Significance
During Alvin and Nautile dives in 1999, samples were collected from water surrounding sulfide chimneys of a hydrothermal vent along the East Pacific Rise and four mesophilic bacteria were isolated, including a novel Vibrio species, Vibrio antiquarius. Genomic, functional, and phylogenetic analyses indicate an intriguing blend of genomic features related to adaptation and animal symbiotic association, and also revealed the presence of virulence genes commonly found in Vibrio species pathogenic for humans. The presence of these virulence genes in an ecologically distinct Vibrio species was surprising. It is concluded that pathogenicity genes serve a far more fundamental ecological role than solely causation of human disease.
Keywords: Vibrio, hydrothermal vent, genomics, EX25
Abstract
Vibrio species are both ubiquitous and abundant in marine coastal waters, estuaries, ocean sediment, and aquaculture settings worldwide. We report here the isolation, characterization, and genome sequence of a novel Vibrio species, Vibrio antiquarius, isolated from a mesophilic bacterial community associated with hydrothermal vents located along the East Pacific Rise, near the southwest coast of Mexico. Genomic and phenotypic analysis revealed V. antiquarius is closely related to pathogenic Vibrio species, namely Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio harveyi, and Vibrio vulnificus, but sufficiently divergent to warrant a separate species status. The V. antiquarius genome encodes genes and operons with ecological functions relevant to the environment conditions of the deep sea and also harbors factors known to be involved in human disease caused by freshwater, coastal, and brackish water vibrios. The presence of virulence factors in this deep-sea Vibrio species suggests a far more fundamental role of these factors for their bacterial host. Comparative genomics revealed a variety of genomic events that may have provided an important driving force in V. antiquarius evolution, facilitating response to environmental conditions of the deep sea.
With more than 110 recognized species, the genus Vibrio comprises a diverse group of heterotrophic bacteria, of which many are known pathogens, causing disease in animals and humans (1, 2). Vibrio cholerae is the most notorious because it is the causative agent of cholera. Vibrio vulnificus and Vibrio parahaemolyticus cause severe illness in humans and are associated with consumption of contaminated seafood (3, 4). Vibrio harveyi (5), Vibrio anguillarum (6, 7), and V. parahaemolyticus (8) continue to cause substantial economic losses to the aquaculture industry worldwide.
Vibrios demonstrate a wide range of niche specialization: for example, free-living, attached to biotic and abiotic surfaces, and resident in both estuarine and marine habitats (9). The deep sea constitutes the largest habitat of the biosphere that supports microbial communities across three domains of life and represents an environment where physiochemical parameters—such as low temperature, high salinity, and high pressure—modulate community structure (10, 11). Several studies have shown the presence of physiologically, metabolically, and phylogenetically diverse mesophilic microbial communities in the deep sea, including Vibrio species (12–15). Barotolerant Vibrio spp. have been isolated from deep-sea sediment and from the gut microflora of invertebrates and fish collected from a variety of deep-sea habitats, including hydrothermal vents (16, 17). For example, strains of Vibrio, Aeromonas, and Pseudomonas spp. were isolated from mud-water samples collected at a depth of 4,940 m, 150 miles east of Cape Canaveral, Florida (18). Several culture-dependent and -independent studies have confirmed the ubiquity of vibrios, and suggested Vibrio populations generally comprise approximately 1% (by molecular techniques) of the total bacterioplankton in estuaries (19), in contrast to culture-based studies demonstrating that vibrios can comprise up to 10% of culturable marine bacteria (20). Clearly, vibrios are ubiquitous and abundant in the aquatic environment on a global scale, including both seawater and sediment (19, 21–25), and repeatedly shown to be present in high densities in and on marine organisms, such as corals (26), fish (27–29), mollusks (30), seagrass, sponges, shrimp (28, 31), and zooplankton (16, 17, 28, 32, 33).
During dives of the deep-sea submersibles Alvin and Nautile in 1999 along the East Pacific Rise, southwest of the Mexico coast, samples of water surrounding sulfide chimneys of a hydrothermal vent community were collected and four mesophilic bacterial isolates were cultured, which were subsequently tested for phenotypic traits, including growth on V. cholerae selective thiosulfate-citrate-bile-salts-sucrose (TCBS) agar (Oxoid). The sampling locations from where these four mesophilic bacteria were isolated are described in Table 1. Using single-gene phylogenies, the four isolates were identified as Shewanella algae, V. harveyi, and two novel Vibrio species, designated Vibrio sp. EX25 and Vibrio sp. EX97. Among them, Vibrio sp. EX25 showed phenotypic and genetic similarity to Vibrio alginolyticus, V. parahaemolyticus, and V. cholerae, all of which are human pathogens. Because it appeared to be a new Vibrio species derived from a novel habitat and closely related to human pathogenic Vibrio spp., we sequenced the whole genome of EX25 to understand its evolutionary lineage and determine its gene content, specifically those genes associated with pathogenicity in humans.
Table 1.
Locations where samples were collected and from which four mesophilic bacterial isolates were obtained
| Sample | Location | Depth (m) | Source | Strain no. |
| 1 | 9°N | 2,520 | Sulfide chimney | EX25 |
| 2 | 9°N | 2,500 | Sulfide chimney | EX97 |
| 3 | 13°N | 2,596 | Sulfide chimney | BB4 |
| 4 | 13°N | 2,602 | Sulfide chimney | A6.mk |
Results and Discussion
Phylogenetic analysis based on 16S rRNA showed that three of the four isolates belonged to the genus Vibrio (BB4, EX25, and EX97) and the fourth isolate, A6, was identified as S. algae. Vibrio BB4 branched with V. harveyi (Fig. 1). Although isolate EX97 was clustered with V. parahaemolyticus, EX25 branched independently of the V. parahaemolyticus, and V. alginolyticus–Vibrio campbellii clade. Both isolates were sucrose-positive, oxidase-positive, and required NaCl for growth. The isolates grew well under both micro aerophilic [Gas Pak anaerobic system (H2 and CO2); Beckton-Dickson] and strict anaerobic conditions (80% N2, 10% CO2, and 10% H2). In 1% tryptone amended with NaCl, EX25 grew at NaCl concentrations up to 10% and in alkaline peptone water visible growth also occurred at temperatures as high as 50 °C. Because protein-encoding genes evolve faster than rRNA genes, additional trees were constructed based on the protein-encoding genes toxR (cholera toxin transcriptional activator), ompW (outer membrane protein W), and other highly conserved genes among Vibrio spp. to yield better resolution in phylogenetic analyses. The constructed phylogenetic trees were in good agreement with 16S tree. DNA-DNA hybridization was also conducted to understand the genomic relatedness of the two isolates to 41 type strains of Vibrionaceae, and the highest relative branching ratio of 42% (Fig. S1) was obtained when EX25 was hybridized with type strains of V. parahaemolyticus and V. alginolyticus.
Fig. 1.
16S rRNA phylogeny of the East Pacific Rise isolates and other Vibrio species by neighbor-joining tree. Bootstrap consensus tree was inferred from 5,000 replicates, representing evolutionary history of the taxa. Scale bar represents 0.01 nucleotide substitutions per sequence position.
Taxonomy of Vibrio EX25 Based on Average Nucleotide Identity.
Species delineation was determined by average nucleotide identity (ANI) between genomes (34), with highest ANI observed between Vibrio EX25 and V. alginolyticus 12G01 (91%) and V. parahaemolyticus RIMD 2210633(84%), both of which is below the hypothesized species demarcation threshold value of 95% (35, 36) (Fig. S2A), indicating EX25 as a separate species closely related to V. alginolyticus and V. parahaemolyticus. A tetra nucleotide signature correlation index, an alignment-free parameter helpful in deciding whether a given pair of organisms belongs to the same species (36), demonstrated EX25 to have highest correlation with V. alginolyticus, yet below the same species threshold (Fig. S2B). These findings, along with16S rDNA and DNA-DNA hybridization results, confirm separate species designation for Vibrio sp. EX25, for which the species name Vibrio antiquarius is proposed (i.e., a bacterium of the aquatic environment derived from the antiquity of the deep sea).
Whole Genome Phylogeny.
The phylogeny of V. antiquarius was inferred by constructing a genome-relatedness neighbor-joining tree, using homologous alignment of 522 orthologous protein-coding genes of 36 Vibrio genomes, as a strict measure of the core Vibrio genome. The evolutionary tree (Fig. 2) showed fully resolved bifurcating patterns, with varying levels of diversity as evidenced by tree branch lengths, placing V. antiquarius, V. alginolyticus, V. parahaemolyticus, Vibrio sp. AND4, and V. harveyi in a monophyletic clade. V. antiquarius EX25 branched with V. alginolyticus, with V. parahaemolyticus as an outgroup to both of them. Strains of V. cholerae and V. vulnificus were each monophyletic within the species. This finding corroborated findings by shared gene content, ANI, and 16S phylogeny, and strongly indicates that at least for the core of these genomes, they share a common ancestry that excludes V. cholerae.
Fig. 2.
Core genome phylogeny of V. antiquarius EX25. Neighbor-joining tree was constructed based on alignment of homologous sequences of 525 conserved ORFs. Scale bar represents 0.05 substitutions per site.
Genome Features.
A combination of Sanger sequencing and 454 pyrosequencing yielded high-quality assembly of the EX25genome, with an asymmetrical, two-chromosome structure observed, consistent for Vibrio genomes (37). The larger (C-I) and smaller (C-II) chromosomes comprised 3.26 and 1.83 Mb, respectively (Fig. S3 and Table S1), with 45% G+C composition. Chromosomal distribution of the genes followed a pattern typical of vibrios, with C-I predominantly carrying genes for viability and growth, and C-II containing genes associated with adaptation to environmental change. A total of 4,529 protein-coding sequences (CDS) were identified (2,846 at C-I and 1,683 at C-II) with 27.5% of CDSs annotated as hypothetical proteins. Additionally, the V. antiquarius EX25 genome contained a superintegron (SI) cassette spanning approximately 113 kb on C-I (Table S1).
Comparative Genomics.
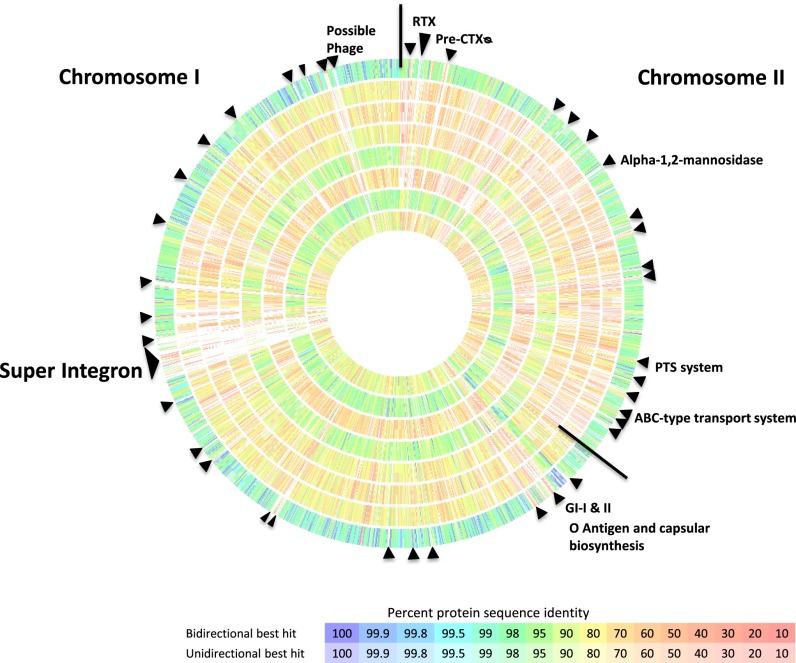
Multigenome comparison (Fig. 3) was carried out using reannotated eight reference Vibrio genomes (38). Pairwise reciprocal BLAST analysis revealed EX25 shared higher predicted CDSs with V. parahaemolyticus (3,973, 87.7%), V. alginolyticus (3,943, 87%), V. harveyi (3,567, 79%), and V. vulnificus (3,309, 73%) (Fig. S4). CDSs shared with V. parahaemolyticus (3,973) and V. vulnificus (3,309) corresponded to 82% and 84.5% of the V. parahaemolyticus and V. alginolyticus genomes. Analyses of shared gene content indicated V. antiquarius, V. alginolyticus, and V. parahaemolyticus are about equidistant (Fig. S4). Additionally, a large number of genomic regions (∼23 on C-I and ∼17 on C-II) were found with significant mismatch (Fig. 2) compared with other reference Vibrio genomes. These mismatches occurred most likely because of the insertion of genomic islands and acquisition of mobile genetic elements in those regions, resulting in strain specific CDSs. Interestingly, C-II contained more strain-specific CDSs than C-I. These chromosomal regions contributed significantly to the 586 CDSs (13% of the V. antiquarius genome), which had no reciprocal match in the V. alginolyticus genome. Approximately 43% (256 CDSs) of these CDSs represent either hypothetical proteins or proteins of unknown function. The presence of an integrase gene, together with a G+C content atypical to the chromosomal G+C, and at least seven major regions of disagreement, including pre-CTX prophage, SI, Vibrio pathogenicity island 2 (VPI-2), ORF: 4331–4339, ORF: 4301–4312, ORF: 3898–3902, and ORF: 1829–1846, suggests these regions are subject to horizontal gene transfer or chromosomal integration via phage, but might have a necessary function in the deep-sea habitat. The V. alginolyticus genome contained 723 CDSs (15% of its genome) without any reciprocal match with V. antiquarius.
Fig. 3.
Genome comparison of V. antiquarius EX25 with other Vibrio genomes. From outer ring to inner ring; V. alginolyticus 12G01, V. cholerae N16961, V. fischeri MJ11, Vibrio furnissii CIP 102972, V. harveyi ATCC BAA-1116, V. hollisae CIP 102972, V. parahaemolyticus RIMD 2210633, V. vulnificus CMCP6. Horizontal lines were drawn to separate chromosomes. Solid black arrows indicate areas of rearrangement or insertion site of genomic islands.
Genome Plasticity.
Genomic islands (GI’s), notably pathogenicity islands, contribute to the evolution and diversification of microbial life. The V. antiquarius genome encoded >70 genomic islands as predicted by Island Viewer (39) (Fig. S5 and Table S2). Among the GIs predicted by multiple methods, 21 were located on C-I and 12 were on C-II. Average size of the islands in C-I was 14 kb, with G+C content ranging from 36 to 40%, which is lower than the overall chromosomal G+C of 45%. The 12 GIs on C-II had a G+C content ranging from 38 to 42%, with 10.7-kb average size. Part of the GI-I (7%) displayed homology (74%) with the O-antigen gene cluster of Escherichia coli serotype O98: K?:H8 and Shigella dysenteriae strain M13547, whereas many of GI-II–encoded proteins are related to capsular polysaccharide biosynthesis and exhibit ∼68% sequence similarity with the E. coli capsule transport proteins. A total of 222 strain-specific CDSs identified in the genome of V. antiquarius (4.9% of the total CDSs) had no reciprocal match in the genomes of V. alginolyticus, V. parahaemolyticus, V. vulnificus, V. harveyi, or V. cholerae. Among V. antiquaries-specific protein-coding sequences, 116 (52.25%) are hypothetical or proteins of unknown function. Linear pairwise comparison of the V. antiquarius EX25 genome demonstrated several intra- and interchromosomal rearrangements compared with V. parahaemolyticus, V. harveyi, V. cholerae, and V. vulnificus (Fig. S6). Compared with V. parahaemolyticus and V. harveyi, C-I of V. antiquarius demonstrated a high degree of synteny, compared with C-II (Fig. S7). Such rearrangements are in agreement with the supposition that extensive genome plasticity is common in Vibrio species, particularly on C-II (40). Additionally, the V. antiquarius genome contains many perfect and approximate tandem repeats. Using the tandem-repeats finder (41), 64 and 20 tandem repeats were identified in the EX25 genome, with period lengths of 6–417 and 6–429 bases in C-I and C-II, respectively (Table S3). Many tandem repeats were in protein-encoding genes exhibiting high mutation rates. The V. antiquarius genome also contained insertion sequences (IS elements) throughout its genome (Table S4). Therefore, these might be important processes in V. antiquarius evolution to facilitate faster adaptation or quicker response to rapidly changing and challenging environmental conditions of the deep sea.
Predicted Biology of V. antiquarius.
The genome of V. antiquarius EX25 encodes a number of genes that are predicted to protect the bacterium from the environmental conditions of the deep sea and are illustrated in Table 2. Functions encoded in the genome include cytochromes for reduction of O2 to H2O2 and cytochrome C551 peroxidase, which detoxifies peroxide, multiple catalase genes, and a superoxide dismutase for tolerating high O2 concentrations, and genes encoding alkyl hydroperoxide reductase to scavenge endogenous hydrogen peroxide (Table 2). The ability to scavenge endogenous hydrogen peroxide was absent in the other Vibrio genomes and is the major antioxidant enzyme of the endosymbiont of the tubeworm, Riftia pachyptila, inhabiting deep-sea vents (42). Metalloendopeptidases and zinc-dependent carboxypeptidases were also present in the genome of V. antiquarius, useful in functioning in high concentrations of heavy metals, including zinc, present in the deep-sea vent environment (43).
Table 2.
Predicted characteristics of V. antiquarius EX25 genome
| Predicted biology | Chromosome (occurrence) | Function |
| Response to environment | ||
| Cytochrome bd | C-II (2) | Protection against O2 and H2O2 |
| Cytochrome C551 peroxidase | C-II | |
| Catalase | C-II (2) | Tolerating high oxygen concentrations |
| Superoxide dismutase | C-I (2) | |
| Alkyl hydroperoxide reductase | C-II | Scavenge endogenous hydrogen peroxide, a trait absent in the other Vibrio genomes |
| Methionine sulfoxide reductases | C-I | Oxidative damage repair |
| Metalloendopeptidases | C-II (2) | Functioning in high concentrations of heavy metals, including zinc |
| Zinc-dependent carboxypeptidases | C-II | |
| Cell sensing system | ||
| LuxP and LuxQ | C-I | Autoinducer-2 (AI-2) mediated quorum sensing, biofilm formation, virulence, and other metabolic functions |
| LuxS and LuxN | C-II | |
| Biofilm-related pathways | ||
| Ornithine and arginine decarboxylase | C-I | Polyamine biosynthesis |
| C-di-GMP phosphodiesterase mbaA | C-I | Norspermidine |
| syp gene cluster | C-I | Gene clusters mediating biofilm formation |
| Polar flagellum cluster, P | C-I | Flagellar clusters |
| Lateral flagellum cluster, LF | C-II | |
| Others | ||
| rmf and sulA | C-I, C-II | Persister cells |
| Multicopper oxidase | C-I | Mn(II) oxidation |
| δ-9 fatty acid desaturase | C-II | Fatty acid unsaturation, essential for growth under high pressure |
| Universal stress proteins | C-II (3) | Function not defined |
Manganese is used as a reliable tracer of hydrothermal vent emissions (44–46) and, unlike the other vibrios, the genome of V. antiquarius contains a gene annotated as multicopper oxidase, an enzyme essential for manganese oxidation, and laccase-like activity (47). V. antiquarius also contains delta-9 fatty acid desaturase. Fatty acid unsaturation is a critical cellular process shown to be essential for growth under high pressure by increasing the rigidity of membranes and genes like delta-9 fatty acid desaturase are presumably up-regulated to increase membrane unsaturation and fluidity (48).
Virulence Factors.
The genome of V. antiquarius encodes a type III secretion system (T3SS), responsible for enabling injection of effector proteins directly into target host eukaryotic cells (49). T3SS genes induce severe diarrhea in models of cholera infection (50) and are frequently found in V. parahaemolyticus (51). Unlike V. parahaemolyticus, V. alginolyticus, V. harveyi, and V. vulnificus, the genome of V. antiquarius also contains two clusters of type VI secretion systems (T6SS) genes on C-II. However, the two effector molecules, VgrG and Hcp, and regulatory proteins, vasK and vasH, are located in different clusters. To date, T6SSs have been defined as required for virulence or survival of a bacterium in a eukaryotic host (52–54). However, Weber et al. (55) reported T6SS in V. anguillarum regulates stress response, suggesting T6SS has an ecological rather than pathological consequence for its host (i.e., an adaptive mechanism). The V. antiquarius genome also contains tight adherence (tad) locus genes (rcp, rcpA, and tadZ), functioning in colonization of surfaces and biofilm formation (56).
V. antiquarius encodes the thermo labile hemolysin (tlh) gene in C-II, with 97% and 84% nucleotide similarity to that of V. alginolyticus and V. parahaemolyticus, respectively, but lacks tdh and trh of V. parahaemolyticus. Multiple putative proteases and other genes, whose products are predicted to encode hemolysins, are present in the genome of V. antiquarius EX25, along with homologs of ToxR, ToxS, and Integron Integrase Intl4, whose role in virulence is well established in V. cholerae and other Vibrio species. Other genes involved in pathogenicity are also present, including type IV pilin, pilA, which encodes proteins expressed during human infection, mannose-sensitive hemagglutinin, and RTX toxin. The genome contains a homolog of vvhA and, in a recent study, Smith and Oliver (57) suggested a role for haemolysin (vvhA) in V. vulnificus to aid in osmoregulation and cold-shock response. The pre-CTX prophage of V. cholerae is indicated by the presence of the accessory cholera enterotoxin, ace, Zonula occludence toxin, zot, RstA phage-related protein, and RstB phage-related integrase genes on C-II. An insertion of bacteriophage genes was noted between rstA and ace, with 96% homology to bacteriophage BfO4K68 (Fig. 4A). This region is most likely horizontally transferrable via bacteriophage BfO4K68. Although the genome of V. antiquarius does not contain the VPI-encoding receptor for CTX prophage, it does have an approximately 27.4-kb contiguous region on C-I (Fig. 4B), with 86% nucleotide sequence similarity to VPI-2, found in both clinical and environmental strains of V. cholerae, a region spanning VC1758 to VC1772 of the canonical VPI-2 and encoding a type I restriction-modification system and five hypothetical proteins. However, sections VC1773 to VC1810 were not found; instead there is a 12-kb region that includes six hypothetical proteins and a phage integrase. The 12-kb insert did not show significant match with any sequences in the National Center for Biotechnology Information GenBank database, except for a 103-bp region with 78% sequence similarity to a transcriptional regulator of V. parahaemolyticus. Recent analyses of the VC1773 to VC1810 region of this island suggest this is a hot-spot for novel DNA insertion (58). The presence of virulence factors, including two reported in V. cholerae, namely pre-CTXΦ and VPI-2 in the noncholera Vibrio antiquarius from a deep-sea environment, suggests their multifaceted role outside the human host; that is, ecological function in the natural habitat and alternate evolutionary origins apart from their core genome.
Fig. 4.
Schematic representation of (A) Pre-CTXphi and (B) VPI-2 like element, identified in the genome of Vibrio antiquarius EX25.
Metagenomic Survey.
To determine whether V. antiquarius may be present in other environmental habitats, V. antiquarius ORFs were queried against publicly available environmental metagenomic datasets. Using a conventional Blast search, V. antiquarius EX25-specific sequences were detected in 89 shotgun metagenomic datasets and comprise saltern (60%), marine (20%), coral (3%), and human gut metagenomes (2%) (Fig. S8). Distribution of V. antiquarius ORFs in these metagenomes suggests ubiquity of V. antiquarius in the natural environment.
Summary
Because vibrios are autochthonous to a diverse and wide range of aquatic niches, it was of interest to investigate their potential presence in the deep-sea hydrothermal vents, the only deep-sea environment having high enough temperature to be supportive of mesophilic growth. Analysis of the samples collected from hydrothermal vents did reveal the presence of vibrios in this environment, and interestingly, isolates demonstrated some phenotypic and genotypic similarity to Vibrio species pathogenic for humans. Whole-genome sequencing and subsequent comparative and phylogenomics analysis of one of the deep-sea Vibrio isolates, EX25, revealed that it belongs to a new Vibrio species, for which we propose the name, Vibrio antiquarius. The genome of V. antiquarius encodes many genes that can be interpreted as contributing to its being native to the deep-sea environment, including genes (i.e., metalloendopeptidases, zinc-dependent carboxypeptidases, and so forth) that are indicative of its potential association with deep-sea animals. Several studies (12–15) in the past have shown the presence of diverse mesophilic microbial communities, including Vibrio species, in various deep-sea environments; however, information on the microbial communities associated with the deep-sea vent animals appeared to be very limited. Our study indicates that mesophilic vibrios are likely to be present in the mesophilic environment of the deep sea, particularly in association with the inhabiting animals. Additionally, the genome contained homologs of many virulence genes that are commonly found in Vibrio species pathogenic to human and other animals. Wide distribution of virulence genes among coastal, estuarine, and riverine Vibrio species, including V. parahaemolyticus, V. cholerae non-O1, Vibrio mimicus, Vibrio hollisae, Vibrio fluvialis, and V. alginolyticus are well known (59–61); however, finding these virulence genes in a deep-sea Vibrio sp. raises a significant question whether pathogenicity genes, in addition to pathogenicity for humans and other animals, are in fact providing ecological functions in the natural environment. Recent studies have shown that several virulence factors and pathways in Vibrio species that have a role in pathogenicity for humans may also have roles in the aquatic environment (62), and some of the virulence genes might be relevant for basic metabolic processes, establishing the symbiosis (63), or modulating prey/predator relationships (64) in their natural ecosystems. For example, the GbpA ligands in V. cholerae, which are involved in the intestinal colonization, have also been reported to mediate bacterial attachment to the chitinous surfaces and biofilm formation in the aquatic environment (65, 66). The tracheal cytotoxin produced by Vibrio fischeri has also been reported to be involved in the symbiotic relationship of V. fischeri with bobtail squid (63). Similarly, the metalloendopeptidases and zinc-dependent carboxypeptidases in V. antiquarius genome also encode genes (i.e., metalloendopeptidases, and zinc-dependent carboxypeptidases, and so forth) that could be indicative of its potential association with deep-sea animals. Therefore, it is possible that some of these pathogenicity genes function in the commensal relationship that V. cholerae and other vibrios have with zooplankton, notably copepods (67), encoding attachment, signaling, and interactions in aquatic communities, including the deep sea, and therefore primarily may play an ecological role in the natural environment. The presence of these genes in ecologically and phylogenetically diverse Vibrio species also suggests that these affiliations between commensals are likely very old and indicate a likely common evolution of Vibrio species into pathogens of humans and marine animals. Clearly, a new perspective is needed for understanding the intersecting roles of Vibrios in the environment and as a pathogen for humans and marine animals.
Materials and Methods
Sample Collection and Isolation of Cultures.
Samples were collected from two sites on the East Pacific Rise, 9°50′N, 104°17′W by Deep Submergence Vehicle Alvin, and 13°N (12°49′N, 103°56′W) at a depth of 2,500 m by Deep Submergence Vehicle Nautile. Further description of sampling procedures and isolation sites has been described elsewhere (68). Samples were inoculated into heterotrophic, anaerobic seawater-based media immediately after sample retrieval onboard the mother ship. Samples collected from 9°N were inoculated into MSH medium (69), with 4 mM FeS, 4 mM H2S, 0.5 g/L yeast extract, 0.5 g/L trypticase peptone, with and without 5 mM acetate. Samples from 13°N were inoculated into MSH medium amended with 0.05 g/L of yeast extract, 0.05 g/L trypticase peptone, 20 mM acetate, 0.3 g/L coenzyme M, and 15 mM of iron pyrophosphate. All samples were incubated at 25 °C (9°N) or 30 °C (13°N). Four pure cultures, obtained by serial dilution and colonies picked from roll tubes, were designated A6 and BB4 (13°N), and EX25 and EX97 (9°N). The cultures were enriched aerobically in alkaline peptone water (APW) and Luria-Bertani broth (LB) and spread on TCBS. Yellow colonies (sucrose positive) on TCBS were streaked on the same agar medium (APW or LB) used for selection. The pure cultures were subjected to biochemical tests using API20E strips (Biomerieux Vitek). Salt tolerance was assayed in nutrient broth containing 3%, 6%, and 8% (wt/vol) NaCl.
DNA–DNA Hybridization.
Relatedness of the isolates to reference strains of Vibrio spp. was also determined by total DNA–DNA hybridization, using random-primer labeling and chemiluminescent detection with DIG High Prime DNA Labeling and Detection Starter Kit II (Roche). Ten replicates of each strain (10 ng) were hybridized with genomic DNA from 41 reference-type strains of Vibrionaceae species. Four additional strains of V. cholerae and two of V. mimicus were included as reference strains and their relative binding ratios calculated. The reference strains were also probed against one another, serving as controls.
Immunological identification, using anti–DIG-AP and CSPD (chemiluminescent substrate) was performed, according to manufacturer’s protocol (Roche). Blots were exposed to X-ray film for 20 min to 4 h, depending on the signal obtained. Developed film was scanned using a densitometer (Personal Densitometer SI) and dots quantitatively evaluated using ImageQuaNT software for Windows NT (Molecular Dynamics, v4.2a).
Genome Sequencing.
The genome of Vibrio sp. EX25 was sequenced by the Joint Genome Institute, and all general aspects of sequencing performed at the Joint Genome Institute can be found at jgi.doe.gov/. Draft sequences were obtained from a blend of Sanger and 454 sequences and involved paired-end Sanger sequencing on 8-kb plasmid libraries to 5× coverage, with 20× coverage of 454 data accomplished and optional paired end Sanger sequencing on 35-kb fosmid libraries to 1–2× coverage (depending on repeat complexity). ThePhred/Phrap/Consed software package (www.phrap.com) was used for sequence assembly and quality assessment (70, 71). After the shotgun stage, reads were assembled with parallel phrap (High Performance Software). Draft assemblies were based on 39,974 total reads. Repeat resolution was performed using Dupfinisher (72). Gaps between contigs were closed by editing in Consed and several targeted finishing reactions, including transposon bombs (73), primer walks on clones, primer walks on PCR products, and adapter PCR reactions. Gene-finding and annotation were achieved using the RAST server (38). The completed genome sequences of Vibrio EX25 contained 42,569 reads, achieving average eightfold sequence coverage, with error rate of less than 1 in 100,000. 16S rRNA GenBank accession numbers for Vibrio sp. BB4, EX25, EX97, and S. algae A6 are AF 319768; AF319769, AF319770, and AF 319767, respectively.
Comparative Genomics.
Genome-to-genome comparison was performed using three approaches. First, nucleotide sequences as whole contigs were directly aligned using the MUMmer (74). Second, ORFs of a given pair of genomes were reciprocally compared with each other, using BLASTN, BLASTP, and TBLASTX (ORF-dependent comparison). Third, a bioinformatic pipeline was developed to identify homologous regions of a given query ORF. Initially, a segment on a target contig homologous to a query ORF was identified using BLASTN. This potentially homologous region was expanded in both directions by 2,000 bp, after which nucleotide sequences of the query ORF and selected target homologous region were aligned, using a pairwise global alignment algorithm (75), and the resultant matched region in the subject contig was extracted and saved as a homolog (ORF-independent comparison). Orthologs and paralogs were differentiated by reciprocal comparison. In most cases, both ORF-dependent and independent comparisons yielded the same orthologs, although the ORF-independent method performed better for draft sequences of low quality where sequencing errors, albeit rare, hampered identification of correct ORFs. Orthologous regions were used to generate phylogenetic trees. The set of orthologous regions for each CDS were aligned using CLUSTALW2. The resultant multiple alignments were concatenated to form genome scale alignments which were then used to generate the neighbor-joining (76) phylogenetic tree.
Supplementary Material
Acknowledgments
We thank Dr. A. L. Reyesenbach and Dr. Krista Longnecker for advice and assistance in collecting isolates and 16S RNA analysis. This study was supported by National Institutes of Health Grant 1R01A139129-01 and National Oceanic and Atmospheric Administration, Oceans and Human Health Initiative Grant S0660009 (to R.R.C.). Funding for genome sequencing was provided by the Office of the Chief Scientist and National Institute of Allergy and Infectious Diseases Microbial Sequencing Centers (N01-AI-30001 and N01-AI-40001).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503928112/-/DCSupplemental.
References
- 1.Farmer JJ, Janda JM, Brenner FW, Cameron DN, Birkhead KM. Genus I. Vibrio pacini 1854. In: Brenner DJ, Kreig NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. 2 Ed. Vol 2. Springer Science Business Media; New York: 2005. pp. 494–546. [Google Scholar]
- 2.Pruzzo C, Huq A, Colwell RR, Donelli G. Pathogenic Vibrio species in the marine and estuarine environment. In: Belkin S, Colwell RR, editors. Ocean and Health Pathogens in the Marine Environment. Springer; New York: 2005. pp. 217–252. [Google Scholar]
- 3.Hülsmann A, et al. RpoS-dependent stress response and exoenzyme production in Vibrio vulnificus. Appl Environ Microbiol. 2003;69(10):6114–6120. doi: 10.1128/AEM.69.10.6114-6120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong HC, Wang P, Chen SY, Chiu SW. Resuscitation of viable but non-culturable Vibrio parahaemolyticus in a minimum salt medium. FEMS Microbiol Lett. 2004;233(2):269–275. doi: 10.1016/j.femsle.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Owens L, Busico-Salcedo N. Vibrio harveyi: Pretty problems in paradise. In: Thompson FL, Austin B, Swings J, editors. The Biology of the Vibrios. ASM Press; Washington, DC: 2006. pp. 266–280. [Google Scholar]
- 6.Crosa JH, Actis LA, Tolmasky ME. The biology and pathogenicity of Vibrio anguillarum and Vibrio ordalii. In: Thompson FL, Austin B, Swings J, editors. The Biology of the Vibrios. ASM Press; Washington, DC: 2006. pp. 251–265. [Google Scholar]
- 7.Miyamoto N, Eguchi M. Response to low osmotic stress in a fish pathogen, Vibrio anguillarum. FEMS Microbiol Lett. 1997;22(3):225–231. [Google Scholar]
- 8.Austin B, Zhang XH. Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol. 2006;43(2):119–124. doi: 10.1111/j.1472-765X.2006.01989.x. [DOI] [PubMed] [Google Scholar]
- 9.Reen FJ, Almagro-Moreno S, Ussery D, Boyd EF. The genomic code: Inferring Vibrionaceae niche specialization. Nat Rev Microbiol. 2006;4(9):697–704. doi: 10.1038/nrmicro1476. [DOI] [PubMed] [Google Scholar]
- 10.Jannasch HW. Microbial interaction with hydrothermal fluids. In: Humphris SE, Zierenberg RA, Mullineaux LS, Thomson RE, editors. Sea Floor Hydrothermal Systems: Physical, Chemical, Biological, and Geological Interactions. American Geophysical Union; Washington, DC: 1995. pp. 273–296. [Google Scholar]
- 11.Karl DM, Wirsen CO, Jannasch HW. Deep-sea primary production at the galapagos hydrothermal vents. Science. 1980;207(4437):1345–1347. [Google Scholar]
- 12.Moyer CL, Dobbs FC, Karl DM. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61(4):1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raguénès G, Christen R, Guezennec J, Pignet P, Barbier G. Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int J Syst Bacteriol. 1997;47(4):989–995. doi: 10.1099/00207713-47-4-989. [DOI] [PubMed] [Google Scholar]
- 14.Reysenbach AL, Longnecker K, Kirshtein J. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl Environ Microbiol. 2000;66(9):3798–3806. doi: 10.1128/aem.66.9.3798-3806.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sievert SM, Kuever J, Muyzer G. Identification of 16S ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos Island (Greece) Appl Environ Microbiol. 2000;66(7):3102–3109. doi: 10.1128/aem.66.7.3102-3109.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohwada K, Tabor PS, Colwell RR. Species composition and barotolerance of gut microflora of deep-sea benthic macrofauna collected at various depths in the atlantic ocean. Appl Environ Microbiol. 1980;40(4):746–755. doi: 10.1128/aem.40.4.746-755.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabor PS, Ohwada K, Colwell RR. Filterable marine bacteria found in the deep sea: Distribution, taxonomy, and response to starvation. Microb Ecol. 1981;7(1):67–83. doi: 10.1007/BF02010479. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz JR, Walder JD, Colwell RR. Deep-sea bacteria: Growth and utilization of hydrocarbons at ambient and in situ pressure. Appl Microbiol. 1974;28(6):982–986. doi: 10.1128/am.28.6.982-986.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg JF, Heidelberg KB, Colwell RR. Bacteria of the gamma-subclass Proteobacteria associated with zooplankton in Chesapeake Bay. Appl Environ Microbiol. 2002;68(11):5498–5507. doi: 10.1128/AEM.68.11.5498-5507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eilers H, Pernthaler J, Glöckner FO, Amann R. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol. 2000;66(7):3044–3051. doi: 10.1128/aem.66.7.3044-3051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbieri E, et al. Occurrence, diversity, and pathogenicity of halophilic Vibrio spp. and non-O1 Vibrio cholerae from estuarine waters along the Italian Adriatic coast. Appl Environ Microbiol. 1999;65(6):2748–2753. doi: 10.1128/aem.65.6.2748-2753.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLoney-Marino CR, Wolfe AJ, Visick KL. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl Environ Microbiol. 2003;69(12):7527–7530. doi: 10.1128/AEM.69.12.7527-7530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reidl J, Klose KE. Vibrio cholerae and cholera: Out of the water and into the host. FEMS Microbiol Rev. 2002;26(2):125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 24.Urakawa H, Kita-Tsukamoto K, Ohwada K. Reassessment of the taxonomic position of Vibrio iliopiscarius (Onarheim et al. 1994) and proposal for Photobacterium iliopiscarium comb. nov. Int J Syst Bacteriol. 1999;49(Pt 1):257–260. doi: 10.1099/00207713-49-1-257. [DOI] [PubMed] [Google Scholar]
- 25.Olive DM, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37(6):1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roque A, Molina-Aja A, Bolán-Mejía C, Gomez-Gil B. In vitro susceptibility to 15 antibiotics of vibrios isolated from penaeid shrimps in Northwestern Mexico. Int J Antimicrob Agents. 2001;17(5):383–387. doi: 10.1016/s0924-8579(01)00308-9. [DOI] [PubMed] [Google Scholar]
- 27.Arias CR, Garay E, Aznar R. Nested PCR method for rapid and sensitive detection of Vibrio vulnificus in fish, sediments, and water. Appl Environ Microbiol. 1995;61(9):3476–3478. doi: 10.1128/aem.61.9.3476-3478.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson JR, et al. Diversity and dynamics of a north Atlantic coastal Vibrio community. Appl Environ Microbiol. 2004;70(7):4103–4110. doi: 10.1128/AEM.70.7.4103-4110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grisez L, Chair M, Sorgeloos P, Ollevier F. Mode of infection and spread of Vibrio anguillarum in turbot Scophthalmus maximus larvae after oral challenge through live feed. Dis Aquat Organ. 1996;26(3):181–187. [Google Scholar]
- 30.Sawabe T, et al. Fluorescent amplified fragment length polymorphism and repetitive extragenic palindrome-PCR fingerprinting reveal host-specific genetic diversity of Vibrio halioticoli-like strains isolated from the gut of Japanese abalone. Appl Environ Microbiol. 2002;68(8):4140–4144. doi: 10.1128/AEM.68.8.4140-4144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandenberghe J, et al. Vibrios associated with Litopenaeus vannamei larvae, postlarvae, broodstock, and hatchery probionts. Appl Environ Microbiol. 1999;65(6):2592–2597. doi: 10.1128/aem.65.6.2592-2597.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol. 1990;56(6):1977–1980. doi: 10.1128/aem.56.6.1977-1980.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konstantinidis KT, Ramette A, Tiedje JM. Toward a more robust assessment of intraspecies diversity, using fewer genetic markers. Appl Environ Microbiol. 2006;72(11):7286–7293. doi: 10.1128/AEM.01398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187(18):6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okada K, Iida T, Kita-Tsukamoto K, Honda T. Vibrios commonly possess two chromosomes. J Bacteriol. 2005;187(2):752–757. doi: 10.1128/JB.187.2.752-757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langille MGI, Brinkman FSL. IslandViewer: An integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25(5):664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han H, et al. Genome plasticity of Vibrio parahaemolyticus: microevolution of the ‘pandemic group’. BMC Genomics. 2008;9(1):570. doi: 10.1186/1471-2164-9-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markert S, et al. Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science. 2007;315(5809):247–250. doi: 10.1126/science.1132913. [DOI] [PubMed] [Google Scholar]
- 43.Karl DM, et al. A microbiological study of Guaymas basin high-temperature hydrothermal vents. Deep-Sea Res. 1988;35(5):777–791. [Google Scholar]
- 44.Webb SM, Dick GJ, Bargar JR, Tebo BM. Evidence for the presence of Mn(III) intermediates in the bacterial oxidation of Mn(II) Proc Natl Acad Sci USA. 2005;102(15):5558–5563. doi: 10.1073/pnas.0409119102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edmonds HN, et al. Discovery of abundant hydrothermal venting on the ultraslow-spreading Gakkel ridge in the Arctic Ocean. Nature. 2003;421(6920):252–256. doi: 10.1038/nature01351. [DOI] [PubMed] [Google Scholar]
- 46.Dick GJ, Lee YE, Tebo BM. Manganese(II)-oxidizing Bacillus spores in Guaymas Basin hydrothermal sediments and plumes. Appl Environ Microbiol. 2006;72(5):3184–3190. doi: 10.1128/AEM.72.5.3184-3190.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ridge JP, et al. A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ Microbiol. 2007;9(4):944–953. doi: 10.1111/j.1462-2920.2006.01216.x. [DOI] [PubMed] [Google Scholar]
- 48.Vezzi A, et al. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science. 2005;307(5714):1459–1461. doi: 10.1126/science.1103341. [DOI] [PubMed] [Google Scholar]
- 49.Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417(6889):656–659. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- 50.Hasan NA, et al. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci USA. 2012;109(29):E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park KS, et al. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun. 2004;72(11):6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: A beginner’s guide. Curr Opin Microbiol. 2008;11(1):3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Cascales E. The type VI secretion toolkit. EMBO Rep. 2008;9(8):735–741. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Filloux A, Hachani A, Bleves S. The bacterial type VI secretion machine: Yet another player for protein transport across membranes. Microbiology. 2008;154(Pt 6):1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 55.Weber B, Hasic M, Chen C, Wai SN, Milton DL. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ Microbiol. 2009;11(12):3018–3028. doi: 10.1111/j.1462-2920.2009.02005.x. [DOI] [PubMed] [Google Scholar]
- 56.Planet PJ, Kachlany SC, Fine DH, DeSalle R, Figurski DH. The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat Genet. 2003;34(2):193–198. doi: 10.1038/ng1154. [DOI] [PubMed] [Google Scholar]
- 57.Smith B, Oliver JD. In situ and in vitro gene expression by Vibrio vulnificus during entry into, persistence within, and resuscitation from the viable but nonculturable state. Appl Environ Microbiol. 2006;72(2):1445–1451. doi: 10.1128/AEM.72.2.1445-1451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy RA, Boyd EF. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol. 2008;190(2):636–647. doi: 10.1128/JB.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishibuchi M. [Origin of the pathogenic vibrios in the environment: inference from the studies on the molecular genetics of Vibrio cholerae and Vibrio parahaemolyticus.] Nihon Saikingaku Zasshi. 1996;51(3):823–832. doi: 10.3412/jsb.51.823. Japanese. [DOI] [PubMed] [Google Scholar]
- 60.Sechi LA, Duprè I, Deriu A, Fadda G, Zanetti S. Distribution of Vibrio cholerae virulence genes among different Vibrio species isolated in Sardinia, Italy. J Appl Microbiol. 2000;88(3):475–481. doi: 10.1046/j.1365-2672.2000.00982.x. [DOI] [PubMed] [Google Scholar]
- 61.Klein SL, Gutierrez West CK, Mejia DM, Lovell CR. Genes similar to the Vibrio parahaemolyticus virulence-related genes tdh, tlh, and vscC2 occur in other Vibrionaceae species isolated from a pristine estuary. Appl Environ Microbiol. 2014;80(2):595–602. doi: 10.1128/AEM.02895-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vezzulli L, Guzmán CA, Colwell RR, Pruzzo C. Dual role colonization factors connecting Vibrio cholerae’s lifestyles in human and aquatic environments open new perspectives for combating infectious diseases. Curr Opin Biotechnol. 2008;19(3):254–259. doi: 10.1016/j.copbio.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 63.McFall-Ngai M. Divining the essence of symbiosis: Insights from the squid-vibrio model. PLoS Biol. 2014;12(2):e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martínez JL. Bacterial pathogens: From natural ecosystems to human hosts. Environ Microbiol. 2013;15(2):325–333. doi: 10.1111/j.1462-2920.2012.02837.x. [DOI] [PubMed] [Google Scholar]
- 65.Chiavelli DA, Marsh JW, Taylor RK. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl Environ Microbiol. 2001;67(7):3220–3225. doi: 10.1128/AEM.67.7.3220-3225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reguera G, Kolter R. Virulence and the environment: A novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J Bacteriol. 2005;187(10):3551–3555. doi: 10.1128/JB.187.10.3551-3555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huq A, et al. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45(1):275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nercessian O, Reysenbach AL, Prieur D, Jeanthon C. Archaeal diversity associated with in situ samplers deployed on hydrothermal vents on the East Pacific Rise (13 degrees N) Environ Microbiol. 2003;5(6):492–502. doi: 10.1046/j.1462-2920.2003.00437.x. [DOI] [PubMed] [Google Scholar]
- 69.Boone DR, Johnson RL, Liu Y. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl Environ Microbiol. 1989;55(7):1735–1741. doi: 10.1128/aem.55.7.1735-1741.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- 71.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8(3):195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 72.Han CS, Chain P. Finishing repeat regions automatically with Dupfinisher. In: Arabnia HR, Valafar H, editors. International Conference on Bioinformatics and Computational Biology. CSREA Press; Livermore, CA: 2006. pp. 141–146. [Google Scholar]
- 73.Goryshin IY, Reznikoff WS. Tn5 in vitro transposition. J Biol Chem. 1998;273(13):7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 74.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 76.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.