Significance
Untargeted metabolomics is rapidly becoming a mainstream technique to discover biomarkers, to study the effects of interventions, and to discover the function of enzymes. Several thousand metabolites can be detected in a single untargeted metabolomics analysis, but state-of-the-art untargeted metabolomics studies still report up to 40% unidentified metabolites, indicating that large parts of the human metabolome are still unexplored. We identified an uncharacterized class of ubiquitous mammalian metabolites: N-lactoyl-amino acids. Using a powerful combination of proteomics and protein fractionation, we unexpectedly discovered that these metabolites are formed from lactate and amino acids by reversed action of the protease cytosolic nonspecific dipeptidase 2. Our approach to identify unknown metabolites and their biosynthesis has general applicability in the further exploration of the human metabolome.
Keywords: unknown metabolites, untargeted metabolomics, ABCC5, MRP5, physical exercise
Abstract
Despite technological advances in metabolomics, large parts of the human metabolome are still unexplored. In an untargeted metabolomics screen aiming to identify substrates of the orphan transporter ATP-binding cassette subfamily C member 5 (ABCC5), we identified a class of mammalian metabolites, N-lactoyl-amino acids. Using parallel protein fractionation in conjunction with shotgun proteomics on fractions containing N-lactoyl-Phe–forming activity, we unexpectedly found that a protease, cytosolic nonspecific dipeptidase 2 (CNDP2), catalyzes their formation. N-lactoyl-amino acids are ubiquitous pseudodipeptides of lactic acid and amino acids that are rapidly formed by reverse proteolysis, a process previously considered to be negligible in vivo. The plasma levels of these metabolites strongly correlate with plasma levels of lactate and amino acid, as shown by increased levels after physical exercise and in patients with phenylketonuria who suffer from elevated Phe levels. Our approach to identify unknown metabolites and their biosynthesis has general applicability in the further exploration of the human metabolome.
Untargeted metabolomics aims to provide a comprehensive snapshot of the metabolome and is becoming a mainstream technique to discover biomarkers, to study the effects of interventions, and to discover the function of enzymes (1).
Recent technical improvements now make it possible to detect several thousand metabolites in a single untargeted metabolomics analysis, but the identity of these metabolites is initially not known beyond their molecular mass (1). Knowing the chemical identity of metabolites is crucial for the proper interpretation of metabolomic studies, however. Online metabolite databases like METLIN (∼240,000 entries) and the Human Metabolome Database (HMDB; ∼42,000 entries) contain vast numbers of metabolites and are extremely useful to annotate the detected molecular masses (2, 3). These databases cover large parts of the metabolome, but significant gaps remain. Due to poorly characterized and promiscuous enzymes, the human metabolome is much larger than initially anticipated (4). State-of-the-art untargeted metabolomics studies still report up to 40% unidentified, but potentially important, metabolites that can be detected reproducibly (5–8). Nevertheless, unknown metabolites are only rarely characterized because of the extensive work required for de novo structure elucidation (9–11). Here, we describe the characterization of a class of mammalian metabolites that we discovered in an untargeted metabolomic screen aimed to identify substrates of the orphan transporter ATP-binding cassette subfamily C member 5 (ABCC5).
ABCC5 is a member of the C-branch of the superfamily of ABC transporters, which use the energy provided by the hydrolysis of ATP to transport substrates across the plasma membrane (12). ABCC5, also known as multidrug resistance-associated protein 5 (MRP5), is ubiquitously expressed, but levels in brain and muscle are especially high (13–15). In most cells, ABCC5 routes to the basolateral plasma membrane (16). Although ABCC5 has been shown to transport nucleoside/nucleotide analogs (17) and antifolates (18) used in cancer chemotherapy, it does so with low affinity, and elevated ABCC5 levels have not been linked to clinical drug resistance (16). Among the few endogenous substrates identified are folates (18) and cyclic nucleotides (19–21). Because the affinity of ABCC5 for these substrates is low, a major function in cGMP excretion is unlikely (16). ABCC5 has been reported to support osteoclast formation in bone metastases (22), to affect chamber depth of the eye (23), and to be involved in heme homeostasis (24), but which ABCC5 substrates might be involved in these processes remains unknown. The Abcc5 KO mouse has no obvious phenotype (25) and has also not been helpful in finding relevant endogenous substrates.
To get more insight into the function of ABCC5, we have started to identify more physiological substrates. In previous experiments, we have used semitargeted metabolomics to discover endogenous substrates of ABCC transporters (26, 27). Because this approach requires some prior knowledge of potential substrates, it is not applicable to ABCC5.
We have therefore used an untargeted metabolomics screen to discover novel substrates of ABCC5. This approach has led to the identification of a ubiquitous class of mammalian metabolites and their unexpected biosynthesis route.
Results
Untargeted Metabolomics Reveals Unknown ABCC5-Related Metabolites.
To identify novel endogenous substrates of ABCC5, we applied comparative untargeted metabolomics to culture medium from HEK 293 parental cells and HEK 293 cells overexpressing human ABCC5 (HEK 293/ABCC5). ABCC5 substrates were expected to accumulate in HEK 293/ABCC5 medium compared with HEK 293/control medium. The liquid chromatography (LC)/MS metabolomic screen revealed several metabolites that were more abundant in HEK 293/ABCC5 medium (Fig. 1), although intracellular levels were generally unaffected (Fig. S1).
Fig. 1.
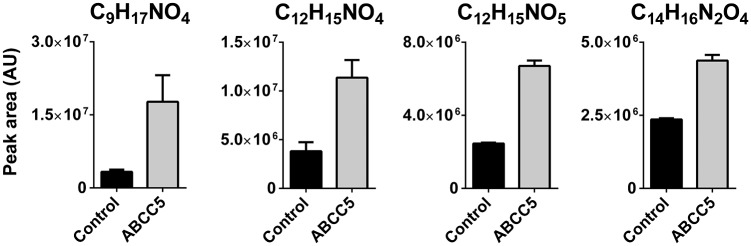
Four unknown metabolites accumulate in culture medium of HEK 293 cells overexpressing ABCC5. HEK 293 control and HEK 293/ABCC5 cells were grown to confluence in six-well plates and cultured for an additional 3 d. Culture medium was analyzed using untargeted LC/MS metabolomics. Comparison of the metabolite profiles revealed four unknown metabolites that were significantly more abundant in medium of HEK 293/ABCC5 cells. Data are presented as mean plus SD (n = 3). AU, arbitrary units. The intracellular levels of the unknowns are depicted in Fig. S1.
Using the accurate mass and isotope pattern of the unknowns, we calculated their chemical formulas, which were used to search metabolite databases. This search did not yield candidate identities because the putative ABCC5 substrates were not present in the large online metabolite databases like METLIN (2) or HMDB (3). Notwithstanding their absence in databases, we also detected the unknowns in many mouse tissues.
Unknown Metabolites Are N-Lactoyl-Amino Acids.
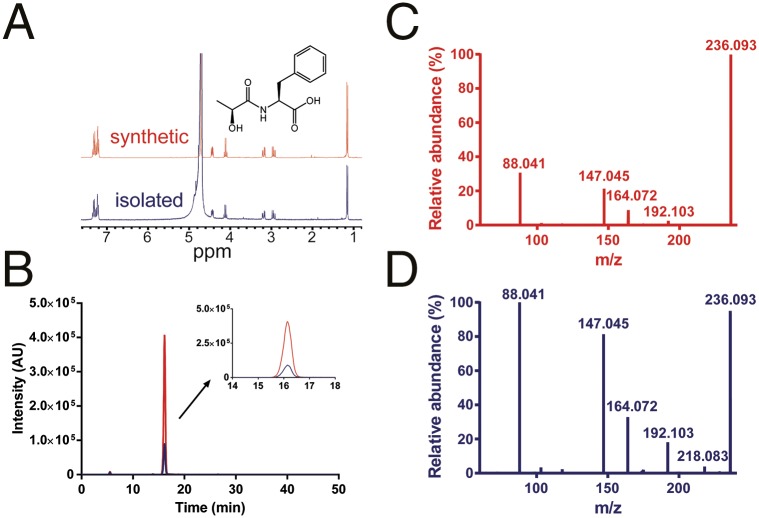
To elucidate the structure of the unknowns, we purified one of the unknown metabolites (C12H15NO4) from a large volume of HEK 293/ABCC5-conditioned culture medium. After purification using C18 flash chromatography and ion-pair C18 and phenyl HPLC, we obtained C12H15NO4 in a sufficient amount and purity for NMR analysis. The 1H-NMR, 13C-NMR, and 1H-COSY NMR spectra indicated that the unknown was N-lactoyl-Phe [2-(2-hydroxypropanamido)-3-phenylpropanoic acid or N-lac-Phe; Fig. S2], which was confirmed by NMR and LC/MS comparison with a reference standard that we synthesized (Fig. 2 and Fig. S2). The retention time on HPLC indicated that the metabolite was composed of l-lactic acid, and not d-lactic acid (Fig. S3).
Fig. 2.
Unknown C12H15NO4 is N-lac-Phe. (A) 1H-NMR spectrum of isolated C12H15NO4 recorded in D2O at 300 MHz matches the 1H-NMR spectrum of the synthesized reference N-lac-Phe. (B) Unknown C12H15NO4 elutes at the same time as N-lac-Phe. Extracted ion chromatogram of the mass corresponding to C12H15NO4 (m/z 236.09) in conditioned medium from HEK 293/ABCC5 cells before (depicted in blue) and after (depicted in red) spiking with synthesized N-lac-Phe (1 μg/mL, 4.2 μM). The high-resolution MS2 fragmentation spectrum of synthesized N-lac-Phe (C) and unknown C12H15NO4 in conditioned medium from HEK 293/ABCC5 (D) match. Differences in background and intensities can be attributed to the lower concentration of unknown C12H15NO4. Additional NMR and MS spectra are presented in Figs. S2 and S4, respectively.
Based on the similarities in the high-resolution MS fragmentation spectra, the other unknowns could subsequently be identified as N-lac-(iso)Leu (C9H17NO4), N-lac-Tyr (C12H15NO5), and N-lac-Trp (C14H16N2O4) (Fig. S4). Although these four N-lactoyl-amino acids were the most abundant of their class, we also detected low levels of ABCC5-dependent metabolites that had masses corresponding to N-lac-Val, N-lac-Met, and N-lac-Gly. N-lactoyl-amino acids are pseudodipeptides consisting of a lactic acid and an amino acid moiety linked by a peptide bond, and they form a previously unidentified class of physiological mammalian metabolites.
N-Lac-Phe Is Transported by ABCC5 in Vitro.
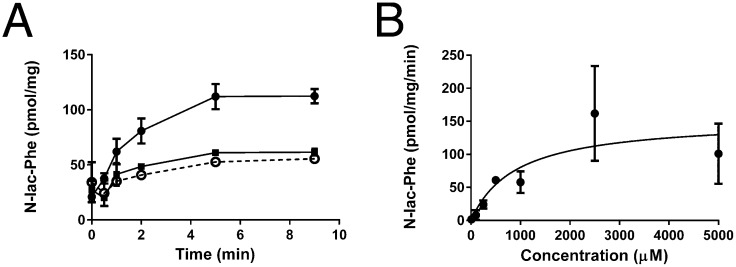
The metabolic screen in which we detected the N-lactoyl-amino acids was started to discover new endogenous substrates of ABCC5. To confirm that N-lactoyl-amino acids are genuine ABCC5 substrates, we studied the uptake of N-lac-Phe into inside-out membrane vesicles. Fig. 3A shows that N-lac-Phe is taken up into vesicles in a time-, ATP-, and ABCC5-dependent fashion, with a Km of 1 mM when fitted to Michaelis–Menten kinetics (Fig. 3B). This estimate is obviously very rough, because considerable background transport into the vesicles made it increasingly difficult to determine the relatively low rate of ABCC5-dependent transport accurately at higher substrate concentrations. Substantial background transport was also observed in culture medium of HEK 293 control cells (Fig. 1), which indicates that other carriers exist for this class of metabolites.
Fig. 3.
N-lac-Phe is transported into inside-out membrane vesicles by ABCC5. (A) Control vesicles (▪) and ABCC5-containing vesicles (○ and ●) were incubated with 100 μM N-lac-Phe at 37 °C in the presence (solid line) and absence (dashed line) of 5 mM ATP. At the indicated time points, a sample containing 75 μg of protein was taken. After washing over a filter, the vesicular content was analyzed by LC/MS (n = 3–6). (B) Concentration dependence was assessed by incubating control and ABCC5-containing vesicles with several concentrations of N-lac-Phe in the presence of ATP and determining ABCC5-dependent uptake after 2 min (n = 3–4 for concentration <1,000 μM and n = 8 for concentration ≥1,000 μM). The data were fitted to Michaelis–Menten kinetics (solid line) using GraphPad Prism. Data are presented as mean ± SEM.
N-Lactoyl-Amino Acids Are Formed by Cytosolic Nonspecific Dipeptidase 2.
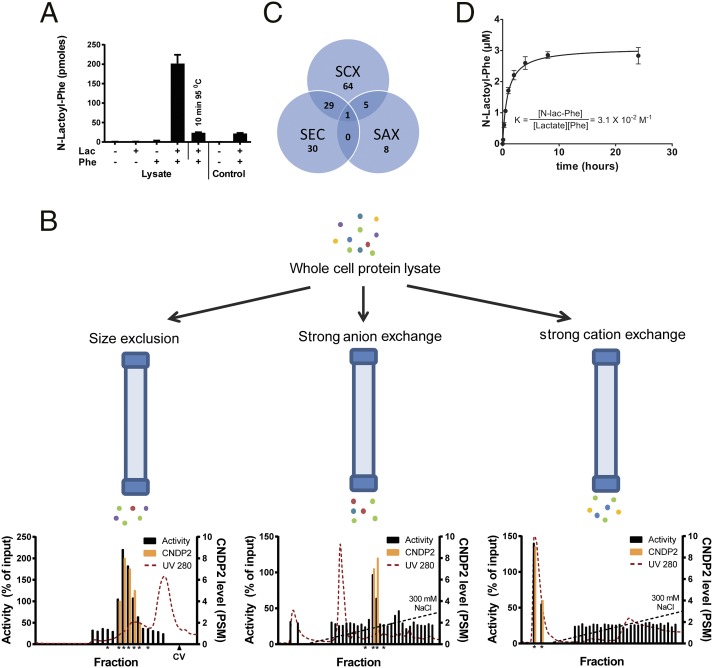
To identify the source of the N-lactoyl-amino acids, we first tested under which conditions these metabolites are formed in vitro. Using whole-cell lysate from HEK 293 cells, we found that N-lac-Phe is formed when both lactate and Phe are incubated in the presence of nondenatured proteins, indicating enzymatic formation (Fig. 4A). Attempts to purify the enzyme from whole-cell lysate by classical sequential protein fractionation were unsuccessful. Although enzyme activity was detected in fractions obtained by size exclusion chromatography, this activity was lost upon subsequent fractionation of active fractions by strong anion exchange chromatography.
Fig. 4.
N-lac-Phe is formed by CNDP2. (A) N-lac-Phe is formed in the presence of Phe, lactate, and intact protein. Whole-cell lysate or control buffer [25 mM Tris⋅HCl (pH 7.4)] was incubated for 30 min at 37 °C in the presence or absence of 10 mM substrates. N-lac-Phe formation was determined by LC/MS and is expressed in arbitrary units. Data are presented as mean (n = 3) plus SD. (B) Whole-cell lysate was fractionated in parallel on three different columns. Enzyme activity was assessed by incubation with 10 mM lactate and Phe (30 min, 37 °C) and is normalized to the activity in unfractionated whole-cell lysate. The levels of CNDP2 were determined in active fractions and neighboring inactive control fractions (all marked by an asterisk) using LC/MS proteomics and are expressed as a peptide spectrum match (PSM) (n = 1). CV, column volume. (C) Although multiple proteins coeluted with enzyme activity for each single fractionation, only a single protein, CNDP2, coeluted with activity in all three fractionations. SAX, strong anion exchange; SCX, strong cation exchange; SEC, size exclusion chromatography. (D) Human recombinant CNDP2 (1 μg) was incubated (37 °C) with 10 mM lactate and Phe in 25 mM Tris⋅HCl (pH 7.4) containing 0.1 mM MnCl2. N-lac-Phe levels were determined by LC/MS and are expressed as arbitrary units. Data are presented as mean (n = 3) plus SD. Additional enzyme kinetics are presented in Figs. S5, S8, and S9.
To bypass this loss of activity over time, we applied size exclusion, strong anion exchange, and strong cation exchange chromatography in parallel and immediately determined the enzymatic activity in the fractions. We analyzed active fractions and neighboring inactive control fractions by LC/MS shotgun proteomics to identify proteins that coeluted with enzymatic activity (Fig. 4B). Although the fractions still contained between 100 and 600 proteins each, only a single protein, cytosolic nonspecific dipeptidase 2 (CNDP2), was present in all active fractions and absent in the negative fractions (Fig. 4C). We confirmed that CNDP2-mediated N-lac-Phe formation, using recombinant human CNDP2 (Fig. 4D). Curve fitting revealed an equilibrium constant K of 3.1 × 10−2⋅M−1. A very similar equilibrium constant (2.7 × 10−2⋅M−1) was obtained by incubating CNDP2 with different concentrations of lactate and Phe for 24 h (Fig. S5).
N-Lactoyl-Amino Acids Are Rapidly Formed in HEK 293 Cells.
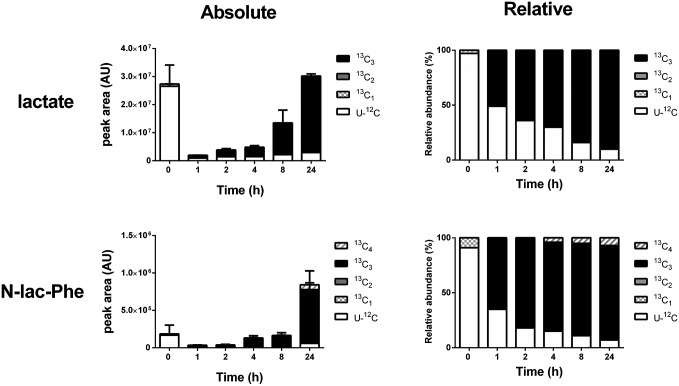
We assessed the speed at which N-lactoyl-amino acids are formed in vitro by metabolic labeling with 13C6-glucose. The 13C6-glucose is rapidly taken up into cells and converted into 13C3-lactate, which can then be incorporated into N-lactoyl-amino acids. Fig. 5 shows that the rate of 13C3-labeling of N-lac-Phe was comparable to formation of 13C3-lactate, its immediate precursor. Similar data were obtained for other N-lactoyl-amino acids. These results indicate very rapid interconversion between amino acids, lactate, and N-lactoyl-amino acids in living cells.
Fig. 5.
13C-Metabolic labeling of lactate and N-lac-Phe occurs at a similar speed in HEK 293 cells. HEK 293 cells were grown to confluence in six-well plates, at which point the medium was replaced with medium containing 13C6-glucose. At several time points, the amount of lactate and N-lac-Phe containing only 12C (U-12C; unlabeled) or one to four 13C atoms per molecule (13C1–4) was determined in lysate by accurate mass LC/MS. Fully 13C-labeled lactate contains three 13C-atoms, which is reflected in the 13C3-labeling of N-lac-Phe. The minor presence of 13C1, 13C2, and 13C4 isotopologs can be explained by the natural occurrence of the 13C isotope (∼1% of all carbon atoms). Levels are expressed as absolute (arbitrary units) and relative values (percentage of isotope total). Data are presented as mean (n = 3) and SD (only for absolute values). Isotope data of corresponding culture medium samples are presented in Fig. S6.
N-Lactoyl-Amino Acid Levels in Human Plasma Depend on the Concentration of Lactate and Amino Acid.
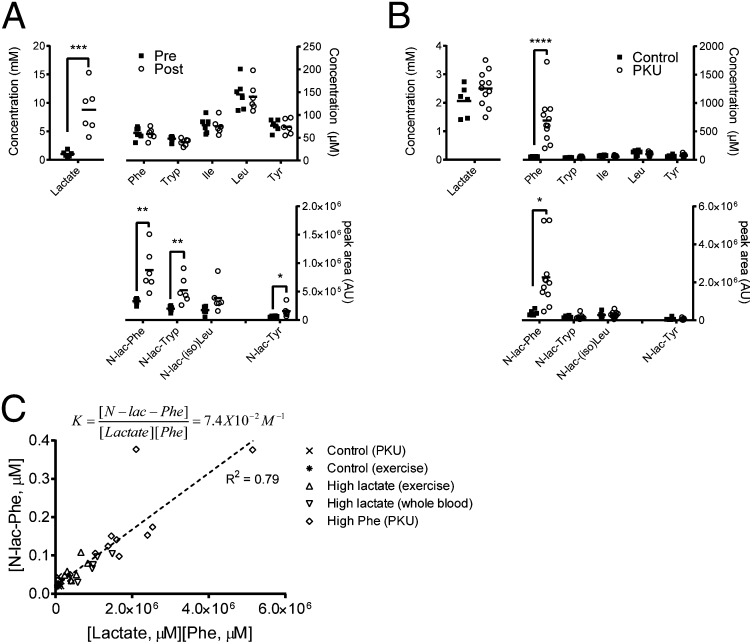
In humans, lactate levels swiftly rise during strenuous physical exercise when insufficient oxygen is available for aerobic glycolysis. To test whether an increase in lactate results in increased N-lactoyl-amino acid levels, we determined the levels of lactate, amino acids, and their corresponding N-lactoyl-amino acids in plasma of six healthy individuals before and immediately after a single bout of strenuous exercise lasting 5–10 min. In line with the rapid formation in vitro, we found that the increased lactate levels coincided with significantly increased levels of N-lactoyl-amino acids in human plasma, whereas amino acid levels remained the same (Fig. 6A).
Fig. 6.
N-lactoyl-amino acid levels in human plasma depend on lactate and amino acid concentrations. (A) Plasma was collected from human volunteers before (pre) and after (post) 5–10 min of strenuous exercise. Lactate and N-lactoyl-amino acid levels in human plasma rapidly increase after physical exercise, whereas the corresponding amino acid levels are unaffected. N-lactoyl-amino acid levels did not increase in resting controls. Horizontal bars represent means (n = 6). (B) Phe and N-lac-Phe levels are significantly increased in plasma of patients with PKU compared with controls, whereas plasma levels of other amino acids, N-lactoyl-amino acids, and lactate are similar. Horizontal bars represent means (n = 6 for controls, n = 11 for patients with PKU). (C) Plasma N-lac-Phe levels are in apparent equilibrium with plasma lactate and Phe levels. Plasma samples obtained from patients with PKU and controls, volunteers before and after exercise, and controls with high lactate due to prolonged whole-blood storage at ambient temperature were analyzed for lactate, Phe, and N-lac-Phe concentration as described in Materials and Methods (n = 34). The dotted line represents a fitted linear function, of which the slope is 7.4 × 10−2⋅M−1. N-lac-Phe is not formed in plasma (Fig. S7).
Patients suffering from the inborn metabolic disorder phenylketonuria (PKU) lack functional Phe hydroxylase and are unable to convert Phe into Tyr. As a result, patients with PKU have increased Phe plasma levels (28). We compared plasma samples of a group of Dutch patients who had PKU with high plasma Phe with control samples and found N-lac-Phe levels that were significantly higher than the N-lac-Phe levels in control samples, whereas lactate levels were comparable (Fig. 6B).
Theoretically, the N-lac-Phe concentration in cells is determined by the concentration of lactate and Phe, and the equilibrium constant K. In Fig. 6C, we plotted the data from the exercise experiment, the patients with PKU and controls, and an additional set of plasma samples with high lactate levels due to prolonged storage as whole blood. Except for a single outlier, the product of the lactate and Phe plasma concentrations showed a good correlation with the plasma N-lac-Phe concentration. The slope of the fitted linear function represents an apparent equilibrium constant K of 7.4 × 10−2⋅M−1, which is in good agreement with the value of 3.1 × 10−2⋅M−1 obtained for CNDP2-mediated N-lac-Phe formation in vitro. Formation of N-lac-Phe required cells, because it was not formed when we incubated plasma with lactate or Phe levels comparable to the lactate or Phe levels found during exercise and in patients with PKU. A modest but significant increase was observed after a 30-min incubation in whole blood (Fig. S7).
Discussion
We have identified an uncharacterized class of mammalian metabolites, N-lactoyl-amino acids. These metabolites are present in many tissues and can approach micromolar concentrations in human plasma. We have also shown that CNDP2 can form N-lactoyl-amino acids in HEK 293 cells.
CNDP2, which is also known as carboxypeptidase of glutamate-like (CPGL) and as carnosine dipeptidase II, belongs to the M20 family of metallopeptidases and requires Mn2+ for full activity (29). Loss of Mn2+ from the enzyme is likely the reason why activity was gradually lost during our initial attempts to isolate the enzyme. CNDP2 is highly conserved across species and is expressed in most tissues (29). As its name indicates, CNDP2 has broad substrate selectivity for dipeptides, but its physiological substrates are not known (29, 30). Cndp2 KO mice do not exist. Several groups reported a role for CNDP2 in tumor suppression, but similar effects were observed with the isoform CPGL-B, which lacks a part of the catalytic domain (31–33). It is unknown whether CPGL-B has any peptidase activity.
The finding that a peptidase was responsible for the biosynthesis of our pseudopeptides was unexpected, because peptidases normally hydrolyze peptide bonds. Although peptidases are known to mediate the formation of peptides through reverse proteolysis, this property has mainly been investigated by organic chemists, who exploit the selectivity and mild reaction conditions for peptide synthesis (34, 35). Peptidases are catalysts, and, as such, they do not change the equilibrium constant between peptide formation and hydrolysis (34). In an aqueous environment, the equilibrium constant strongly favors peptide hydrolysis over peptide formation. Although formation of N-lactoyl-amino acids from lactic acid is thermodynamically very unfavorable, the formation from lactic acid esters is not. To rule out the possibility that the N-lactoyl-amino acids we detected were formed from traces of lactic acid esters present in our commercial lactic acid preparation (≥99% purity), we tested the rate of CNDP2-mediated N-lac-Phe formation from methyl-lactate and found that it is a relatively poor CNDP2 substrate (Fig. S8). Lactic acid esters are therefore a highly unlikely precursor for N-lac-Phe. Importantly, we found that N-lac-Phe is quickly hydrolyzed by CNDP2, like its model substrate Cys-Gly (Fig. S9), one of the best CNDP2 substrates identified thus far (30).
Because peptide bond hydrolysis is strongly favored over peptide bond formation in an aqueous environment, reverse proteolysis is considered to be negligible in vivo by many (34), although some have proposed a role for reverse proteolysis in antigen formation (36). The apparent equilibrium constants of 3.1 × 10−2 M−1, 2.7 × 10−2 M−1, and 7.4 × 10−2 M−1 calculated from our data are higher than the values reported for reverse proteolysis of regular amino acids (10−3 to 10−4 M−1) (34). As a result, the high intracellular levels of lactate and amino acids, which are in the high micromolar to millimolar range, allow N-lactoyl-amino acid levels in plasma to approach micromolar levels (Fig. 6C). Contrary to common conviction, our data show that reverse proteolysis is not negligible in vivo, as long as the substrates are present in considerable concentrations.
Because lactate, amino acids, and CNDP2 are ubiquitous, N-lactoyl-amino acids are present in many tissues. Interestingly, N-lac-Val, N-lac-Leu, and N-lac-Ile have been identified in the urine of a patient with elevated branched-chain amino acids due to maple syrup urine disease (37). Another lactic acid amide, N6-lac-Lys, has been detected in human plasma and was found to be increased in the plasma of hemodialytic patients (38). As opposed to the biosynthesis described here, N6-lac-Lys is an amide-advanced glycation end product, formed nonenzymatically through the Maillard reaction (39). Although similar, N6-lac-Lys does not belong to the class of N2-lactoyl-amino acids identified here, because its lactate moiety is conjugated to the N6-amino group in the Lys side chain.
N2-lactoyl-amino acids have recently also been identified in soy sauce (40) and Parmigiano–Reggiano cheese (41). In cheese, N-lactoyl-amino acids were found to be formed by an enzyme present in lactic acid bacteria (42, 43). Although the enzyme was not identified, Bottesini et al. (43) showed that N-lac-Phe could be formed in vitro by a yeast peptidase, carboxypeptidase Y. Of note, all except one of the N-lactoyl-amino acids reported thus far contain a hydrophobic amino acid moiety, indicating enzyme selectivity.
The metabolic labeling experiments and in vitro enzyme kinetics show that interconversion between lactate, amino acids, and N-lactoyl-amino acids is very rapid. This fast interconversion explains why N-lactoyl-amino acid levels, except those N-lactoyl-amino acid levels of N-lac-Trp, did not change intracellularly upon ABCC5 overexpression: N-lactoyl-amino acids effluxed by ABCC5 are continuously replenished from the large pool of lactate and amino acids.
The fast interconversion is also in line with the rapid increase of N-lactoyl-amino acids upon physical exercise. Although many studies have assessed the effect of strenuous exercise on the metabolite profile in blood, none identified the N-lactoyl-amino acids (44–46). Nevertheless, many of the studies report multiple unidentified exercise-dependent metabolites, most likely including N-lactoyl-amino acids.
We show that N-lac-Phe plasma levels are increased in patients with PKU with increased plasma Phe levels. The N-lac-Phe levels in plasma were highly correlated with the product of the lactate and Phe concentration. N-lac-Phe is of cellular origin, because it is not formed in plasma (Fig. S7), which apparently does not contain CNDP2. The formation of N-lac-Phe in whole blood, however, indicates that blood cells are also capable of forming N-lactoyl-amino acids, which is in line with the reported CNDP2 expression in peripheral blood leukocytes (29) and its likely presence in erythrocytes (47). The modest rate of formation we observed in whole blood cannot account for the quick formation during physical exercise, but it does explain the high levels of N-lac-Phe found in whole-blood samples that were stored at ambient temperature for prolonged times.
Using vesicular transport assays, we confirmed that ABCC5 transports N-lac-Phe, and therefore most likely also transports the other N-lactoyl-amino acids. ABCC5 transports N-lac-Phe with a low affinity (Km of ∼1 mM), which is comparable to the Km of the physiological ABCC5 substrate folate (1 mM) (18). This Km in the low millimolar range exceeds the submicromolar plasma and, presumably intracellular, levels by far.
Currently, N-lactoyl-amino acids do not have a known function. The chemical structure of lactate resembles a small amino acid-like Ala. Most likely, N-lactoyl-amino acids are nonspecific byproducts of the promiscuous peptidase CNDP2. Many enzymes promiscuously catalyze reactions, and even relatively specific enzymes will produce large absolute amounts of side products as soon as substrate levels and turnover rates are high (48, 49). Interestingly, CNDP1 was recently identified as an enzyme that degrades nonspecific dipeptide byproducts formed during carnosine synthesis (50). Even if N-lactoyl-amino acids are byproducts of CNDP2, they might represent useful extracellular biomarkers for intracellular amino acid concentrations, because they are only formed inside cells.
Our results underline that large parts of the metabolome remain to be discovered, even in a well-defined in vitro cell system (4). Untargeted metabolomics provides an obvious method to address this gap in our knowledge. The parallel proteomics approach that we have developed to identify CNDP2 as the enzyme responsible for N-lactoyl amino acid formation is a powerful approach for the identification of any other enzyme of which the activity can be measured. We envision that this approach has broad applicability and is particularly useful for the identification of unstable enzymes that cannot be purified by classical sequential protein fractionation without losing activity.
Materials and Methods
Cell Lines and Culture Conditions.
HEK 293/ABCC5 (51) and control cells were cultured as described (52), seeded in six-well plates (Costar; Corning) at a density of 7.5 × 105 cells per well, and allowed to grow overnight. At this point, the medium was replaced with 2 mL of fresh medium and the cells were incubated for 3 d. Culture medium was collected on ice, whereas cultured cells were washed with cold PBS and lysed in 1 mL of 70% (vol/vol) cold methanol.
Untargeted Metabolomics.
Untargeted metabolomics was performed as described (52), but with a scan range of m/z 50–250 and m/z 250–1,500 for the primary metabolic screen and a scan range of m/z 150–750 for all other experiments. Differentially present metabolites were detected with the XCMS platform (53). The accurate mass of differentially present compounds was queried in the online HMDB (3) and METLIN (2) databases. The accurate mass and isotope distribution were used to determine the chemical formula with SIRIUS software (54).
Purification of C12H15NO4 from Culture Medium.
The unknown C12H15NO4 was purified from ∼1 L of HEK 293/ABCC5 medium in four consecutive chromatography steps that are specified in SI Materials and Methods. Fractions containing C12H15NO4 were pooled, freeze-dried three times to remove ammonium acetate and water residues, and dissolved in D2O for NMR analysis.
Structure Elucidation.
NMR spectra [1H-NMR, 13C-NMR (attached proton test), and 1H-COSY] were determined in D2O using a Bruker Avance 300 spectrometer (1H, 300 MHz; 13C, 100 MHz) at 298 K. Fragmentation spectra (MS2 and MS3) were acquired at a normalized collision energy of 35.
N-l-Lac-l-Phe Synthesis.
Reference N-l-Lac-l-Phe was synthesized from l-Phe ethyl ester and (−)-ethyl-l-lactate by carbodiimide/1-hydroxybenzotriazole coupling as described in SI Materials and Methods. N-l-lac-l-Phe was obtained as a white foam (783 mg, 66%) of which the chemical structure was confirmed by NMR and MS (SI Materials and Methods).
Vesicular Transport.
Inside-out membrane vesicles were prepared from HEK 293 cells transiently transduced with human ABCC5 or GFP (control) as described (55). Transport of N-lac-Phe into vesicles was assessed in a 96-well format (27). Filtered vesicles (75 μg of protein) were lysed, evaporated to dryness, and reconstituted in 50 μL of mobile phase A before analysis by LC/MS (Materials and Methods, Untargeted Metabolomics).
Enzyme Activity Assay.
HEK 293 cell lysate [in 25 mM Tris⋅HCl (pH 7.4) containing 1 mM DTT] was clarified by incubation with protamine sulfate (1%, 30 min at 4 °C) and stored at −80 °C. Lysate containing ∼50 μg of protein was incubated (30 min at 37 °C) in a final volume of 500 μL of 25 mM Tris⋅HCl (pH 7.4), with or without 10 mM l-(+)-lactate (≥99% pure; Sigma) or l-Phe (Sigma). Recombinant human CNDP2 (1 μg, C-terminal HIS-tagged; Prospec) was incubated (37 °C) with l-lactate and l-Phe in 25 mM Tris⋅HCl (pH 7.4), containing 0.1 mM MnCl2. The reactions were quenched by the addition of 50 μL of acetic acid, followed by centrifugation (5 min, 21,800 × g, 4 °C). The supernatant was applied to a Strata-X solid phase extraction column (33-μm polymeric reversed phase, 30 mg/1 mL; Phenomenex) to remove the lactate, after which N-lac-Phe levels were determined by LC/MS, as described in Materials and Methods, Untargeted Metabolomics.
Protein Fractionation and Proteomics.
Samples of cell lysate containing ∼250 μg of total protein were fractionated on an AKTAmicro system (GE Healthcare) using a Superdex 200 5/150 size exclusion column, a mini Q PC 3.2/3 strong anion exchange column, or a mini S PC 3.2/3 strong cation exchange column (all from GE Healthcare), as detailed in SI Materials and Methods. A volume corresponding to 20% of the collected fractions of interest was immediately used to test enzyme activity.
Tryptic digests of selected fractions were analyzed using nano-LC coupled to a Thermo Orbitrap Fusion mass spectrometer (Thermo Scientific) as detailed in SI Materials and Methods.
13C6-Glucose Metabolic Labeling.
HEK 293 cells were seeded in poly-d-Lys–coated six-well plates (Corning) and grown to confluence. At time 0, the medium was replaced with 1 mL of DMEM without pyruvate and glucose, with 10% (vol/vol) FCS and 4.64 g/mL 13C6-d-glucose (Aldrich). After 0, 1, 2, 4, 8, and 24 h, the culture medium and cells were collected and processed as described in SI Materials and Methods. N-lactoyl-amino acids were detected as described in Materials and Methods, Untargeted Metabolomics. Lactate labeling was determined by LC/MS, using a ZIC-HILIC column (150 × 0.5 mm, 3.5 μm; SeQuant), as detailed in SI Materials and Methods.
Human Exercise and N-Lactoyl-Amino Acids.
We collected blood samples from six human volunteers immediately before and after a single bout of strenuous exercise lasting 5–10 min. Blood was collected after informed consent in lithium heparin vacutainer tubes (BD) and immediately placed on ice. Within 15 min, the samples were centrifuged (10 min, 3,000 × g, 4 °C), after which the plasma layer was collected and stored at −20 °C. Plasma lactate levels were determined on a Cobas C501 analyzer (Roche), whereas amino acids were determined by ion-exchange chromatography with ninhydrin detection using an Aminotac JLC-500V analyzer (JEOL). To determine N-lactoyl-amino acid levels, 500 μL of plasma was mixed with 500 μL of water and 100 μL of acetic acid. The acidified plasma was processed and analyzed as described in Materials and Methods, Enzyme Activity Assay. Our study was sent to the Patient Trial Committee of the Netherlands Cancer Institute for review, and they determined that the study was exempt from approval.
PKU and N-Lactoyl-Amino Acids.
Plasma samples (heparin) were collected from patients with PKU and controls with unrelated metabolic diseases as a part of routine clinical chemistry testing. Samples were anonymized before analysis. Patients with PKU were under dietary control, but samples were selected based on high plasma Phe levels. Amino acid levels were determined with ninhydrin detection, as described above.
Lactate was quantified in plasma using an ultraperformance LC/tandem MS instrument (Waters Quattro Premier XE) and a bridged ethyl hybrid-amide column (100 × 2.1 mm, 1.7 μm; Waters) by multiple reaction monitoring acquisition in the negative ionization mode. N-lactoyl-amino acid levels were determined as described for the human exercise plasma samples.
Supplementary Material
Acknowledgments
We thank Liesbeth Hoekman and Enver Delic for their technical assistance, and Alfred Schinkel and Huib Ovaa for a critical reading of our manuscript. We also thank an anonymous reviewer who made very helpful suggestions for improving the manuscript. Part of this work was performed within the framework of the project “Proteins at Work,” financed by the Netherlands Organization for Scientific Research as part of the National Roadmap Large-Scale Research Facilities of the Netherlands (Project 184.032.201).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424638112/-/DCSupplemental.
References
- 1.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tautenhahn R, et al. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30(9):826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wishart DS, et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiehn O, Barupal DK, Kind T. Extending biochemical databases by metabolomic surveys. J Biol Chem. 2011;286(27):23637–23643. doi: 10.1074/jbc.R110.173617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin S-Y, et al. Multiple Tissue Human Expression Resource (MuTHER) Consortium An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langley RJ, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. 2013;5(195):195ra95. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Psychogios N, et al. The human serum metabolome. PLoS ONE. 2011;6(2):e16957. doi: 10.1371/journal.pone.0016957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouatra S, et al. The human urine metabolome. PLoS ONE. 2013;8(9):e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalisiak J, et al. Identification of a new endogenous metabolite and the characterization of its protein interactions through an immobilization approach. J Am Chem Soc. 2009;131(1):378–386. doi: 10.1021/ja808172n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, et al. Practical approach for the identification and isomer elucidation of biomarkers detected in a metabonomic study for the discovery of individuals at risk for diabetes by integrating the chromatographic and mass spectrometric information. Anal Chem. 2008;80(4):1280–1289. doi: 10.1021/ac702089h. [DOI] [PubMed] [Google Scholar]
- 11.Dunn WB, et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted. Metabolomics. 2013;9(Suppl 1):S44–S66. [Google Scholar]
- 12.Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters: The multidrug resistance-associated proteins. J Natl Cancer Inst. 2000;92(16):1295–1302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- 13.Kool M, et al. Analysis of expression of cMOAT (MRP2), MRP3, MRP4, and MRP5, homologues of the multidrug resistance-associated protein gene (MRP1), in human cancer cell lines. Cancer Res. 1997;57(16):3537–3547. [PubMed] [Google Scholar]
- 14.Belinsky MG, Bain LJ, Balsara BB, Testa JR, Kruh GD. Characterization of MOAT-C and MOAT-D, new members of the MRP/cMOAT subfamily of transporter proteins. J Natl Cancer Inst. 1998;90(22):1735–1741. doi: 10.1093/jnci/90.22.1735. [DOI] [PubMed] [Google Scholar]
- 15.McAleer MA, Breen MA, White NL, Matthews N. pABC11 (also known as MOAT-C and MRP5), a member of the ABC family of proteins, has anion transporter activity but does not confer multidrug resistance when overexpressed in human embryonic kidney 293 cells. J Biol Chem. 1999;274(33):23541–23548. doi: 10.1074/jbc.274.33.23541. [DOI] [PubMed] [Google Scholar]
- 16.Borst P, de Wolf C, van de Wetering K. Multidrug resistance-associated proteins 3, 4, and 5. Pflugers Arch. 2007;453(5):661–673. doi: 10.1007/s00424-006-0054-9. [DOI] [PubMed] [Google Scholar]
- 17.Reid G, et al. Characterization of the transport of nucleoside analog drugs by the human multidrug resistance proteins MRP4 and MRP5. Mol Pharmacol. 2003;63(5):1094–1103. doi: 10.1124/mol.63.5.1094. [DOI] [PubMed] [Google Scholar]
- 18.Wielinga P, et al. The human multidrug resistance protein MRP5 transports folates and can mediate cellular resistance against antifolates. Cancer Res. 2005;65(10):4425–4430. doi: 10.1158/0008-5472.CAN-04-2810. [DOI] [PubMed] [Google Scholar]
- 19.Jedlitschky G, Burchell B, Keppler D. The multidrug resistance protein 5 functions as an ATP-dependent export pump for cyclic nucleotides. J Biol Chem. 2000;275(39):30069–30074. doi: 10.1074/jbc.M005463200. [DOI] [PubMed] [Google Scholar]
- 20.Wielinga PR, et al. Characterization of the MRP4- and MRP5-mediated transport of cyclic nucleotides from intact cells. J Biol Chem. 2003;278(20):17664–17671. doi: 10.1074/jbc.M212723200. [DOI] [PubMed] [Google Scholar]
- 21.Laue S, et al. cCMP is a substrate for MRP5. Naunyn Schmiedebergs Arch Pharmacol. 2014;387(9):893–895. doi: 10.1007/s00210-014-1018-9. [DOI] [PubMed] [Google Scholar]
- 22.Mourskaia AA, et al. ABCC5 supports osteoclast formation and promotes breast cancer metastasis to bone. Breast Cancer Res. 2012;14(6):R149. doi: 10.1186/bcr3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nongpiur ME, et al. 2014. ABCC5, a gene that influences the anterior chamber depth, is associated with primary angle closure glaucoma. PLoS Genet 10(3):e1004089, and correction (2014) 10(4):e1004352.
- 24.Korolnek T, Zhang J, Beardsley S, Scheffer GL, Hamza I. Control of metazoan heme homeostasis by a conserved multidrug resistance protein. Cell Metab. 2014;19(6):1008–1019. doi: 10.1016/j.cmet.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Wolf CJF, et al. cGMP transport by vesicles from human and mouse erythrocytes. FEBS J. 2007;274(2):439–450. doi: 10.1111/j.1742-4658.2006.05591.x. [DOI] [PubMed] [Google Scholar]
- 26.van de Wetering K, Feddema W, Helms JB, Brouwers JF, Borst P. Targeted metabolomics identifies glucuronides of dietary phytoestrogens as a major class of MRP3 substrates in vivo. Gastroenterology. 2009;137(5):1725–1735. doi: 10.1053/j.gastro.2009.06.052. [DOI] [PubMed] [Google Scholar]
- 27.Krumpochova P, et al. Transportomics: Screening for substrates of ABC transporters in body fluids using vesicular transport assays. FASEB J. 2012;26(2):738–747. doi: 10.1096/fj.11-195743. [DOI] [PubMed] [Google Scholar]
- 28.Blau N, van Spronsen FJ, Levy HL. Phenylketonuria. Lancet. 2010;376(9750):1417–1427. doi: 10.1016/S0140-6736(10)60961-0. [DOI] [PubMed] [Google Scholar]
- 29.Teufel M, et al. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J Biol Chem. 2003;278(8):6521–6531. doi: 10.1074/jbc.M209764200. [DOI] [PubMed] [Google Scholar]
- 30.Kaur H, Kumar C, Junot C, Toledano MB, Bachhawat AK. Dug1p Is a Cys-Gly peptidase of the gamma-glutamyl cycle of Saccharomyces cerevisiae and represents a novel family of Cys-Gly peptidases. J Biol Chem. 2009;284(21):14493–14502. doi: 10.1074/jbc.M808952200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, et al. Underexpressed CNDP2 participates in gastric cancer growth inhibition through activating the MAPK signaling pathway. Mol Med. 2014;20:17–28. doi: 10.2119/molmed.2013.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J-H, et al. Loss of 18q22.3 involving the carboxypeptidase of glutamate-like gene is associated with poor prognosis in resected pancreatic cancer. Clin Cancer Res. 2012;18(2):524–533. doi: 10.1158/1078-0432.CCR-11-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang P, et al. Identification of carboxypeptidase of glutamate like-B as a candidate suppressor in cell growth and metastasis in human hepatocellular carcinoma. Clin Cancer Res. 2006;12(22):6617–6625. doi: 10.1158/1078-0432.CCR-06-1307. [DOI] [PubMed] [Google Scholar]
- 34.Bordusa F. Proteases in organic synthesis. Chem Rev. 2002;102(12):4817–4868. doi: 10.1021/cr010164d. [DOI] [PubMed] [Google Scholar]
- 35.Liebscher S, et al. N-terminal protein modification by substrate-activated reverse proteolysis. Angew Chem Int Ed Engl. 2014;53(11):3024–3028. doi: 10.1002/anie.201307736. [DOI] [PubMed] [Google Scholar]
- 36.Berkers CR, de Jong A, Ovaa H, Rodenko B. Transpeptidation and reverse proteolysis and their consequences for immunity. Int J Biochem Cell Biol. 2009;41(1):66–71. doi: 10.1016/j.biocel.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 37.Hagenfeldt L, Naglo AS. New conjugated urinary metabolites in intermediate type maple syrup urine disease. Clin Chim Acta. 1987;169(1):77–83. doi: 10.1016/0009-8981(87)90395-0. [DOI] [PubMed] [Google Scholar]
- 38.Henning C, Smuda M, Girndt M, Ulrich C, Glomb MA. Molecular basis of maillard amide-advanced glycation end product (AGE) formation in vivo. J Biol Chem. 2011;286(52):44350–44356. doi: 10.1074/jbc.M111.282442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smuda M, Glomb MA. Fragmentation pathways during Maillard-induced carbohydrate degradation. J Agric Food Chem. 2013;61(43):10198–10208. doi: 10.1021/jf305117s. [DOI] [PubMed] [Google Scholar]
- 40.Frerot E, Chen T. Identification and quantitation of new glutamic acid derivatives in soy sauce by UPLC/MS/MS. Chem Biodivers. 2013;10(10):1842–1850. doi: 10.1002/cbdv.201300150. [DOI] [PubMed] [Google Scholar]
- 41.Sforza S, Cavatorta V, Galaverna G, Dossena A, Marchelli R. Accumulation of non-proteolytic aminoacyl derivatives in Parmigiano-Reggiano cheese during ripening. Int Dairy J. 2009;19(10):582–587. [Google Scholar]
- 42.Sgarbi E, et al. Microbial origin of non proteolytic aminoacyl derivatives in long ripened cheeses. Food Microbiol. 2013;35(2):116–120. doi: 10.1016/j.fm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Bottesini C, Tedeschi T, Dossena A, Sforza S. Enzymatic production and degradation of cheese-derived non-proteolytic aminoacyl derivatives. Amino Acids. 2014;46(2):441–447. doi: 10.1007/s00726-013-1637-3. [DOI] [PubMed] [Google Scholar]
- 44.Nieman DC, Shanely RA, Gillitt ND, Pappan KL, Lila MA. Serum metabolic signatures induced by a three-day intensified exercise period persist after 14 h of recovery in runners. J Proteome Res. 2013;12(10):4577–4584. doi: 10.1021/pr400717j. [DOI] [PubMed] [Google Scholar]
- 45.Pohjanen E, et al. A multivariate screening strategy for investigating metabolic effects of strenuous physical exercise in human serum. J Proteome Res. 2007;6(6):2113–2120. doi: 10.1021/pr070007g. [DOI] [PubMed] [Google Scholar]
- 46.Krug S, et al. The dynamic range of the human metabolome revealed by challenges. FASEB J. 2012;26(6):2607–2619. doi: 10.1096/fj.11-198093. [DOI] [PubMed] [Google Scholar]
- 47.Lewis WH, Truslove GM. Electrophoretic heterogeneity of mouse erythrocyte peptidases. Biochem Genet. 1969;3(5):493–498. doi: 10.1007/BF00485609. [DOI] [PubMed] [Google Scholar]
- 48.Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 49.Linster CL, Van Schaftingen E, Hanson AD. Metabolite damage and its repair or pre-emption. Nat Chem Biol. 2013;9(2):72–80. doi: 10.1038/nchembio.1141. [DOI] [PubMed] [Google Scholar]
- 50.Veiga-da-Cunha M, Chevalier N, Stroobant V, Vertommen D, Van Schaftingen E. Metabolite proofreading in carnosine and homocarnosine synthesis: Molecular identification of PM20D2 as β-alanyl-lysine dipeptidase. J Biol Chem. 2014;289(28):19726–19736. doi: 10.1074/jbc.M114.576579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wijnholds J, et al. Multidrug-resistance protein 5 is a multispecific organic anion transporter able to transport nucleotide analogs. Proc Natl Acad Sci USA. 2000;97(13):7476–7481. doi: 10.1073/pnas.120159197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jansen RS, et al. ABCC6 prevents ectopic mineralization seen in pseudoxanthoma elasticum by inducing cellular nucleotide release. Proc Natl Acad Sci USA. 2013;110(50):20206–20211. doi: 10.1073/pnas.1319582110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 54.Böcker S, Letzel MC, Lipták Z, Pervukhin A. SIRIUS: Decomposing isotope patterns for metabolite identification. Bioinformatics. 2009;25(2):218–224. doi: 10.1093/bioinformatics/btn603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El-Sheikh AAK, van den Heuvel JJMW, Koenderink JB, Russel FGM. Interaction of nonsteroidal anti-inflammatory drugs with multidrug resistance protein (MRP) 2/ABCC2- and MRP4/ABCC4-mediated methotrexate transport. J Pharmacol Exp Ther. 2007;320(1):229–235. doi: 10.1124/jpet.106.110379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.