Significance
Developing sensors to image cellular metabolites and signaling molecules in living cells is challenging. Here we describe Spinach riboswitches, a novel class of genetically encoded metabolite sensor based on riboswitches, a group of naturally occurring ligand-binding RNAs. Spinach riboswitches use Spinach, an RNA aptamer that binds and activates the fluorescence of an otherwise nonfluorescent small-molecule fluorophore. Drawing upon structural insights into the mechanism of structural switching in riboswitches, we show that Spinach can be swapped into various riboswitches, allowing metabolite binding to induce Spinach fluorescence directly. Expression of Spinach riboswitches in cells allows metabolite levels to be imaged in real time in live bacterial cells. Spinach riboswitches thus provide a novel approach to image cellular metabolites in living cells.
Keywords: RNA probes, cellular imaging, riboswitch, fluorescence, metabolite
Abstract
Riboswitches are natural ligand-sensing RNAs typically that are found in the 5′ UTRs of mRNA. Numerous classes of riboswitches have been discovered, enabling mRNA to be regulated by diverse and physiologically important cellular metabolites and small molecules. Here we describe Spinach riboswitches, a new class of genetically encoded metabolite sensor derived from naturally occurring riboswitches. Drawing upon the structural switching mechanism of natural riboswitches, we show that Spinach can be swapped for the expression platform of various riboswitches, allowing metabolite binding to induce Spinach fluorescence directly. In the case of the thiamine 5′-pyrophosphate (TPP) riboswitch from the Escherichia coli thiM gene encoding hydroxyethylthiazole kinase, we show that insertion of Spinach results in an RNA sensor that exhibits fluorescence upon binding TPP. This TPP Spinach riboswitch binds TPP with affinity and selectivity similar to that of the endogenous riboswitch and enables the discovery of agonists and antagonists of the TPP riboswitch using simple fluorescence readouts. Furthermore, expression of the TPP Spinach riboswitch in Escherichia coli enables live imaging of dynamic changes in intracellular TPP concentrations in individual cells. Additionally, we show that other riboswitches that use a structural mechanism similar to that of the TPP riboswitch, including the guanine and adenine riboswitches from the Bacillus subtilis xpt gene encoding xanthine phosphoribosyltransferase, and the S-adenosyl-methionine-I riboswitch from the B. subtilis yitJ gene encoding methionine synthase, can be converted into Spinach riboswitches. Thus, Spinach riboswitches constitute a novel class of RNA-based fluorescent metabolite sensors that exploit the diversity of naturally occurring ligand-binding riboswitches.
Among the most important tools for studying the dynamic changes in the intracellular level of signaling molecules and metabolites in living cells are genetically encoded fluorescent sensors. Most sensors comprise two fluorescent proteins flanking a central target-binding domain (1, 2). Binding of the target molecule induces conformational changes that reposition the fluorescent proteins. This repositioning results in changes in FRET between the fluorescent proteins, which can be imaged by microscopy in living cells (3). These sensors have provided fundamentally novel insights into the dynamic changes in calcium, cyclic nucleotides, glucose, and other molecules during cellular signaling and disease states (4).
Although genetically encoded sensors are powerful tools for imaging cellular metabolites, only a small number of sensors have been created because these sensors require a protein that binds the target molecule of interest and which undergoes a target-induced conformational change that is sufficient to induce a change in FRET. However, proteins with these properties are not readily available for most physiologically important metabolites. As a result, sensors have not been generated for many important metabolites and signaling molecules.
Recently we described an alternative class of genetically encoded sensors composed of RNA (5). These sensors are based on Spinach, an “RNA mimic of green fluorescent protein” (6). Spinach is an RNA aptamer that binds fluorophores resembling the hydroxybenzylidene imidazolinone (HBI) fluorophore in GFP, including 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI) (6, 7). Similar to the manner by which GFP induces fluorescence in HBI, Spinach binds to the otherwise nonfluorescent DFHBI and switches it to a highly fluorescent state. This property has allowed Spinach to be used to tag and image specific RNAs in living cells (6).
We recently converted the constitutively fluorescent Spinach–DFHBI complex into sensors that fluoresce in proportion to the concentration of specific metabolites (5). These sensors comprise two domains: Spinach and a target-binding aptamer. The target-binding aptamer is inserted into a structurally critical stem of Spinach (5). Because the target-binding aptamer is unstructured without the target molecule, the critical stem is disrupted, preventing Spinach from folding and binding DFHBI. However, when the aptamer binds its target, the aptamer folds, enabling the folding of the critical stem in Spinach (Fig. S1A). This folding leads to Spinach fluorescence that can be detected in vitro or in cells. Using this approach, we created sensors by fusing Spinach to aptamers that bind S-adenosyl-methionine (SAM), ADP, and other metabolites (5). These sensors allowed imaging of the dynamics and turnover of SAM and ADP in live bacterial cells (5). This approach was applied subsequently to detect and image cyclic di-GMP and various proteins in living cells (8, 9). These allosteric Spinach-based sensors demonstrate the versatility of RNA as a platform for creating genetically encoded sensors for imaging cellular metabolites.
There are important technical challenges that must be addressed when creating allosteric Spinach-based sensors. These sensors rely on metabolite-binding aptamers, which typically are generated using approaches such as systematic evolution of ligands by exponential enrichment (SELEX) (10, 11). SELEX-generated aptamers typically exhibit poor selectivity and affinity that are not desirable for sensors. Selectivity is important because the cellular environment is highly complex and can contain unknown metabolites with structures similar to that of the desired target molecule. In principle, aptamer selectivity can be achieved using negative selection during SELEX (12); however, it is difficult to perform negative selection against all the diverse, structurally related metabolites in the cell, especially if some of them are unknown.
Aptamer affinity also is critical for sensor function (13). If the aptamer has too high affinity for the target, the target will be fully bound to the aptamer regardless of how its concentration changes in cells. If the affinity is too low, very little target will be bound, resulting in minimal fluorescence activation in cells. In most cases, aptamer binding affinities that are similar to the cellular metabolite concentration will produce sensors exhibiting fluorescence that corresponds directly to the metabolite concentration (13). However, it is difficult to obtain aptamers with a predetermined affinity for a target molecule using SELEX (14). Thus, obtaining aptamers that have a suitable target-binding affinity remains a challenge.
In contrast, riboswitches have evolved to have the type of in vivo binding affinity and selectivity needed to function as highly precise sensors for physiologically important intracellular metabolites. Riboswitches are RNA regulatory elements found primarily in bacterial mRNAs that switch conformations upon binding small molecules (15). These conformational switches typically activate or inhibit protein synthesis but can regulate other aspects of mRNA metabolism (16). Computational comparative genomics-based approaches have identified more than 100 different putative riboswitches (17–22), most of which are found in transcripts encoding metabolic enzymes or regulators. Riboswitches appear to have evolved to allow highly precise control of protein expression in response to dynamic changes in cellular metabolite concentrations.
Here we describe Spinach riboswitches, a new type of genetically encoded sensor for imaging metabolites based on natural riboswitches. We show that Spinach can be incorporated into the thiamine 5′-pyrophosphate (TPP) riboswitch from the Escherichia coli thiM gene encoding hydroxyethylthiazole kinase, resulting in an RNA-based sensor that is fluorescently activated upon binding TPP. This TPP Spinach riboswitch binds TPP with affinity and selectivity similar to that of the endogenous riboswitch and enables the discovery of agonists and antagonists of the TPP riboswitch through simple fluorescence readouts. Furthermore, expression of the TPP Spinach riboswitch in E. coli enables imaging of dynamic changes in intracellular TPP concentrations that occur in response to extracellular thiamine (23). Last, we show that other riboswitches that use a structural mechanism similar to that of the TPP riboswitch, including the guanine and adenine riboswitches from the Bacillus subtilis xpt gene encoding xanthine phosphoribosyltransferase, and the S-adenosyl-methionine-I riboswitch from the B. subtilis yitJ gene encoding methionine synthase, can be converted into Spinach riboswitches. Thus, Spinach riboswitches constitute a novel class of RNA-based fluorescent metabolite sensors that take advantage of the evolutionarily optimized affinity and selectivity seen in natural riboswitches.
Results
Structure-Guided Design and Optimization of a Spinach Riboswitch.
We wondered if the ligand-induced conformational switching mechanisms seen in riboswitches could be used to switch Spinach from a nonfluorescent to a fluorescent state. We were struck by the structural similarity of a portion of the thiM TPP riboswitch and Spinach. The TPP riboswitch contains a TPP-binding aptamer domain and a second expression platform that regulates the translation of the thiM transcript (Fig. 1A) (24). This expression platform constitutes an ∼40-Å-long helical domain (25, 26). Importantly, a short transducer sequence in the riboswitch can adopt two positions within the riboswitch (Fig. 1A). In the absence of TPP, the transducer sequence binds to the switching sequence in the TPP-binding domain. However, TPP binding to the riboswitch displaces the transducer sequence, allowing the transducer sequence to form one of the strands of the expression platform helix (Fig. 1A). As a result of helix formation, the ribosome’s access to the Shine–Dalgarno (SD) element is blocked, thereby inhibiting translation (25–28).
Fig. 1.
Structure-guided design of a TPP Spinach riboswitch. (A) Metabolite-dependent natural switching mechanism of TPP riboswitches. Riboswitches contain an aptamer domain (blue) and an expression platform (black). In the absence of TPP binding, the transducer sequence (gray) is hybridized to the switching sequence (yellow) in the aptamer domain, leaving the SD sequence accessible. When TPP binds, the aptamer domain undergoes a structural rearrangement, which disrupts this strand hybridization. The transducer sequence is displaced from the switching sequence and hybridizes to the SD sequence, forming the helical structure of the expression domain. (B) Secondary structure and design of TPP Spinach riboswitch. The Spinach riboswitch is generated by replacing the expression platform of the thiM TPP riboswitch with Spinach (green). TPP binding releases the transducer sequence (gray) from the switching sequence (yellow), forming a critical helix in Spinach. Formation of this helix is critical for Spinach to bind and activate the fluorescence of DFHBI (green ball). A linker sequence (dark gray) is used to position the aptamer domain and Spinach optimally. (C) Structural model of DFHBI binding to Spinach. The figure was prepared with PyMOL software based on previously reported Spinach structure (29). The portion of Spinach that contains the transducer sequence that is modified to form the TPP Spinach riboswitch is shown. The structural model highlights the DFHBI-binding pocket (green ball-and-stick model), which is formed by a U⋅A⋅U base triple (teal; bases are labeled with letters) and a G-quadruplex (deep violet). The portion of the H1 helix (5′-UCCA-3′) that provides the U in the U⋅A⋅U base triple is indicated in light gray. This sequence is modified to form the transducer sequence in the TPP Spinach riboswitch.
Spinach has a structure similar to that of the helical expression platform of the TPP riboswitch. The fluorophore-binding domain of Spinach comprises a coaxial helical stack composed of two A-form helices separated by an atypical G-quadruplex motif (29, 30). The DFHBI-binding pocket is formed by the junction between the upper helix (H1) and the G-quadruplex (29, 30). The helical structure is necessary to stabilize a U⋅A⋅U base triple that serves as the upper surface of the DFHBI-binding pocket (Fig. 1C). Because the helical structure is required for DFHBI binding to Spinach, we reasoned the TPP riboswitch could regulate Spinach fluorescence if the transducer sequence controls the formation of the H1 helix.
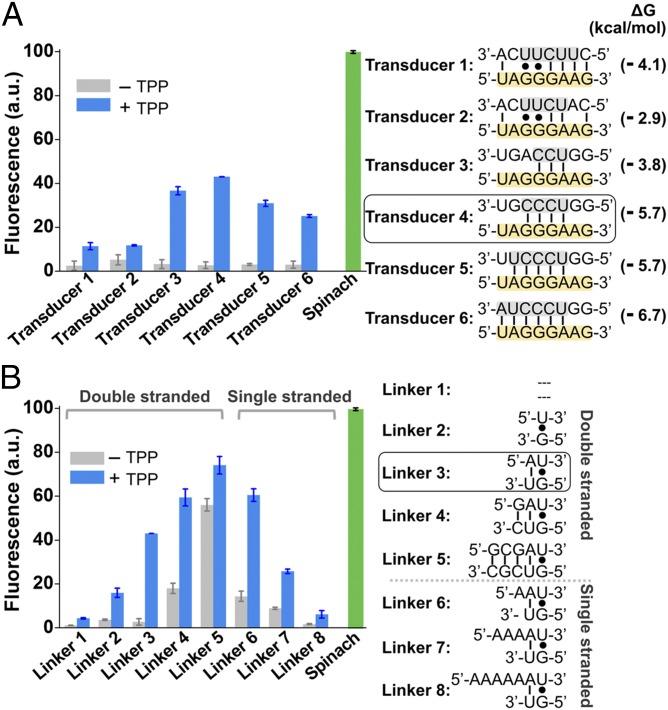
We therefore decided to engineer the TPP riboswitch so that TPP binding would cause the transducer sequence to reposition itself into the H1 helix of Spinach and thereby activate Spinach fluorescence (Fig. 1B). In these experiments, we did not want to alter the switching sequence, to preserve the naturally evolved affinity and specificity of TPP binding in the riboswitch. Therefore we focused on the transducer sequence. In the thiM TPP riboswitch the transducer sequence (5′-CUUCUU-3′) hybridizes to the switching sequence (5′-GGGAAG-3′). We noticed that a portion of the H1 helix (5′-UCCA-3′) that abuts the DFHBI-binding pocket has near base complementarity to the switching sequence (Fig. 1C). We therefore designed the sensors so that these H1 helix sequences would serve as the transducer sequence, hybridizing to different portions of the switching sequence in the TPP-binding aptamer domain (Table S1). The number and composition of nucleotides in the transducer sequence that hybridizes with the switching sequence were varied, and in each case base complementarity within the H1 helix was maintained (Fig. 2A and Fig. S2).
Fig. 2.
Optimization of Spinach riboswitch performance. (A) Optimization of transducer sequences. Six transducer sequences (transducers 1–6, gray) were tested for their ability to mediate TPP-induced fluorescence of the TPP Spinach riboswitch. In each case, the transducer sequence is similar to the transducer sequence found in the wild-type thiM TPP riboswitches. In each case, the switching sequence (yellow) in the aptamer domain is unchanged. The optimal sensor (transducer 4) contained a transducer sequence helix comprising only four Watson–Crick base pairs. The 5′-U base of this transducer sequence is positioned to form the U⋅A⋅U base triple that serves as the upper surface of the DFHBI-binding pocket. Shown are mean and SEM values of three independent replicates. The Gibbs free energy change (∆G) of hybridization events between each transducer sequence and the switching sequence was calculated using Mfold online software. (B) Optimization of linker sequences. Spinach was connected with the aptamer domain by different linker sequences. The linker position is highlighted in dark gray in Fig. 1B. The linker was one of eight double-stranded or single-stranded sequences. The double-stranded linkers of different lengths (linkers 1–5) extend the lower helix (H2) and rotate Spinach. The different single-stranded linkers (linkers 6–8) provide flexibility of relative domain positions that could facilitate helix formation. Shown are mean and SEM values of three independent replicates. The optimal linker (linker 3) displayed a 15.9-fold increase in fluorescence signal upon the addition of 100 µM TPP. a.u., arbitrary units.
We next asked if these RNAs function as TPP sensors. Each RNA contained a different H1 helix sequence adjacent to the DFHBI-binding pocket (Fig. 2A and Fig. S2). The fluorescence spectrum of each RNA was measured in a solution containing 100 nM RNA and 10 µM DFHBI. The RNAs exhibited between 1.3- and 15.9-fold increase in fluorescence upon addition of 100 µM TPP (Fig. 2A). The optimal sensor (transducer 4) contained a transducer sequence helix of only four Watson–Crick base pairs. The 5′-U base of this transducer sequence is positioned to form the U⋅A⋅U base triple that serves as the upper surface of the DFHBI-binding pocket (Fig. 1C). Thus, TPP riboswitches can be fashioned into TPP-sensing Spinach riboswitches.
Optimizing Spinach Riboswitch Performance.
We next asked if the Spinach riboswitch performance could be optimized further. The Spinach riboswitch contains a two-nucleotide-long single-stranded linker between the TPP-binding aptamer domain and Spinach. To test if changing its length and flexibility enhances sensor function, we made variants of optimal sensor (transducer 4) in which the linker was one of eight double-stranded or single-stranded sequences (Fig. 2B). The double-stranded linkers of different lengths (linkers 1–5) are predicted to extend the lower helix (H2) and rotate Spinach, thereby facilitating or disturbing the ability of the transducer sequence to access its binding site in the H1 helix. The different single-stranded linkers (linkers 6–8) change the distance that the transducer sequence must traverse to complete the H1 helix in Spinach and provide additional flexibility that could facilitate helix formation.
We first examined the double-stranded linkers. For all experiments, we used the optimal transducer and switching sequence identified in Fig. 2A. A short double-stranded sequence (linker 3) produced optimal signal (Fig. 2B). Sensors containing this linker exhibited low background fluorescence and a 15.9-fold increase in fluorescence upon incubation with 100 µM TPP (Figs. 2B and 3A). The effects of these linkers were not caused by improved folding of Spinach because the addition of these double-stranded linkers to Spinach did not influence Spinach fluorescence (29, 30). Because of the optimal performance of linker 3, sensors with this linker were used to generate the data presented in Fig. 2A.
Fig. 3.
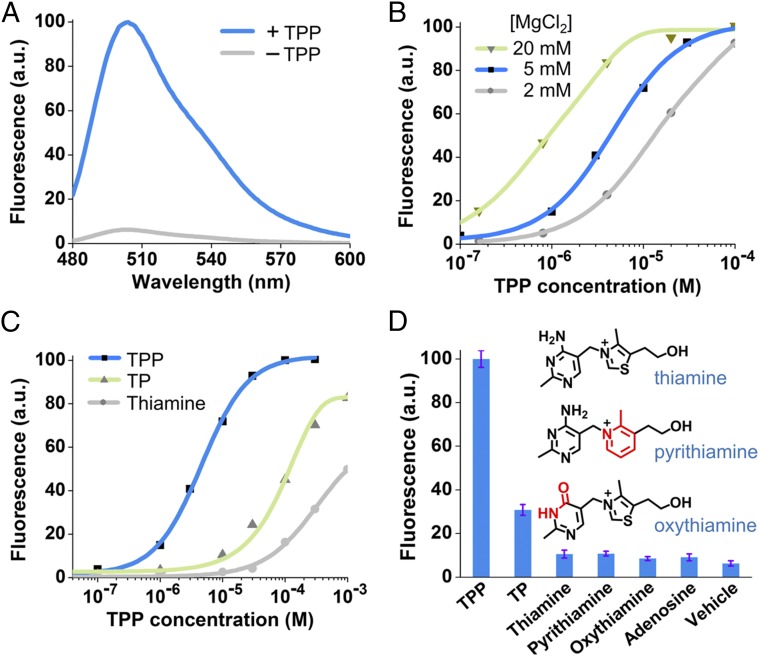
In vitro characterization of the Spinach riboswitch. (A) Fluorescence emission spectra of the Spinach riboswitch in the presence or absence of TPP. Spectra were measured in a solution containing 100 nM RNA, 10 µM DFHBI, and either 0 or 100 µM TPP. (B) Dose–response curve for fluorescence detection of TPP by the Spinach riboswitch at different concentrations of MgCl2. The maximum fluorescence signal of the Spinach riboswitch at each concentration of MgCl2 was set to 100. Half-maximal fluorescence is reached at 9.0 µM, 4.4 µM, and 0.77 µM TPP at 2 mM (gray), 5 mM (blue), and 20 mM (green) concentrations of MgCl2, respectively. The EC50 of the Spinach riboswitch is influenced by magnesium concentration, as is consistent with the known effect of magnesium on the binding affinity of the TPP riboswitch (32). In live-cell experiments, 2 mM MgCl2 was used to image TPP Spinach riboswitch fluorescence because 2 mM represents a physiological magnesium level often seen in bacterial cells (Fig. S4D) (59). (C) Dose–response curve for fluorescence activation of the TPP Spinach riboswitch by TPP and TPP analogs. Half-maximal fluorescence is reached at 4.4 µM TPP (blue). The dose–response curves for thiamine (gray) and thiamine monophosphate (TP, green) were measured also; half-maximal fluorescence is reached at 955 µM and 116 µM, respectively. The Spinach riboswitch is highly specific for TPP but also is activated by thiamine monophosphate or thiamine, as is consistent with the known binding specificity of the TPP riboswitch (32). (D) Selectivity of the Spinach riboswitch. Fluorescence emission was measured in the presence of 30 µM of the indicated compounds. Shown are mean and SEM values of three independent replicates. Chemical structures of pyrithiamine and oxythiamine are drawn with differences from thiamine indicated in red.
We next examined the effect of single-stranded linkers added to linker 3. Each of these linkers (linkers 6–8) exhibited either high background fluorescence or low fluorescence activation (Fig. 2B). Interestingly, a structural model of the Spinach riboswitch containing linker 3 suggests that the transducer sequence is positioned in space so that it can interact readily with the aptamer domain in the riboswitch (Fig. S3). Thus, the improved performance of linker 3 may reflect optimized positioning of the transducer sequence relative to the switching sequence.
In Vitro Characterization of the Spinach Riboswitch.
We next asked if the Spinach riboswitch retains the moderate binding affinity of the thiM TPP riboswitch. A dose–response curve for TPP was measured with the optimized Spinach riboswitch. The EC50 of the Spinach riboswitch measured at physiological magnesium levels (2 mM) was 9.0 µM. The Spinach riboswitch detected TPP throughout the concentration range (5–100 µM) that TPP is known to exhibit normally in bacteria (31). Half-maximal fluorescence measured at 20 mM MgCl2 was reached at 770 nM TPP (Fig. 3B). This EC50 is similar, although slightly weaker, than the Kd (∼600 nM) measured for the thiM TPP riboswitch at 20 mM MgCl2 (24, 32). Taken together, these data suggest that the Spinach riboswitch preserves the affinity seen in the natural riboswitch.
We next asked if the Spinach riboswitch retains the selectivity of the natural riboswitch. The thiM TPP riboswitch binds both thiamine and thiamine monophosphate, albeit at a lower affinity than TPP (24). The Spinach riboswitch showed binding to both thiamine and thiamine monophosphate, with an EC50 of 955 µM and 116 µM, respectively (Fig. 3C). These 216-fold and 25.4-fold decreases in affinity relative to TPP are consistent with the previously reported selectivity of the E. coli TPP riboswitch (24). Additionally, as expected, pyrithiamine and oxythiamine, which show minimal binding to the TPP riboswitch (24), induce nearly undetectable levels of fluorescence in the TPP Spinach riboswitch (Fig. 3D). To rule out the possibility that these compounds simply inhibit Spinach fluorescence or DFHBI binding to Spinach, we incubated these compounds with Spinach. None of these compounds affected Spinach fluorescence (Fig. S4A). These results suggest that the binding selectivity of the Spinach riboswitch matches that of the parental TPP riboswitch.
Using the Spinach Riboswitch to Identify Agonists and Antagonists of Riboswitch.
Riboswitches, including the TPP riboswitch, are promising antimicrobial drug targets (33, 34). To identify small molecules that bind and activate or inhibit riboswitches, the riboswitches have been placed in front of GFP or other reporters to monitor riboswitch activity in cells (35). However, these approaches miss potentially valuable hits that are not cell permeable but which could be the basis for further lead development. Thus, there is considerable interest in simple in vitro assays that can be used to identify and characterize riboswitch-binding molecules directly (36). We therefore asked if Spinach riboswitches can be used to identify antagonists and agonists of the TPP riboswitch using simple fluorescence readouts.
The antimicrobial drug pyrithiamine pyrophosphate (PTPP) is a known agonist of the TPP riboswitch (35). The Spinach riboswitch exhibited a 14.8-fold increase in fluorescence upon addition of PTPP, whereas its prodrug pyrithiamine only induced background fluorescence signal (Fig. 4A). Several other thiamine analog drug candidates, including oxythiamine, hypoxanthine, 6-aminoquinoxaline, and benzothiadiazol-5-ylmethanol (36–38), also were incubated with the Spinach riboswitch. Only minor increases in fluorescence were observed with these compounds (Fig. 4A), consistent with the reported poor TPP riboswitch activation by these drug candidates (36). Thus, the Spinach riboswitch is capable of identifying a known agonist of the TPP riboswitch.
Fig. 4.
Using the Spinach riboswitch to identify agonists and antagonists of the TPP riboswitch. (A) Agonist activity of thiamine analogs on the Spinach riboswitch. Fluorescence emission was measured by incubating the Spinach riboswitch (100 nM) with 100 µM of each thiamine analog. Only PTPP, a known TPP riboswitch agonist, induced substantial fluorescence (14.8-fold increase). Shown are mean and SEM values of three independent replicates. The chemical structures of all compounds are indicated. (B) Characterization of thiamine analogs for TPP riboswitch antagonist activity. To identify antagonists, each candidate was incubated in the presence of the known riboswitch activator, TPP. Fluorescence emission was measured in the presence of 100 nM sensor, 2 µM TPP, and 100 µM of each thiamine analog drug candidate. An identical mixture of each drug candidate with TPP and 100 nM Spinach was used to normalize the fluorescence signal. Shown are mean and SEM values of three independent replicates. The chemical structures of all of the compounds are indicated.
We next asked if the Spinach riboswitch also can be used to identify antagonists of the TPP riboswitch. In these experiments, the Spinach riboswitch (100 nM) was incubated with 2 µM TPP and 100 µM of each potential antagonist (Fig. 4B). These experiments showed that pyrithiamine and thiamine could inhibit TPP-mediated activation of the Spinach riboswitch, as is consistent with the known ability of these compounds to block activation of the TPP riboswitch (24). As a control, we tested each compound with Spinach to confirm that they do not inhibit Spinach fluorescence or DFHBI binding to Spinach. None of these compounds affected Spinach fluorescence (Fig. S4A). Taken together, these data indicate that the Spinach riboswitch can be used to identify and characterize agonists and antagonists of the TPP riboswitch readily.
Kinetics of Spinach Riboswitch Activation and Deactivation.
We next asked if the Spinach riboswitch could be used as a genetically encoded sensor to image TPP levels in E. coli in real time. For the Spinach riboswitch to be useful, it must have rapid TPP-induced fluorescence activation relative to the timescale of TPP biosynthesis in cells. Thus, we first wanted to assess the kinetics of Spinach riboswitch activation and deactivation. Mixing TPP with the Spinach riboswitch led to half the maximal fluorescence signal in 150 s, with >90% of maximal levels within ∼10 min (Fig. S4B).
In addition, we measured how quickly the Spinach riboswitch fluorescence is lost upon TPP removal. In these experiments, the Spinach riboswitch was incubated with TPP (100 µM) for 1 h, and then the TPP was removed by rapid gel filtration. Spinach riboswitch fluorescence was deactivated by >90% within 8 min (Fig. S4C). The rates of fluorescence activation and deactivation are similar to the switching kinetics of the endogenous thiM TPP riboswitch (39). These results indicated that the kinetics of fluorescence activation and deactivation occur within a timescale that is significantly faster than the previously described 1-hr time courses for changes in endogenous TPP levels in bacteria (40).
Live-Cell Imaging of TPP Biosynthesis with the Spinach Riboswitch.
We next used the sensor to monitor TPP synthesis dynamics in living Rosetta2 (DE3) E. coli cells (Novagen). Thiamine can be actively transported into cells and converted to TPP by thiamine kinase (23, 34). Thus, we added thiamine (10 µM) to Spinach riboswitch-expressing cells cultured in thiamine-free medium for 2 h. Cells were imaged every 15 min over 3 h (Fig. 5A). The fluorescence level per cell from 250 cells was measured at each time point in three independent experiments. Under these conditions, we observed a time-dependent increase in the mean cellular fluorescence at each time point (Fig. 5A). These data indicate thiamine treatment increases Spinach riboswitch fluorescence.
Fig. 5.
Imaging of TPP biosynthesis in living cells using the Spinach riboswitch. (A) Kinetics of TPP biosynthesis in E. coli. Cells expressing the Spinach riboswitch were cultured briefly in thiamine-free medium for 2 h and then imaged every 15 min for 3 h after the addition of 10 µM thiamine. Adenosine is an inhibitor of de novo TPP biosynthesis. Adenosine (300 μg/mL) was preadded to the thiamine-free medium to reduce basal TPP levels. Spinach riboswitch activation was monitored as an increase in fluorescence signal over time. Images are pseudocolored to show the fold increase in fluorescence at each time point relative to 0 min. The color scale represents 0- to 10.0-fold changes (black to yellow) in fluorescence signal. (Scale bar, 5 µm.) (B) The fluorescence increase in Spinach riboswitch-expressing cells is caused by TPP biosynthesis. In this experiment, we used pharmacologic treatments to determine if Spinach riboswitch fluorescence reflects binding to TPP and not to other cell metabolites. In each experiment, Spinach-riboswitch–expressing cells were cultured in thiamine-free medium for 2 h and then were treated with 10 µM thiamine and the indicated compounds. Adenosine is an inhibitor of de novo TPP biosynthesis. Cells pretreated for 2 h with adenosine (300 μg/mL) exhibited markedly reduced Spinach riboswitch fluorescence at 0 min, reflecting reduced basal TPP levels. Adenosine induces the expression of thiamine kinase (40); the increase in initial fluorescence was 2.3-fold faster in adenosine-treated cells than in cells not treated with adenosine. In another experiment, 1 mM oxythiamine was added 10 min before the addition of 10 µM thiamine and was present throughout imaging. Oxythiamine is a competitive inhibitor of thiamine kinase; the increase in initial fluorescence was 3.1-fold slower in cells treated with oxythiamine than in cells treated with thiamine only. As a control, the addition of 500 µM oxythiamine did not activate the fluorescence of the Spinach riboswitch (gray line, pretreated for 2 h with 300 μg/mL adenosine). Each data point represents the average fluorescence value of 250 cells from three experimental replicates. (C) Distribution of Spinach riboswitch fluorescence levels within a population of E. coli cells. The fluorescence in each cell was normalized to control for cell-to-cell variability in Spinach riboswitch expression levels. To do so, we used a plasmid that expresses both the Spinach riboswitch and the eqFP670 far-red fluorescent protein. Cells were imaged after 3 h of treatment with 10 µM thiamine, and the fluorescence of the Spinach riboswitch in individual cells was normalized for the fluorescence signal from eqFP670 (red) in the same cells. The average normalized fluorescence value (blue) was set to 100%, and individual cells were binned according to relative brightness. The percentage of cells in each bin is plotted. A total of 250 cells were quantified from three experimental replicates. (Scale bar, 10 µm.) (D) Representative examples of fast, moderate, and slow TPP-synthesizing cells. Imaging of Spinach riboswitch fluorescence revealed different cellular populations based on the rate of TPP biosynthesis. Cells that have a rate of increase within two SDs of the average (black) are considered to have moderate biosynthesis rates (gray). Cells that have a rate of increase greater or less than two SDs from the average are termed “fast” (red) or “slow” (blue), respectively. Time points after the addition of thiamine are indicated. The color scale represents 0- to 10.0-fold changes (black to yellow) in fluorescence signal. (Scale bar, 3 µm.)
We next asked if the increase in Spinach riboswitch fluorescence correlates linearly with the actual TPP levels in cells. To answer this question, we measured the intracellular TPP concentration at each time point using an HPLC assay (41). In these experiments the increase in fluorescence from the Spinach riboswitch correlated directly with the measured cellular TPP level (Fig. S5). Thus, the Spinach riboswitch fluorescence corresponds directly to the intracellular TPP concentration in E. coli.
We next wanted to confirm that Spinach riboswitch fluorescence reflects binding to TPP but not to other cellular components. To do so, we used adenosine, which inhibits the synthesis of thiazole phosphate, a TPP precursor in the de novo TPP biosynthesis pathway (42). Cells were cultured in thiamine-free medium, resulting in TPP levels generated exclusively through the de novo pathway. In these cells, adenosine treatment (300 μg/mL) markedly reduced Spinach riboswitch fluorescence (0 min time point of Fig. 5B). These data are consistent with the idea that Spinach riboswitch fluorescence is regulated by endogenous TPP levels.
A consequence of the reduced TPP levels in adenosine-treated cells is an up-regulation of thiamine kinase, a critical component of the TPP salvage pathway (40). The increase in thiamine kinase is expected to allow bacteria to convert thiamine to TPP more efficiently. Indeed, in cells treated with adenosine (300 μg/mL) for 2 h, thiamine treatment resulted in a rate of fluorescence increase that was 2.3-fold faster than in cells not treated with adenosine (Fig. 5B). However, pretreatment of cells with the thiamine kinase inhibitor oxythiamine (1 mM) (43) for 10 min before the addition of thiamine prevented this accelerated increase in Spinach riboswitch fluorescence (Fig. 5B). Taken together, these results indicate that the Spinach riboswitch fluorescence derives from intracellular TPP.
Population Analysis Reveals Cell-to-Cell Variation in TPP Biosynthesis.
We next asked if there is cell-to-cell variability in TPP biosynthesis. The intracellular level of TPP has not been measured previously at the single-cell level because of the absence of imaging tools. Previous methods have used HPLC and therefore represent population-wide measurements of TPP (42). To test the cell-to-cell variability in TPP biosynthesis, we modified the Spinach riboswitch expression plasmid to contain a separate promoter expressing the eqFP670 far-red fluorescent protein (44). Thus, eqFP670 expression can be used to normalize for variations in plasmid expression level between cells. A similar plasmid has been used previously to normalize aptamer expression for directed evolution experiments (45).
We next asked if all cells exhibit identical responses to thiamine treatment. In these experiments, thiamine was added to cells cultured in thiamine-free medium for 3 h. The Spinach riboswitch fluorescence was measured at a single-cell level in E. coli and normalized to eqFP670 fluorescence. The distribution of these fluorescence measurements showed a largely symmetric distribution; however, a prominent population of outliers exhibited high levels of Spinach riboswitch fluorescence (Fig. 5C). This variation does not reflect variation in plasmid expression, because the eqFP670 expression lacked this variability (Fig. 5C).
To understand further the cell-to-cell variation in TPP biosynthesis, we monitored TPP biosynthesis over time in individual cells. Spinach fluorescence was measured every 30 min, and the trajectory of TPP biosynthesis was plotted for each cell. Examination of these trajectories indicated that the cells differed in the initial rate of TPP biosynthesis—slow, moderate, or fast—resulting in different final steady-state levels of TPP measured at 3 h (Fig. 5D). The majority (74%) of cells exhibited moderate TPP biosynthesis rates, but 12% and 14% of cells exhibited slow and fast biosynthesis rates, respectively. Although other aspects of TPP biosynthesis could influence the cell-to-cell variability in TPP levels, these data suggest that initial TPP biosynthesis rates likely have an important role.
We next wanted to obtain further evidence that the variation in Spinach riboswitch fluorescence reflects changes in the cellular TPP concentration and not in the Spinach riboswitch expression. Therefore we expressed the Spinach aptamer in the same expression plasmid and monitored the variability in Spinach expression (Fig. S5C). In these experiments, 84% of cells exhibited Spinach fluorescence within (±) 10% of the mean Spinach fluorescence. The distribution of fluorescence in these cells followed a Gaussian distribution (Fig. S5D). Furthermore, thiamine treatment caused very little change in Spinach fluorescence over the time course of the 3-h treatment. Taken together, these results support the idea that the variability in Spinach riboswitch fluorescence reflects the cell-to-cell variation in TPP biosynthesis.
Modular Generation of Spinach Riboswitch Activated by Other Metabolites.
We next asked if this new Spinach riboswitch sensor design can be applied to other riboswitch classes. We focused on the Bacillus subtilis xpt guanine riboswitch, a mutant xpt riboswitch that binds adenine, and the B. subtilis yitJ SAM-I riboswitch (Fig. S6) (46–48). Each riboswitch is similar to the TPP riboswitch in that ligand binding displaces the transducer sequence, resulting in the formation of a helix in the expression platform. However, each of these riboswitches contains an additional 5′ competitor sequence that binds the switching sequence after ligand binding. Thus, these riboswitches have a switching mechanism similar to that in the TPP riboswitch.
To convert these riboswitches to Spinach riboswitches, we first replaced their expression platforms with Spinach. Next, we focused on the switching sequence and the transducer sequence. Unlike the TPP riboswitch, the switching sequences in these riboswitches are fairly degenerate across species (46–48). We therefore changed the switching sequence in these Spinach riboswitches to the core 5′-GGGA-3′ switching sequence in the TPP Spinach riboswitch. We next converted the transducer sequences in these riboswitches to the 5′-UCCC-3′ that was used in the optimal TPP Spinach riboswitch.
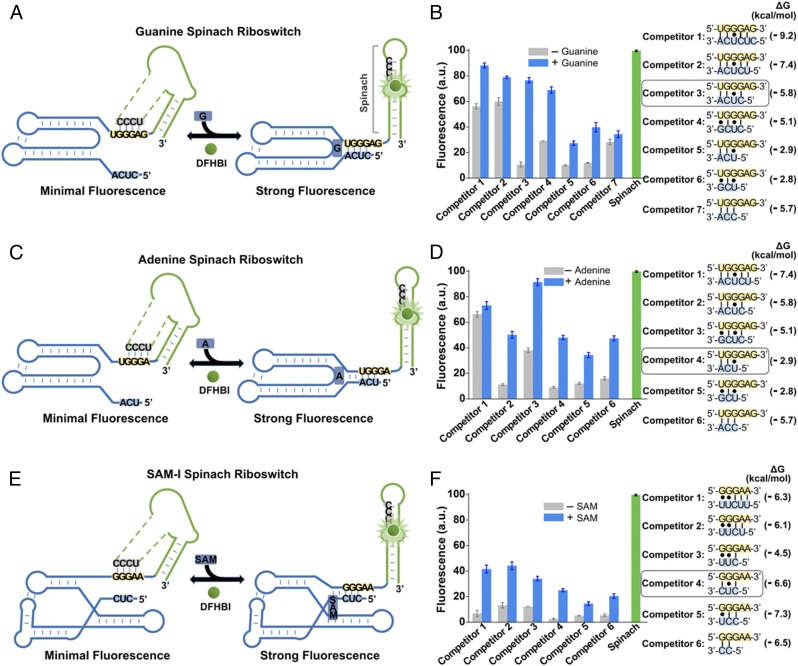
We next tested each candidate Spinach riboswitch after the addition of 10 µM guanine, 100 µM adenine, or 100 µM SAM, respectively (Fig. 6). In the guanine and adenine Spinach riboswitches, metabolite binding induced 57% and 10% increases in fluorescence, respectively, with high basal fluorescence seen in the absence of metabolite. However, in the SAM-I Spinach riboswitch, SAM induced a much more prominent (5.1-fold) increase in fluorescence.
Fig. 6.
Generation of Spinach riboswitches activated by diverse metabolites. (A) Secondary structure and design of the optimal guanine Spinach riboswitch inspired by the natural switching mechanism of the xpt guanine riboswitches. The guanine Spinach riboswitch is generated by swapping Spinach (green) into the expression platform of the xpt guanine riboswitch. Spinach binds and activates the fluorescence of DFHBI only after the target metabolite induces the release of the transducer sequence (gray) from the aptamer domain (blue) so it can bind to the H1 helix in Spinach. The switching sequence and the competitor sequence are highlighted in yellow and blue, respectively. (B) Optimization of the competitor sequence in the guanine Spinach riboswitch. Seven competitor sequences based on the xpt guanine riboswitches (competitors 1–7) were incorporated to transduce guanine binding into a Spinach riboswitch fluorescence signal. The optimal sensor (competitor 3) displayed a 6.2-fold increase in fluorescence signal upon the addition of 10 µM guanine. Shown are mean and SEM values of three independent replicates. The Gibbs free energy change (∆G) of hybridization events between each competitor sequence and the switching sequence was calculated using Mfold online software. (C) Secondary structure and design of the optimal adenine Spinach riboswitch inspired by the switching mechanism of the xpt adenine riboswitches. The adenine Spinach riboswitch is generated by swapping Spinach (green) into the expression platform of the xpt adenine riboswitch. Spinach binds and activates the fluorescence of DFHBI only after the target metabolite induces the release of the transducer sequence (gray) from the aptamer domain (blue) so it can bind to the H1 helix in Spinach. The switching sequence and the competitor sequence are highlighted in yellow and blue, respectively. (D) Optimization of the competitor sequence in the adenine Spinach riboswitch. Six competitor sequences based on the xpt adenine riboswitch (competitors 1–6) were incorporated to transduce adenine binding into a Spinach riboswitch fluorescence signal. The optimal sensor (competitor 4) displayed a 4.3-fold increase in fluorescence signal upon the addition of 100 µM adenine. Shown are mean and SEM values of three independent replicates. The Gibbs free energy change (∆G) of hybridization events between each competitor sequence and the switching sequence was calculated using Mfold online software. (E) Secondary structure and design of the optimal SAM-I Spinach riboswitch inspired by the natural switching mechanism of the yitJ SAM-I riboswitches. The SAM-I Spinach riboswitch is generated by swapping Spinach (green) into the expression platform of the yitJ SAM-I riboswitch. Spinach binds and activates the fluorescence of DFHBI only after the target metabolite induces the release of the transducer sequence (gray) from the aptamer domain (blue) so it can bind to the H1 helix in Spinach. The switching sequence and the competitor sequence are highlighted in yellow and blue, respectively. (F) Optimization of the competitor sequence in the SAM-I Spinach riboswitch. Six competitor sequences from the yitJ SAM-I riboswitch (competitors 1–6) were incorporated to transduce SAM binding into a Spinach riboswitch fluorescence signal. The optimal sensor (competitor 4) displayed an 8.8-fold increase in fluorescence signal upon the addition of 100 µM SAM. Shown are mean and SEM values of three independent replicates. The Gibbs free energy change (∆G) of hybridization events between each competitor sequence and the switching sequence was calculated using Mfold online software.
To improve sensor performance, we modified the competitor sequence. The competitor sequence influences the riboswitch by binding the switching sequence and displacing the transducer sequence (46–48). We reasoned that this interaction may be too strong in the guanine and adenine riboswitches, resulting in displaced transducer sequence and high basal fluorescence. Thus, we tested seven competitor sequences comprising different lengths and predicted degrees of hybridization with the switching sequence. In these experiments, a shorter competitor sequence resulted in markedly improved performance, with a 6.4-fold increase in fluorescence seen for the guanine Spinach riboswitch (Fig. 6B). Similarly, a short competitor sequence resulted in optimal performance of the adenine and SAM-I riboswitch (4.3-fold and 8.8-fold increases, respectively; Fig. 6 D and F).
Lasy we examine the specificity of these Spinach riboswitches (Table S1). In each case, the Spinach riboswitch maintained the high selectivity for the target molecule that is present in the parental riboswitch (Fig. S7). Together, these data suggest that a variety of naturally occurring riboswitches can be converted to Spinach riboswitches.
Discussion
Here we describe a strategy for swapping Spinach into certain naturally occurring riboswitches, allowing the riboswitches to function as genetically encoded fluorescent metabolite sensors. Drawing upon structural insights into the mechanism of structural switching in riboswitches, we show that Spinach can be swapped for the expression platform of various riboswitches, allowing metabolite binding to induce Spinach fluorescence directly. These Spinach riboswitches markedly simplify screening for riboswitch agonists and antagonists and can be used for fluorescence imaging of metabolite dynamics in living cells. Spinach riboswitches therefore are a novel class of RNA-based genetically encoded metabolite sensors.
Spinach riboswitches have advantages over other riboswitch-based methods for imaging metabolite levels. Riboswitches have been inserted into transcripts encoding GFP, so that changes in metabolite levels lead to changes in cellular fluorescence (49, 50). However, because GFP accumulates in cells, it is difficult to observe fluctuations in metabolite levels using this method. Temporal resolution also may be limited because of the time required for nascent GFP to mature to a fluorescent form (49, 50). Riboswitches also have been modified to contain fluorescent dyes, enabling FRET signals to occur in response to changes in metabolite concentration (51). This approach requires chemical modification of the RNA, which is much more challenging than using the Spinach riboswitches described here. Furthermore, these chemically tagged riboswitches are not genetically encodable, so they are difficult to use in living cells.
Using naturally occurring riboswitches for the design of sensors takes advantage of the affinity and selectivity that have been optimized by evolutionary selection. A major challenge with SELEX-generated aptamers is achieving the desired affinity. These aptamers typically bind target in the 0.1–10 µM range (52). Because many metabolites are present in the 10–1,000 µM range (53), aptamers with these affinities would be needed for sensor development. This requirement is complicated further because the binding affinity usually is measured in an artificial solution and can be highly influenced by ion concentrations, as is the case with the TPP riboswitch. Thus, it is difficult to know the binding affinity for a SELEX-derived aptamer in the cytosol of a living cell. This problem can be overcome by using naturally occurring riboswitches. Naturally occurring riboswitches have evolved to exhibit in vivo binding affinities that correspond to the cellular concentrations of their targets. Thus, using riboswitches rather than SELEX-derived aptamers provides an opportunity to achieve affinity and selectivity tuned for live bacterial cells.
The mechanism of Spinach riboswitches is different from that of the initial allosteric Spinach sensors (Fig. S1) (5, 8, 9, 54). Spinach riboswitches switch between two folded conformations in which the transducer sequence is associated with either the ligand-binding pocket or Spinach. In contrast, allosteric Spinach sensors switch from an unfolded to a folded state. In both cases, the sensor relies on stabilizing the helix that terminates in a U⋅A⋅U base triple that forms the surface of the DFHBI-binding pocket in Spinach. Although the two types of sensors use different mechanisms to switch from nonfluorescent to fluorescent states, the signal-to-noise ratio and kinetics of activation/deactivation of allosteric Spinach sensors (5) are similar to those of the Spinach riboswitches reported here. Thus, the primary advantage of Spinach riboswitches is that they retain the affinity and selectivity of the parental riboswitch.
In principle, the aptamer domains of riboswitches can be fused directly to the Spinach to make allosteric Spinach sensors (5, 8, 9, 54). Indeed, the aptamer domain of the SAM-III and GEMM-I riboswitches were used to generate SAM and cyclic di-GMP sensors, respectively (5, 8, 9, 54). However, this approach cannot be used if the aptamer domain does not have a stem at its base, as is the case in the pseudoknot-containing SAM-II, preQ1-II, and S-adenosylhomocysteine (SAH) riboswitches (55–57). Additionally, this approach can be used only if the aptamer domains undergo metabolite-induced folding upon binding the metabolites. Thus, the Spinach riboswitch has important advantages over allosteric Spinach sensors that use aptamer domains of riboswitches.
Riboswitches are seen primarily in bacteria and fungi but are rarely seen in mammalian cells. As a result, riboswitches have evolved to detect critical microbial metabolites that may not be as important for mammalian physiology. Additionally, the affinity and selectivity that evolved in bacterial riboswitches may not be suitable for use in mammalian cells. Also, because riboswitches are highly structured, they may be highly susceptible to degradation pathways in mammalian cells. Thus, selectivity, affinity, and RNA stability would be need to be optimized to use these sensors in mammalian cells.
Currently, many riboswitches remain as “orphans” with no known target. Various computational approaches have been described to predict potential metabolites that bind to the riboswitch, often based on the identity of the gene products that are regulated by the riboswitch (17–22). Because these riboswitches potentially can be converted into Spinach riboswitches, it can be valuable to deorphanize riboswitches to facilitate the development of novel genetically encoded sensors for diverse metabolites.
Materials and Methods
Cloning Sensor for Expression in E. coli.
RNA sensors were expressed in E. coli using pET28c (EMD Biosciences). The Spinach riboswitch was PCR amplified with primers containing either BglII or XhoI restriction sites on the 5′ or 3′ ends of the sensor sequence, respectively. This PCR product then was cloned into a pET28c-based vector (45) directly upstream of the T7 terminator sequence with BglII and XhoI restriction sites.
A pETDuet-1–based vector was engineered (45) to allow expression of the TPP Spinach riboswitch sensor under a T7 promoter and the eqFP670 far-red fluorescent protein under a second T7 promoter, which was used for expression normalization. The TPP Spinach riboswitch sensor was PCR amplified with primers containing either EagI or SacII restriction sites on the 5′ or 3′ ends of the sensor sequence, respectively. This PCR product then was cloned into the previously described (45) pETDuet-6-eqFP670 vector directly upstream of the T7 terminator sequence with EagI and SacII restriction sites.
Live-Cell Imaging of TPP Accumulation.
Rosetta2 (DE3) E. coli cells (Novagen) were transformed to express the Spinach riboswitch from pET28c under the control of a T7 promoter as previously described (5). Control cells were transformed with pET28c or pET28c containing the Spinach2 sequence (58). Cells were plated and grown overnight, and single colonies were picked for inoculation overnight in LB medium supplemented with 50 μg/mL kanamycin (LB-Kan). At OD600 = 0.4, 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to the culture alone or with 300 μg/mL adenosine, and shaking at 37 °C was continued for another 2 h. After induction, 100 μL of the culture was removed, spun down to pellet the culture, and resuspended in 1 mL of M9 minimal medium (pH 6.0). A 200-μL aliquot of resuspended culture then was plated on poly-d-lysine–coated, 24-well, glass-bottom dishes (MatTek) and incubated for 45 min at 37 °C. Adherent cells were washed twice and then incubated with 200 μM DFHBI in M9 minimal medium (pH 6.0) for 1 h at 25 °C. Then cells were treated with a 10-μM final concentration of thiamine or pyrithiamine alone or thiamine with 1 mM oxythiamine. Live fluorescence images were taken with a CoolSNAP HQ2 CCD camera (Photometrics) through a 60× oil objective mounted on a Nikon TE2000 microscope and were analyzed with the NIS-Elements software (Nikon), as described previously (5). The filter set was a sputter-coated filter cube with excitation filter 470/40 nm, dichroic mirror 495 nm (long pass), and emission filter 525/50 nm (Chroma Technology). The background signal for DFHBI alone was determined by imaging cells expressing an empty control vector under identical conditions and was subtracted using NIS-Elements software.
The eqFP670 fluorescent protein gene and the Spinach riboswitch coexpressing experiments were carried out from the pETDuet-5-TPP-eqFP670 plasmid described above. After induction, cells were adhered and imaged as described above.
Population Analysis of TPP Synthesis.
Population analysis was carried out using NIS-Elements software. Individual cells in a captured field were traced manually as regions of interest (ROIs) and were tracked through the different time points of image capture. Then fluorescence intensity was calculated by dividing the total fluorescence by the calculated volume for each ROI to give the mean intensity per unit volume. Volume was estimated using the equation V = πr2[(4/3)r + a], where V is volume, r is radius, and a is the side length of the E. coli cell. The fold increase in fluorescence over time was calculated as the ratio of mean intensity at a desired time point to that at time 0. After this value was calculated, cells were binned according to their percentage of total signal relative to the average signal, which was normalized to 100%.
Supplementary Material
Acknowledgments
We thank members of the S.R.J. laboratory and especially Grigory S. Filonov, Wenjiao Song, Karen Wu, Matteo Bolomini-Vittori, Nina Svensen, and Hyaeyeong Kim for useful comments and suggestions. This work was supported by NIH Grants R01 NS064516 and R01 EB010249 (to S.R.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504354112/-/DCSupplemental.
References
- 1.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3(12):906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 2.Frommer WB, Davidson MW, Campbell RE. Genetically encoded biosensors based on engineered fluorescent proteins. Chem Soc Rev. 2009;38(10):2833–2841. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd Ed Springer; New York: 2006. [Google Scholar]
- 4.Lindenburg L, Merkx M. Engineering genetically encoded FRET sensors. Sensors (Basel) 2014;14(7):11691–11713. doi: 10.3390/s140711691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335(6073):1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333(6042):642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song W, Strack RL, Svensen N, Jaffrey SR. Plug-and-play fluorophores extend the spectral properties of Spinach. J Am Chem Soc. 2014;136(4):1198–1201. doi: 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song W, Strack RL, Jaffrey SR. Imaging bacterial protein expression using genetically encoded RNA sensors. Nat Methods. 2013;10(9):873–875. doi: 10.1038/nmeth.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kellenberger CA, Wilson SC, Sales-Lee J, Hammond MC. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. J Am Chem Soc. 2013;135(13):4906–4909. doi: 10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 11.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 12.Stoltenburg R, Reinemann C, Strehlitz B. SELEX—a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Strack RL, Jaffrey SR. New approaches for sensing metabolites and proteins in live cells using RNA. Curr Opin Chem Biol. 2013;17(4):651–655. doi: 10.1016/j.cbpa.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klug SJ, Famulok M. All you wanted to know about SELEX. Mol Biol Rep. 1994;20(2):97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 15.Nahvi A, et al. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9(9):1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 16.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152(1-2):17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg Z, et al. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11(3):R31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrick JE, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101(17):6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker JL, et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335(6065):233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinberg Z, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35(14):4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandal M, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306(5694):275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 22.Nelson JW, et al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9(12):834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurgenson CT, Begley TP, Ealick SE. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419(6910):952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 25.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441(7097):1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards TE, Ferré-D’Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14(9):1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Serganov A, Patel DJ. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys. 2012;41:343–370. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceres P, Garst AD, Marcano-Velázquez JG, Batey RT. Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth Biol. 2013;2(8):463–472. doi: 10.1021/sb4000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warner KD, et al. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat Struct Mol Biol. 2014;21(8):658–663. doi: 10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, et al. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 2014;10(8):686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama H, Hayashi R. Biosynthesis of thiamine pyrophosphate in Escherichia coli. J Bacteriol. 1972;109(2):936–938. doi: 10.1128/jb.109.2.936-938.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulshina N, Edwards TE, Ferré-D’Amaré AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 2010;16(1):186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deigan KE, Ferré-D’Amaré AR. Riboswitches: Discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011;44(12):1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du Q, Wang H, Xie J. Thiamin (vitamin B1) biosynthesis and regulation: A rich source of antimicrobial drug targets? Int J Biol Sci. 2011;7(1):41–52. doi: 10.7150/ijbs.7.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12(12):1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Elena C, Liuhong C, Chris A, Finian LJ, Smith GA. Fragment screening against the thiamine pyrophosphate riboswitch thiM. Chem Sci. 2011;2:157–165. [Google Scholar]
- 37.Chen L, Cressina E, Leeper FJ, Smith AG, Abell C. A fragment-based approach to identifying ligands for riboswitches. ACS Chem Biol. 2010;5(4):355–358. doi: 10.1021/cb9003139. [DOI] [PubMed] [Google Scholar]
- 38.Warner KD, et al. Validating fragment-based drug discovery for biological RNAs: Lead fragments bind and remodel the TPP riboswitch specifically. Chem Biol. 2014;21(5):591–595. doi: 10.1016/j.chembiol.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35(16):5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonardi R, Roach PL. Thiamine biosynthesis in Escherichia coli: In vitro reconstitution of the thiazole synthase activity. J Biol Chem. 2004;279(17):17054–17062. doi: 10.1074/jbc.M312714200. [DOI] [PubMed] [Google Scholar]
- 41.Gerrits J, Eidhof H, Brunnekreeft JW, Hessels J. Determination of thiamin and thiamin phosphates in whole blood by reversed-phase liquid chromatography with precolumn derivatization. Methods Enzymol. 1997;279:74–82. doi: 10.1016/s0076-6879(97)79011-6. [DOI] [PubMed] [Google Scholar]
- 42.Kawasi T, Iwashima A, Nose Y. Regulation of thiamine biosynthesis in Escherichia coli. J Biochem. 1969;65(3):407–416. doi: 10.1093/oxfordjournals.jbchem.a129028. [DOI] [PubMed] [Google Scholar]
- 43.Cerecedo LP, Soodak M, Eusebi AJ. Studies on thiamine analogues. I. Experiments in vivo. J Biol Chem. 1951;189(1):293–299. [PubMed] [Google Scholar]
- 44.Shcherbo D, et al. Near-infrared fluorescent proteins. Nat Methods. 2010;7(10):827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136(46):16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113(5):577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 47.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11(12):1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10(9):701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 49.Topp S, et al. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl Environ Microbiol. 2010;76(23):7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wachter A, et al. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell. 2007;19(11):3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haller A, Altman RB, Soulière MF, Blanchard SC, Micura R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc Natl Acad Sci USA. 2013;110(11):4188–4193. doi: 10.1073/pnas.1218062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKeague M, Derosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012;2012:748913. doi: 10.1155/2012/748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5(8):593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kellenberger CA, Hammond MC. In vitro analysis of riboswitch-Spinach aptamer fusions as metabolite-sensing fluorescent biosensors. Methods Enzymol. 2015;550:147–172. doi: 10.1016/bs.mie.2014.10.045. [DOI] [PubMed] [Google Scholar]
- 55.Corbino KA, et al. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6(8):R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA. 2008;14(4):685–695. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell. 2008;29(6):691–702. doi: 10.1016/j.molcel.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods. 2013;10(12):1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alatossava T, Jütte H, Kuhn A, Kellenberger E. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol. 1985;162(1):413–419. doi: 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.