of differential activity for a number of other small molecules between the cells. As shown in Fig. 4, a total of 27 different compounds between D11/V11, 15 between D11/V21, and 20 between V11/V21 had different concentrations in the animal–dorsal, animal–ventral, or vegetal–ventral quadrants. For example, the D11 blastomeres were markedly approximately three to seven times richer in serine, threonine, cis-urocanate, and trans-urocanate than V11 and V21 and contained approximately eight times more glycine than V11. These data suggest that the Ser–Gly–Thr pathway (SI Appendix, Fig. S2) is the most active in the D11 blastomeres, in the animal–dorsal quadrant. Interestingly, the V11 and V21 cells accumulated more hypoxanthine than D11, suggesting up-regulation of pathways producing this metabolite on the ventral side of the embryo. V11 cells also accumulated approximately two to five times higher amounts of choline, acetylcholine (Fig. 3, Right), acetylcarnitine, N6,N6,N6-trimethyllysine, and glycine betaine than V21 and D11, showing up-regulation of pathways producing these metabolites in the animal–ventral quadrant. Furthermore, V21 blastomeres contained approximately three times more S-adenosylmethionine and ∼4–16 times more Ser–Arg (Fig. 3, Right) than the other two cell types, revealing up-regulation of pathways producing these small molecules in the vegetal-ventral quadrant. In contrast, tyrosine (Fig. 3, Right), triethanolamine, and ornithine exhibited biregional enrichment, giving higher signal intensity in the D11 and V21 blastomeres. These data capture surprising chemical cell heterogeneity in 16-cell embryos, which may be driven by a single or multiple factors such as cell type (Fig. 1), location in an electrochemical field (32), or physical properties, such as cell size (V21 is larger than D11 or V11), amount and size of yolk platelets (V21 contains more and larger yolk platelets compared with D11 and V11), and pigmentation (V11 is darkly pigmented, D11 is moderately pigmented, and V21 does not contain pigment granules).
Fig. 4.
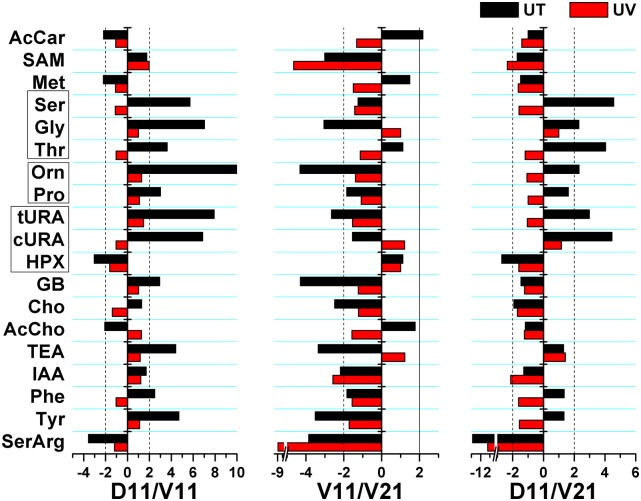
Functional evaluation of small-molecular differences between D11, V11, and V21 blastomeres. Differential metabolic activity between the D11/V11 and D11/V21 blastomere types that is characteristic of the untreated (UT, black) embryo was lost for most metabolites in UV light-ventralized (UV, red) embryos. These results establish that the small-molecular cell heterogeneity in the 16-cell embryo is driven by blastomere type rather than the location, size, or pigmentation of the cells. Small molecules are listed in SI Appendix, Table S2. Statistical and biological significances are tabulated in SI Appendix, Table S3.
To test whether the different metabolomes of the D11, V11, and V21 blastomeres result from their different sizes, locations, or pigmentation, or from their different contents of the original egg cytoplasm, we performed a similar analysis on embryos that were ventralized by exposure to UV light. It is well established in Xenopus that the entry of the sperm into the oocyte at fertilization causes the rotation of the cortical cytoplasm in a ventral–animal to dorsal–vegetal direction (33). Exposing the vegetal pole of eggs to UV light within 40 min of fertilization inhibits the cortical cytoplasmic rotation that is required for dorsal axis formation, resulting in embryos that are missing dorsal–anterior structures, particularly those derived from the D11 blastomere (33). We exposed several hundred embryos derived from the same parents to UV irradiation according to standard protocols (SI Appendix). When they reached the 16-cell stage, D11, V11, and V21 blastomeres were dissected from different embryos and processed as detailed earlier. Their siblings, both UV-treated and untreated controls from the same fertilization, were raised to larval stages to monitor the degree of ventralization. Most of the UV-treated embryos (>90%; SI Appendix) were missing head structures and were composed of a characteristically large belly piece and minimal dorsal trunk tissues; none of the sibling controls showed this phenotype. However, at the 16-cell-stage, UV-irradiated embryos and their dissected blastomeres were indistinguishable from those of their untreated siblings. For both sets of embryos, V21 blastomeres were not pigmented, V11 blastomeres were more pigmented than D11, and V21 blastomeres were larger than D11 or V11. Hence, any small-molecular differences measured between blastomeres in the UV-treated embryos compared with the control would not result from differences in size, position, or pigmentation.
After UV-treated embryos reached the 16-cell stage, a total of n = 5–7 blastomeres (biological replicates) were isolated for each of the D11, V11, and V21 cell types from different embryos (without distinguishing the left–right origin of the cells), and their small-molecule composition was determined using the CE-µESI-MS instrument. Unlike in the untreated blastomeres, HCA calculated on the relative abundance of the ∼40 identified metabolites found random clustering between the samples based on cell type or molecular features, essentially painting a heat map with no discernable features (SI Appendix, Fig. S5). Fig. 4 compares the relative abundance of a subset of these identified small molecules between the untreated- and UV light-treated blastomeres that were differentially active between the D11/V11, D11/V21, or V11/V21 cell types (SI Appendix, Table S2). The data reveal that differential regulation of these small molecules that naturally dominated the D11/V11 and D11/V21 blastomere pairs in the untreated embryos were lost in the ventralized embryos beyond the limit of detection of CE-µESI-MS. Most notable are the Ser–Gly–Thr and the urocanate–hypoxanthine pathways that were expressed to a similar extent in all blastomeres following UV treatment. Interestingly, ventralization further increased the abundance of S-adenosylmethionine, hypoxanthine, and serine–arginine between the V11/V21 cell types. This suggests that small-molecular polarity was enhanced in the animal–vegetal axis on the ventral side of the embryo, which correlates with the lack of formation of dorsal tissues. As the 16-cell embryos were visually indistinguishable based on physical appearance, this experimental manipulation confirms that differential small-molecular activity between the D11, V11, and V21 cells is due to cell type rather than their size, location, or extent of pigmentation.
Developmental Significance of Metabolite Differences.
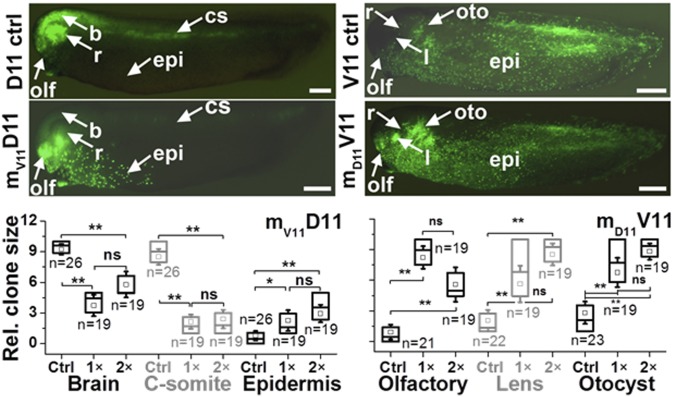
To determine whether the small-molecular differences quantified between the different blastomere types have a developmental significance, we tested the effect of a subset of metabolites on their tissue fate via microinjection of metabolite standards and tracing their cell lineage for more than 100 different (untreated) Xenopus embryos. Three additional blastomeres were selected for the V11 and D11 blastomere types, cells V116–8 and D116–8, to quantify the native concentration of cell type-specific metabolites, specifically threonine and histidine in the D11 and acetylcholine, methionine, and alanine in the V11 blastomeres; external calibration is discussed in SI Appendix with calibration curves shown in SI Appendix, Fig. S1. Results from these quantitative experiments, shown in SI Appendix, Table S4, are in good agreement with independent measurements on whole Xenopus embryos; for example, alanine was measured at ∼360 nM in an average, 90-nL-volume V11 blastomere, and this compound has been reported at 100 µM to 2 mM in the whole embryo (12). Threonine, which accumulated in the D11 blastomeres endogenously at a concentration of 10 pmol per cell (±∼50% SD) (SI Appendix, Table S4), was injected with equal amount of serine as a mixture of standard l-amino acids (mD11) into V11 blastomeres at equivalent (1×, n = 19) and twofold (2×, n = 19) final concentration compared with the D11 cells, giving rise to a clone of mD11V11 cells as the embryo develops. Likewise, methionine and acetylcholine, which V11 blastomeres naturally accumulated at 6.5 pmol/cell (±60% SD) and 600 fmol/cell (±∼30% SD) concentration (SI Appendix), respectively, were injected as a mixture of standard l-amino acids (mV11) into D11 blastomeres in 1× (n = 19) and 2× abundance (n = 19), giving rise to the mV11D11 clone. Additionally, the microinjected blastomeres and the corresponding controls for D11 (n = 26) and V11 (n = 23) blastomeres were also injected with lineage tracers to detect the location of the clone at later stages of development (SI Appendix). Bright-field and fluorescence microscopy revealed that for embryos fixed at larval stages when all of the main organs have formed, cells derived from mV11D11 blastomeres contributed significantly less to brain and central somite and significantly more to ventral trunk epidermis compared with control embryos injected only with lineage tracer (Fig. 5). This change in cell fate likely is due to the much broader distribution of the clone across the dorsal midline at gastrulation stages (SI Appendix, Fig. S6). Cells derived from mD11V11 contributed significantly more to anterior head structures including cement gland, olfactory placode, lens, retina, and otocyst (Fig. 5). This change in cell fate likely is due to the expansion of the clone across the animal hemisphere at gastrulation stages (SI Appendix, Fig. S6). These results demonstrate that the differential distribution of even a few of the blastomere-specific metabolites that were uncovered (Figs. 2 and 3) can alter how the descendant cells are distributed in the embryo, ultimately affecting their tissue fates.
Fig. 5.
Assessment of developmental significance for blastomere type-characteristic metabolites by microinjection and cell lineage tracking. Larval stages (side views, dorsal to the top) injected with gfp mRNA alone (D11 ctrl; V11 ctrl) or metabolites plus gfp mRNA (mV11D11; mD11V11). Green fluorescent cells are descended from the injected blastomere. (Left) In D11 ctrl embryos, there are large numbers of cells in the brain (b), retina (r), olfactory pit (olf), and central somite (cs). In V11-metabolite-injected D11 embryos (mV11D11), the numbers of descendants in brain and central somite are significantly reduced, and those in the ventral epidermis (epi) are increased. (Right) In V11 ctrl embryos, there are only small numbers of cells in the olfactory pit, lens (l), and otocyst (oto). In D11-metabolite-injected V11 embryos (mD11V11), the numbers of descendants in these head structures are increased. Quantitation of the number of cells in selected tissues show significant differences between control and metabolite-injected clones. Statistical significance is marked at *P < 0.05 and **P < 0.005. (Scale bars, 275 μm for all images.)
The data provided herein demonstrate surprising heterogeneity of small-molecular activity between blastomeres at an early stage of embryonic development when there are no other known indications of cellular diversity or differentiation. Although they are consistent with previously demonstrated protein synthesis differences, which also correlate with cell fates (34), we do not yet know their physiological significance. It is possible that some of these differences may prefigure the contributions of these cells to different tissues [i.e., their cell fate (35, 36)]. For example, D11 blastomeres are major precursors of the nervous system (16, 37). In our measurements, they were found to contain higher levels of serine, consistent with serine acting as a vital neurotrophic factor in the developing nervous system (38). Furthermore, the accumulation of serine in D11 blastomeres was lost after UV light treatment, consistent with these embryos not developing head structures, including brain, whereas microinjection of serine into V11 blastomeres led to a change in cell fate to form several head structures. Other differences may reflect different rates of cell division in the untreated blastomeres. For example, accumulation of threonine has been linked to high-flux metabolism, such as in differentiating embryonic stem cells (39), purposed to feed the machinery of protein synthesis, one-carbon metabolism via glycine (SI Appendix, Fig. S2), and the tricarboxylic acid cycle. This is consistent with our observation that D11 blastomeres, which divide slightly faster than V21 cells, contained significantly more threonine and glycine (Figs. 2 and 3). Also in agreement, D11 cells were found to contain more alanine, and this metabolite has recently been proposed to be the energy source of the developing Xenopus embryo (12). Another interesting difference between D11 and the other blastomeres is the production of higher amounts of cis- and trans-urocanate. The formation and proliferation of mammalian and amphibian adult stem cells is affected by the histidine ammonia lyase gene that encodes histidinase, the enzyme that catalyzes deamination of histidine to urocanate (SI Appendix, Fig. S2) (40). In D11 cells, the total urocanate (cis and trans)/histidine ratio was approximately three times higher than both the V11 and V21 cells (P < 0.005), perhaps indicating an advanced state of differentiation even at these early cleavage stages. Alternatively, the role of urocanate in D11 may lie in photoprotection, because this molecule efficiently converts from cis to trans form by UV irradiation and is known to accumulate to a high concentration (0.3–20 mM) in the uppermost layers of the mammalian epidermis (41). Supporting this notion, D11 and V11 blastomeres are located in the animal (upper) portion of the embryo, which naturally is exposed to sunlight, and contained up to 7.0 times more total urocanate on average (P < 0.005) than the V21 blastomeres, which are unpigmented and naturally face away from sunlight. Although the full extent of the physiological roles of these metabolite differences remains to be revealed, our data demonstrate that changing the concentrations of even a few metabolites can ultimately affect blastomere cell fate.
In agreement, many of the differences observed between blastomeres dissected from untreated, control embryos were lost in the UV-treated embryos in our measurements. For example, the accumulations of serine, threonine, glycine, histidine, and total urocanate in D11 blastomeres in the control embryos were not observed in the ventralized embryos, which shared the same relative cell sizes, positions, and pigmentation as the untreated blastomeres. These differences suggest that cortical cytoplasmic rotation is required for the D11 blastomere to acquire its distinct metabolic characteristics. However, because UV irradiation affects the cis–trans conversion of urocanate, it will be important to use other techniques for altering cell fates to further evaluate the mechanism by which the metabolic activity of these early cells are regulated. Investigations targeting these small molecules during embryonic development, particularly during the early stages that are transcriptionally silent, will help establish the physiological importance of our measurements and elucidate their relationships to the known transcriptome and proteome.
Conclusions
In this work we have addressed a bottleneck in cell and developmental biology, the measurement of small-molecular activity in single embryonic cells using MS. We have demonstrated that single-cell CE-µESI-MS makes it possible to capture a snapshot of small-molecular activity in individual cells at the early cleavage stage of the 16-cell Xenopus embryo. The results reported herein are the first example to our knowledge of unbiased, large metabolite data on more than 80 distinct small molecules, including 40 identified metabolites, between different cells. We find that D11, V11, and V21 blastomeres exhibit characteristic small-molecular activity that reproducibly contributes to their commitment to neuronal, epidermal, and hindgut tissues in the adult organism, as validated by microinjection and cell-fate tracing experiments. The metabolomic differences uncovered in this work complement recent transcriptomic asymmetry (9) in the animal–vegetal axis of the early embryo and provide previously unidentified findings for asymmetry in the dorso-ventral axis, underscoring the importance of performing metabolic measurements on the level of the single cell. These small-molecular differences between single embryonic cells and different regions of the embryo’s body have not been technologically discernable by classical, cell-averaging measurements thus far.
We believe that single-cell MS, including CE-µESI as presented here, heralds new investigative possibilities for cell and developmental biology. To this end, micromanipulation raises great benefits to tailor single-cell MS to a broader range of applications. Direct sampling of single cells by microcapillaries, as recently demonstrated for neurons (42) and mammalian cells (25), offers an attractive means to enhance the throughput and the precision of measurements on single cells in an embryo using the technique presented here. By providing complementary information to already available genomic, transcriptomic, and proteomic data, we anticipate that small-molecular measurements by single-cell CE-µESI-MS will facilitate basic and translational research in cell and developmental biology to help develop a holistic understanding of how cells acquire their differentiated state.
Methods
SI Appendix provides a detailed account of standard protocols that were developed to maintain the Xenopus animal colony (approved by The George Washington University Animal Care and Use Committee, IACUC A311), perform ventralization by UV light irradiation, microinject individual blastomeres, perform cell fate analyses, and dissect single identified blastomeres. Single blastomere extracts were measured by a single-cell capillary electrophoresis μ-ESI-MS platform constructed as described earlier (29). Statistical and multivariate data analysis was performed as detailed in SI Appendix. Student’s t test (P < 0.05) was used to indicate statistical significance. A fold change of two or more marked biological significance.
Supplementary Material
Acknowledgments
We thank William F. Rutkowski (Department of Mechanical and Aerospace Engineering, The George Washington University) for machining the sample-loading components of the capillary electrophoresis-microflow electrospray ionization MS system. This work was supported by National Institutes of Health Grant R21 GM114854 (to P.N.), The George Washington University Department of Chemistry Start-Up Funds (to P.N.), and National Science Foundation Grant MCB-1121711 (to S.A.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423682112/-/DCSupplemental.
References
- 1.Macaulay IC, Voet T. Single cell genomics: Advances and future perspectives. PLoS Genet. 2014;10(1):e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14(9):618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 3.Grant PA, Yan B, Johnson MA, Johnson DLE, Moody SA. Novel animal pole-enriched maternal mRNAs are preferentially expressed in neural ectoderm. Dev Dyn. 2014;243(3):478–496. doi: 10.1002/dvdy.24082. [DOI] [PubMed] [Google Scholar]
- 4.Xanthos JB, Kofron M, Wylie C, Heasman J. Maternal VegT is the initiator of a molecular network specifying endoderm in Xenopus laevis. Development. 2001;128(2):167–180. doi: 10.1242/dev.128.2.167. [DOI] [PubMed] [Google Scholar]
- 5.White JA, Heasman J. Maternal control of pattern formation in Xenopus laevis. J Exp Zoolog B Mol Dev Evol. 2008;310(1):73–84. doi: 10.1002/jez.b.21153. [DOI] [PubMed] [Google Scholar]
- 6.Hainski AM, Moody SA. Xenopus maternal RNAs from a dorsal animal blastomere induce a secondary axis in host embryos. Development. 1992;116(2):347–355. doi: 10.1242/dev.116.2.347. [DOI] [PubMed] [Google Scholar]
- 7.Yan B, Moody SA. The competence of Xenopus blastomeres to produce neural and retinal progeny is repressed by two endo-mesoderm promoting pathways. Dev Biol. 2007;305(1):103–119. doi: 10.1016/j.ydbio.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collart C, et al. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141(9):1927–1939. doi: 10.1242/dev.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flachsova M, Sindelka R, Kubista M. Single blastomere expression profiling of Xenopus laevis embryos of 8 to 32-cells reveals developmental asymmetry. Scientific Reports. 2013;3:2278. doi: 10.1038/srep02278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sindelka R, Sidova M, Svec D, Kubista M. Spatial expression profiles in the Xenopus laevis oocytes measured with qPCR tomography. Methods. 2010;51(1):87–91. doi: 10.1016/j.ymeth.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Sun LL, et al. Quantitative proteomics of Xenopus laevis embryos: Expression kinetics of nearly 4000 proteins during early development. Scientific Reports. 2014;4:4365. doi: 10.1038/srep04365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vastag L, et al. Remodeling of the metabolome during early frog development. PLoS ONE. 2011;6(2):e16881. doi: 10.1371/journal.pone.0016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha B, et al. Subcellular metabolite and lipid analysis of Xenopus laevis eggs by LAESI mass spectrometry. PLoS ONE. 2014;9(12):e115173. doi: 10.1371/journal.pone.0115173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson JL, Carten JD, Farber SA. 2011. Zebrafish lipid metabolism: From mediating early patterning to the metabolism of dietary fat and cholesterol. Zebrafish: Cellular and Developmental Biology, Methods in Cell Biology, eds Detrich HW, Westerfield M, Zon LI (Elsevier, San Diego), Vol 101, 3rd Ed, pp 111–141.
- 15.Baskin JM, Dehnert KW, Laughlin ST, Amacher SL, Bertozzi CR. Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proc Natl Acad Sci USA. 2010;107(23):10360–10365. doi: 10.1073/pnas.0912081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moody SA. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev Biol. 1987;119(2):560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- 17.Rubakhin SS, Romanova EV, Nemes P, Sweedler JV. Profiling metabolites and peptides in single cells. Nat Methods. 2011;8(4) Suppl:S20–S29. doi: 10.1038/nmeth.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svatoš A. Single-cell metabolomics comes of age: New developments in mass spectrometry profiling and imaging. Anal Chem. 2011;83(13):5037–5044. doi: 10.1021/ac2003592. [DOI] [PubMed] [Google Scholar]
- 19.Ye H, Greer T, Li L. Probing neuropeptide signaling at the organ and cellular domains via imaging mass spectrometry. J Proteomics. 2012;75(16):5014–5026. doi: 10.1016/j.jprot.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zenobi R. Single-cell metabolomics: Analytical and biological perspectives. Science. 2013;342(6163):1243259. doi: 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- 21.Rubakhin SS, Lanni EJ, Sweedler JV. Progress toward single cell metabolomics. Curr Opin Biotechnol. 2013;24(1):95–104. doi: 10.1016/j.copbio.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ibáñez AJ, et al. Mass spectrometry-based metabolomics of single yeast cells. Proc Natl Acad Sci USA. 2013;110(22):8790–8794. doi: 10.1073/pnas.1209302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolee JA, Shrestha B, Mengistu G, Vertes A. Observation of subcellular metabolite gradients in single cells by laser ablation electrospray ionization mass spectrometry. Angew Chem Int Ed Engl. 2012;51(41):10386–10389. doi: 10.1002/anie.201205436. [DOI] [PubMed] [Google Scholar]
- 24.Date S, Mizuno H, Tsuyama N, Harada T, Masujima T. Direct drug metabolism monitoring in a live single hepatic cell by video mass spectrometry. Anal Sci. 2012;28(3):201–203. doi: 10.2116/analsci.28.201. [DOI] [PubMed] [Google Scholar]
- 25.Pan N, et al. The single-probe: A miniaturized multifunctional device for single cell mass spectrometry analysis. Anal Chem. 2014;86(19):9376–9380. doi: 10.1021/ac5029038. [DOI] [PubMed] [Google Scholar]
- 26.Hofstadler SA, Severs JC, Smith RD, Swanek FD, Ewing AG. Analysis of single cells with capillary electrophoresis electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun Mass Spectrom. 1996;10(8):919–922. doi: 10.1002/(SICI)1097-0231(19960610)10:8<919::AID-RCM597>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 27.Valaskovic GA, Kelleher NL, McLafferty FW. Attomole protein characterization by capillary electrophoresis-mass spectrometry. Science. 1996;273(5279):1199–1202. doi: 10.1126/science.273.5279.1199. [DOI] [PubMed] [Google Scholar]
- 28.Mellors JS, Jorabchi K, Smith LM, Ramsey JM. Integrated microfluidic device for automated single cell analysis using electrophoretic separation and electrospray ionization mass spectrometry. Anal Chem. 2010;82(3):967–973. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nemes P, Rubakhin SS, Aerts JT, Sweedler JV. Qualitative and quantitative metabolomic investigation of single neurons by capillary electrophoresis electrospray ionization mass spectrometry. Nat Protoc. 2013;8(4):783–799. doi: 10.1038/nprot.2013.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M, et al. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database issue) D1:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukumoto T, Kema IP, Levin M. Serotonin signaling is a very early step in patterning of the left-right axis in chick and frog embryos. Curr Biol. 2005;15(9):794–803. doi: 10.1016/j.cub.2005.03.044. [DOI] [PubMed] [Google Scholar]
- 32.Adams DS, et al. Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development. 2006;133(9):1657–1671. doi: 10.1242/dev.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harland R, Gerhart J. Formation and function of Spemann’s organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 34.Klein SL, King ML. Correlations between cell fate and the distribution of proteins that are synthesized before the midblastula transition in Xenopus. Rouxs Arch Dev Biol. 1988;197(5):275–281. doi: 10.1007/BF00380021. [DOI] [PubMed] [Google Scholar]
- 35.Dale L, Slack JMW. Fate map for the 32-cell stage of Xenopus laevis. Development. 1987;99(4):527–551. doi: 10.1242/dev.99.4.527. [DOI] [PubMed] [Google Scholar]
- 36.Moody SA, Kline MJ. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat Embryol (Berl) 1990;182(4):347–362. doi: 10.1007/BF02433495. [DOI] [PubMed] [Google Scholar]
- 37.Moody SA. Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev Biol. 1987;122(2):300–319. doi: 10.1016/0012-1606(87)90296-x. [DOI] [PubMed] [Google Scholar]
- 38.Kalhan SC, Hanson RW. Resurgence of serine: An often neglected but indispensable amino Acid. J Biol Chem. 2012;287(24):19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325(5939):435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luu N, et al. Differential regulation of two histidine ammonia-lyase genes during Xenopus development implicates distinct functions during thyroid hormone-induced formation of adult stem cells. Cell Biosci. 2013;3(1):43. doi: 10.1186/2045-3701-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laihia JK, et al. Urocanic acid binds to GABA but not to histamine (H1, H2, or H3) receptors. J Invest Dermatol. 1998;111(4):705–706. doi: 10.1046/j.1523-1747.1998.00351.x. [DOI] [PubMed] [Google Scholar]
- 42.Aerts JT, et al. Patch clamp electrophysiology and capillary electrophoresis-mass spectrometry metabolomics for single cell characterization. Anal Chem. 2014;86(6):3203–3208. doi: 10.1021/ac500168d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.