Abstract
Background
The secreted Meloidogyne javanica fatty acid- and retinol-binding (FAR) protein Mj-FAR-1 is involved in nematode development and reproduction in host tomato roots. To gain further insight into the role of Mj-FAR-1 in regulating disease development, local transcriptional changes were monitored in tomato hairy root lines with constitutive mj-far-1 expression compared with control roots without inoculation, and 2, 5 and 15 days after inoculation (DAI), using mRNA sequencing analysis.
Results
Gene-expression profiling revealed a total of 3970 differentially expressed genes (DEGs) between the two lines. Among the DEGs, 1093, 1039, 1959, and 1328 genes were up- or downregulated 2-fold with false discovery rate < 0.001 in noninoculated roots, and roots 2, 5, and 15 DAI compared with control roots, respectively. Four main groups of genes that might be associated with Mj-FAR-1-mediated susceptibility were identified: 1) genes involved in biotic stress responses such as pathogen-defense mechanisms and hormone metabolism; 2) genes involved in phenylalanine and phenylpropanoid metabolism; 3) genes associated with cell wall synthesis, modification or degradation; and 4) genes associated with lipid metabolism. All of these genes were overrepresented among the DEGs. Studying the distances between the treatments, samples from noninoculated roots and roots at 2 DAI clustered predominantly according to the temporal dynamics related to nematode infection. However, at the later time points (5 and 15 DAI), samples clustered predominantly according to mj-far-1 overexpression, indicating that at these time points Mj-FAR-1 is more important in defining a common transcriptome.
Conclusions
The presence of four groups of DEGs demonstrates a network of molecular events is mediated by Mj-FAR-1 that leads to highly complex manipulation of plant defense responses against nematode invasion. The results shed light on the in vivo role of secreted FAR proteins in parasitism, and add to the mounting evidence that secreted FAR proteins play a major role in nematode parasitism.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1426-3) contains supplementary material, which is available to authorized users.
Keywords: Fatty acid- and retinol-binding protein, Nematode–host interaction, RNA-Seq, Plant defense mechanism, Meloidogyne, Parasitism, Effector
Background
Among the most devastating plant-parasitic nematodes are the sedentary Meloidogyne root-knot nematodes (RKNs), which are obligate biotrophs [1]. These parasites interact with their hosts in a subtle and sophisticated manner that is achieved by sustaining a constitutive dialog with select host cells in the vascular cylinder. These cells are the nematode feeding sites, termed giant cells (GCs), upon which nematode development and reproduction rely [2-4]. Although the mechanism by which RKNs establish the GC system is unknown, increasing evidence indicates that glandular secretions (effectors) injected into plant cells by the nematodes interact directly or indirectly with essential plant components, leading to the establishment and maintenance of nematode feeding sites [5-9]. Two esophageal gland types are involved in producing effectors: two subventral glands and one dorsal gland [6]. Other organs in contact with the external environment that produce secretory proteins include the amphids and cuticle. In the last two decades, several cuticle proteins from plant-parasitic nematodes have been identified, including some that are important for parasitism [10-13]. To ensure successful nematode development and reproduction, the nematode-induced feeding-site structure must be maintained for up to 6 weeks, which requires continuous suppression of the plant defense response throughout this period. Suppression of plant defense genes after RKN infection has been demonstrated in microarray studies [14], but the mechanism governing this suppression remains elusive. Two major pathogen-defense-signaling pathways that have been extensively studied are the salicylic acid (SA)-dependent pathway and a SA-independent pathway that involves jasmonic acid (JA) and ethylene (ET) [15,16]. These pathways crosstalk via a complex network of regulatory interactions, and are susceptible to continuous manipulation by plant pathogens for the promotion of virulence and disease production [16].
A group of proteins termed fatty acid- and retinol-binding (FAR) proteins, secreted by all trophic groups of nematodes, have long been acknowledged for their potential function in host immunomodulation [17-23]. FAR proteins are of major interest for several reasons. i) They may play an important role in scavenging fatty acids and retinol for the survival of the parasite [20]. ii) They may induce localized depletion of essential lipids such as oxylipins, thereby compromising the host’s defensive immune response [11,24]. iii) They are located at the host–parasite interface [18]. iv) Their structure is unique to the nematode phylum and is unlike that of any other known family of lipid-binding proteins [18,19,21,25]. Together with their presence in multiple families of parasitic nematodes, these findings lend support to the notion that this nematode-restricted family of proteins plays a crucial role in host parasitism [18].
The role played by plant-parasitic FAR proteins in negating the plant’s defense response was first studied for the potato cyst nematode Globodera pallida FAR (Gp-FAR-1) demonstrating lipid-binding activity of Gp-FAR-1 to linoleic and linolenic acids, and inhibition of lipoxygenase (LOX)-mediated modification of these substrates in vitro [11]. More recently, a functional analysis of the role of Meloidogyne javanica FAR (Mj-FAR-1) in RKN–plant interactions was performed [26]. The spike in expression of mj-far-1 by the parasitic nematode M. javanica second-stage juveniles (J2) at 3–5 days after inoculation (DAI), together with its abundant deposition in the apoplast during the sedentary stages, suggests a primary role for this effector protein in the early and late stages of the host–parasite interaction. Moreover, constitutive expression of mj-far-1 in tomato (Solanum lycopersicum) hairy roots renders plants more susceptible to infection by M. javanica [26]. Increased host susceptibility to nematode infection following the overexpression of nematode parasitism genes has been documented [27,28], suggesting that an excess of some effector proteins enhances a compatible host–parasite interaction via modulation of the plant stress [28] and defense [27] responses. Despite extensive research into the functional role of plant-parasitic FAR proteins [11,26], little is known about the molecular mechanisms underlying the increased susceptibility response in mj-far-1-expressing roots. To further clarify the increased susceptibility in a root line expressing mj-far-1 in response to M. javanica infection, we analyzed gene expression in roots of transgenic tomato differing in their constitutive expression of the nematode mj-far-1. Many of the genes that were differentially regulated in mj-far-1-expressing roots were tomato genes known to play important roles in pathogen-mediated defense responses. These responses involve physicochemical processes, such as cell wall regulation and modification, and biochemical responses such as biosynthesis and regulation of compounds associated with fatty acids and the phenylpropanoid-signaling pathways. Our results provide insights into the transcription-regulation events, driven by Mj-FAR-1 secreted by the invading nematode, that facilitate nematode development and disease production in the host plant.
Results
Transcriptomic data collection and analysis
Our experimental system exploited the higher susceptibility of roots overexpressing mj-far-1 upon M. javanica infection [26] to characterize mj-far-1-mediated differences in gene expression during the M. javanica infection process. Root samples of vector 11.5 carrying the kanamycin-resistance gene (Kan control roots) and mj-far-1.1 lines overexpressing mj-far-1 (OE roots) from in vitro-infected tomato root cultures were harvested at 2, 5, and 15 DAI. Equivalent root segments from noninoculated root cultures of both lines were used as reference root tissues. At 5 DAI, the harvested samples of root tips or segments showed prominent swelling, an indication of nematode invasion and establishment (Figure 1A,B). At 15 DAI, mature galls on primary roots were hand-dissected (Figure 1C,D). As reported previously [26], accelerated disease development was observed for OE roots compared with Kan roots, as indicated by increased gall incidence (Figure 1C,D). To monitor the expression levels of differentially expressed genes (DEGs) with disease progression, and to evaluate the effect of mj-far-1 overexpression, which underlies the increase in susceptibility, changes in gene expression were investigated by directly comparing noninoculated OE and Kan roots (Figure 1E, comparison 1). Similarly, expression profiles of OE and Kan roots were compared at designated time points after inoculation (Figure 1E, comparisons 2–4). In addition, noninoculated Kan and OE root gene-expression profiles were compared to those upon inoculation of the same root line at 2, 5, and 15 DAI (Figure 1E, comparisons 5–10). RNA was extracted for transcriptome analysis as described in Experimental procedures and RNA-Seq was performed on the Illumina HiSeq™ 2000 platform, yielding an average of 26.6 million high-quality reads per sample (Table 1). Paired-end transcript sequences were mapped against the International Tomato Annotation Group (ITAG) Solanum lycopersicum protein reference version 2.3 (http://solgenomics.net) with SOAPaligner/SOAP2 [29]. Gene expression was quantified as the total number of reads (paired-end) from each sample that uniquely align to the transcriptome reference of ITAG2.3 using the aligner SOAP2. An average 20.1 million reads from paired-end sequencing uniquely aligned to the reference sample, and represented an average of 75.6% of the total reads (Table 1) used in the bioinformatics analysis.
Figure 1.
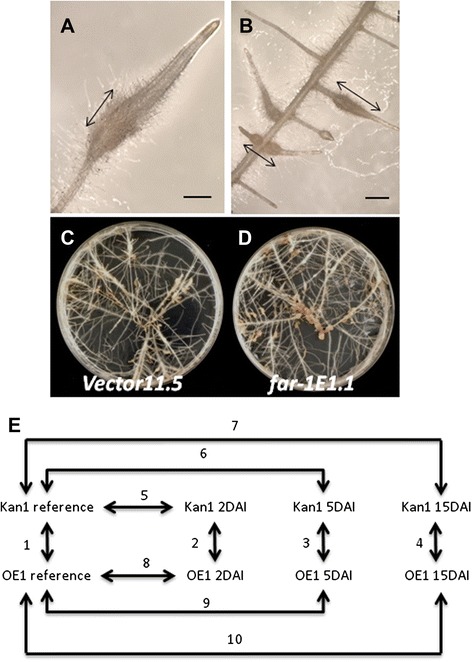
Plant material used for RNA-Seq analysis and the experimental design for complete tomato RNA-Seq profiling of tomato root lines carrying the kanamycin-resistance gene (Kan) or overexpressing mj-far-1 (OE) inoculated with Meloidogyne javanica . A representative 5 DAI gall used for RNA extraction on roots of the tomato control Kan (A) and OE line (B) following inoculation with Meloidogyne javanica. Arrows indicate segments collected for RNA extraction. Bars = 150 μm. Late infection stage of the tomato control Kan line (C) and OE line (D) as shown at 15 DAI. Note the increased size and density of galls on the OE vs. Kan roots. (E) Schematic representation of the experimental procedure. Four comparisons (1–4) between control Kan and OE lines were performed at each time point: 0, 2, 5, and 15 DAI. In addition, the reference tissue of each root line was directly compared with the transcript profile of the same root line at 2, 5, and 15 DAI: comparisons 5–7 for Kan and 8–10 for OE lines.
Table 1.
Alignment of RNA-Seq reads to the ITAG2.3 reference transcriptome
| Treatment | Non-inoculated | 2 DAI | 5 DAI | 15 DAI | ||||
|---|---|---|---|---|---|---|---|---|
| Root lines | OE | KAN | OE | KAN | OE | KAN | OE | KAN |
| High Quality Paired-End Reads | 27,267,694 | 27,277,056 | 25,641,052 | 26,805,144 | 26,170,776 | 26,498,432 | 26,382,764 | 26,932,422 |
| Uniquely Aligned Reads | 20,798,101 | 20,941,168 | 19,677,396 | 20,722,903 | 19,444,292 | 20,150,007 | 19,296,181 | 20,207,686 |
| Total Unmapped Reads | 6,071,686 | 5,911,916 | 5,568,661 | 5,679,513 | 6,306,052 | 5,877,328 | 6,581,004 | 6,168,739 |
The number of paired-end and uniquely aligned sequence reads analyzed from Illumina sequencing runs of all eight samples are arranged by treatment type (noninoculated, and 2, 5, and 15 DAI) and root line.
mj-far-1-mediated differences in gene expression
To assess the regulation of tomato transcripts by Mj-FAR-1, differential expression analysis (see Figure 1E for all comparisons conducted) was performed between inoculated and noninoculated OE and Kan root lines. For the calling of differentially regulated genes, the false discovery rate (FDR) threshold was set to ≤0.001 and log2 ratio ≥ 1. The number of DEGs, both upregulated (>1-fold) and downregulated (<1-fold), increased with time after inoculation for both OE and Kan root lines (Figure 2A). Based on Venn diagrams (Figure 2B–D), a total of 3970 DEGs were identified in OE compared with Kan root lines using FDR ≤ 0.001. The number of DEGs common to the OE root line alone and the OE root–nematode interactions is indicated in the overlapping portions of the circles. Of the 3970 genes, 2069 were upregulated and 2205 were downregulated in OE vs. Kan lines for all inoculated and noninoculated samples. The numbers of upregulated genes in noninoculated OE samples and those at 2, 5, and 15 DAI compared with the Kan roots at the same time points were 324, 225, 1241, and 707, respectively (Figure 2C). The numbers of downregulated genes of noninoculated OE samples and those inoculated at 2, 5, and 15 DAI compared with Kan roots were 769, 814, 718, and 621, respectively (Figure 2D). A total of 61 upregulated and downregulated genes overlapped between all noninoculated and inoculated OE root samples (Figure 2B, Table 2). These genes might contribute to the mj-far-1-associated increase in susceptibility in the mj-far1.1 root line. Among this group were genes involved in fatty acid metabolism, such as those encoding the long-chain fatty acid-CoA ligase (Solyc08g008310.2.1) and the lipid-modification enzyme lipase (Solyc05g018770.1.1). In addition, a group of hormone signal-related genes that were differentially regulated in OE roots included JA-related genes, such as a gene encoding a proteinase inhibitor (Solyc03g098710.1.1), and auxin-related genes, such as the gene encoding indole-3-acetic acid-amido synthetase (Solyc02g092820.2.1). Other genes involved in plant defense that encoded the WRKY transcription factor (Solyc05g053380.2.1), a pathogenesis-related (PR) gene (Solyc09g007020.1.1), and a gene involved in the phenylalanine pathway (Solyc02g081800.1.1) showed consistent differential regulation among the treatments (Table 2). An additional group of genes that might shed light on mj-far-1 regulation of gene expression constituted DEGs unique to inoculated samples, in which a total of 52 upregulated and downregulated genes overlapped only among inoculated OE roots (Table 3). This group included genes involved in cell wall modification and remodeling, such as those encoding expansin-like proteins (Solyc08g07790 0.2.1and Solyc03g093390.2.1) and cell wall protein (CWP) (Solyc09g097770.2), hormone-related genes such as those encoding auxin-responsive protein (Solyc08g021820.2.1) and gibberellin synthesis (Solyc12g042980.1.1), a gene of the phenylpropanoid pathway encoding chalcone synthase (CHS) (Solyc05g053550.2.1), and defense-related genes such as those encoding pathogenesis-related proteins (Solyc07g006710.1.1 and Solyc01g106640.2.1). Changes associated with fatty acid metabolism that were restricted to the inoculated root samples included genes encoding the fatty acid elongase 3-ketoacyl-CoA synthase (Solyc03g005320.2.1) and the nonspecific lipid-transfer protein (Solyc06g054070.2.1) (Table 3).
Figure 2.
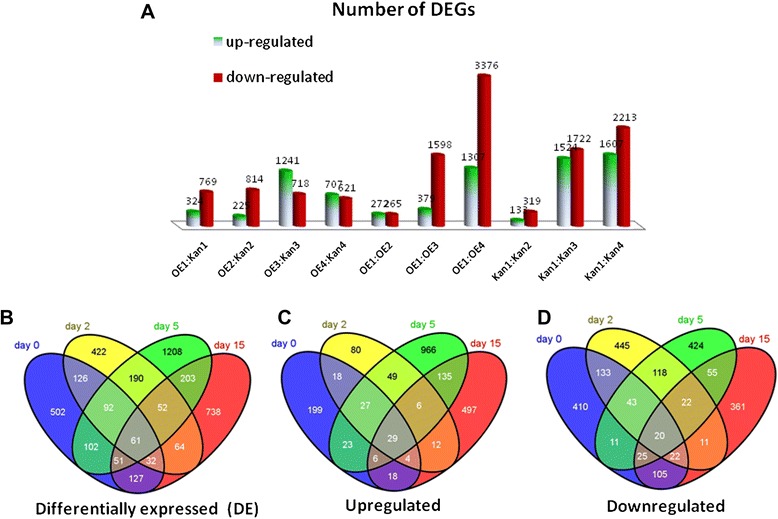
Regulation of differentially expressed genes among inoculated, noninoculated of mj-far-1 overexpressing and control roots. (A) Comparison of differentially regulated genes from RNA-Seq data between tomato root lines at each designated time point. Numbers of up- and downregulated genes are indicated from 10 comparisons. (B) A generalized Venn diagram showing intersection of genes that are differentially regulated (up- and downregulated) in mj-far1.1 root line compared with vector 11.5 control roots among noninoculated, 2, 5, and 15 DAI samples. (C) Intersection of genes that are upregulated in the mj-far1.1 root line compared with vector 11.5 control roots among noninoculated, 2, 5, and 15 DAI samples. (D) Intersection of genes that are downregulated in mj-far1.1 root line compared with vector 11.5 control roots among noninoculated, 2, 5, and 15 DAI samples. Genes present in two sets are shown in the intersection, so that the sum of the numbers within a circle is the total number of genes in that set. The size of the circles is not representative of the quantity of probe sets. Overlapping areas represent common probe sets. Fold change with an absolute value >2 and P value ≤ 0.05 was used for the analyses.
Table 2.
Differentially expressed genes common to noninoculated and inoculated mj-far-1 -overexpressing root line and their functional categories
| Functional categories | Sub-category | Gene ID | OE1-KAN1 | OE2-KAN2 | OE3-KAN3 | OE4-KAN4 | Annotation |
|---|---|---|---|---|---|---|---|
| Cell Wall | Proteins LLR | Solyc07g053840.1.1 | -2.44 | -2.20 | -9.83 | -2.24 | LRP receptor-like serine/threonine-protein kinase , RLP |
| Cellulose synthesis COBRA | Solyc03g114900.2.1 | -1.22 | -1.19 | -2.29 | -1.46 | COBRA-like protein | |
| Degradation pectate lyase and polygalacturonases | Solyc12g096750.1.1 | -2.32 | -3.37 | -1.90 | -7.01 | Polygalacturonase 4 | |
| Modification | Solyc10g086520.1.1 | -1.23 | -1.49 | -1.81 | -1.02 | Expansin-1 | |
| UDP glucosyl and glucoronyl transferase | Solyc05g053400.1.1 | 2.20 | 2.57 | 1.15 | 3.53 | Glucosyltransferase | |
| Cell organization | Chloroplast location | Solyc06g062400.1.1 | 2.12 | 1.65 | 2.26 | 1.70 | Cloroplast unusual positioning 1A |
| Development | Nodule formation | Solyc05g055540.1.1 | 2.94 | 1.64 | 2.54 | 1.99 | Nodulin family protein |
| unkown | Solyc06g082460.1.1 | -1.90 | -1.29 | -1.11 | -1.46 | Plant-specific domain TIGRO1568 family protein | |
| Hormone metabolism | Auxin induced/regulated/responsive | Solyc02g092820.2.1 | -2.99 | -1.80 | -4.29 | -5.59 | Indole-3-acetic acid-amido synthetase GH3.8 |
| Solyc12g005310.1.1 | -2.75 | -1.18 | -3.60 | -4.05 | Auxin-responsive GH3-like | ||
| Ethylene synthesis/degradation | Solyc09g010000.2.1 | 1.42 | 1.80 | 1.87 | 1.11 | 1-aminocyclopropane-1-carboxylate oxidase-like protein | |
| Solyc11g072310.1.1 | 1.51 | 1.45 | 1.41 | 1.72 | Gibberellin 20 -oxidase-3 | ||
| Solyc06g073580.2.1 | -3.78 | -1.50 | -1.01 | 1.16 | 1-aminocyclopropane-1-carboxylate oxidase-like protein | ||
| Jasmonic acid induced/regulated/responsive | Solyc03g098710.1.1 | -4.38 | -1.50 | -2.09 | 2.49 | Kunitz-type proteinase inhibitor A4 (Fragment) | |
| Solyc03g098720.2.1 | -10.39 | -1.72 | -10.48 | 4.26 | Kunitz trypsin inhibitor | ||
| Lipid metabolism | FA Synthesis and FA Elongation | Solyc08g008310.2.1 | -3.21 | -2.93 | -3.52 | -3.27 | Long-chain-fatty-acid-CoA ligase |
| Phospholipid synthesis | Solyc12g040790.1.1 | -1.07 | -1.19 | -1.40 | 2.19 | Menaquinone biosynthesis methyltransfere ubiE | |
| Lipid modification | Solyc05g018770.1.1 | -2.58 | -12.08 | -10.93 | -4.19 | Esterase/lipase/thioesterase (Fragment) | |
| N-metabolism/degradation | Glutamate dehydrogenase | Solyc05g052100.2.1 | -5.33 | -11.68 | -2.12 | -1.56 | Glutamate dehydrogenase |
| Phosphotransfer and Pyrophoshatases guanylate kinase | Solyc10g79140.1.1 | 1.30 | 1.75 | 2.57 | 2.48 | Guanylate kinase | |
| NUDIX hydrolases | Solyc07g045430.2.1 | 2.34 | 2.61 | 1.62 | -2.97 | Nudix hydrolase 2 | |
| Polyamine metabolism | SAM/decarboxylase | Solyc06g054460.1.1 | -1.37 | -2.12 | -1.17 | -1.96 | S-adenosylmethionine decarboxylase proenzyme |
| Spermidine/synthase | Solyc06g053510.2.1 | -5.21 | -2.87 | -1.88 | 1.21 | Spermidine synthase | |
| Protein metabolism | Protease | Solyc05g016250.2.1 | -11.21 | -11.18 | -11.08 | -9.72 | Cysteine-type peptidase |
| Solyc07g066500.1.1 | 11.02 | 9.87 | 4.88 | 11.14 | U1p1 protease family C-terminal catalytic domain containing protein | ||
| Solyc01g057880.1.1 | -4.10 | -5.48 | -3.35 | -4.10 | U1p1 protease family C-terminal catalytic domain containing protein | ||
| Postranslational modification | Solyc08g066400.1.1 | -10.91 | -10.97 | -10.39 | -10.64 | Protein kinase (Fragment) | |
| Solyc03g083800.1.1 | 5.01 | 10.74 | 5.59 | 11.45 | Serine/threonine-protein phosphatase 7 long form homolog | ||
| Targeting | Solyc12g096550.1.1 | 1.20 | 1.10 | 1.23 | 1.11 | Pheophorbide a oxygenase family | |
| Transcription factor | GeBP like | Solyc07g052900.1.1 | 1.21 | 1.55 | 1.66 | 1.96 | Os09g0451700 protein (Fragment) |
| Solyc07g052700.2.1 | 2.84 | 2.57 | 4.21 | 2.30 | MADS-box transcription factor 1 | ||
| MADS box transcription factor/family | Solyc02g089200.2.1 | 4.79 | 3.11 | 5.74 | 4.14 | MADS-box transcription factor | |
| Solyc07g052720.2.1 | 3.67 | 2.31 | 4.65 | 2.99 | MADS-box protein AGL66 | ||
| Putative transcription regulator | Solyc10g051140.1.1 | -1.94 | -2.58 | -1.90 | -1.81 | Genomic DNA chromosome 5 P1 clone MTE17 | |
| WRKY domain transcription factor family | Solyc05g053380.2.1 | -2.30 | -1.99 | 1.60 | -2.02 | WRKY transcription factor 31 | |
| unknown | Solyc01g100440.1.1 | 2.07 | 1.33 | 1.89 | 2.42 | Transcription regulatory protein SNF5 | |
| Secondary metabolism | Isoflavone reductase | Solyc10g052500.1.1 | -4.36 | -510 | -3.32 | -4.16 | Phenylcoumaran benzylic ether reductase 3 |
| Isoprenoids/terpenoids | Solyc10g005390.2.1 | 1.86 | 1.47 | 2.96 | 2.67 | Linalool synthase | |
| Phenylpropanoids | Solyc02g08100.1.1 | 2.24 | 2.86 | 3.13 | 1.41 | Acyltransferase (Fragment) | |
| Signaling | Sugar and nutrien phisiology | Solyc12g099780.1.1 | -1.69 | -1.48 | 1.91 | 1.05 | Unknown Protein |
| Receptor kinase leucine rich repeat XI | Solyc12g009780.1.1 | 8.74 | 3.37 | 3.18 | 8.43 | LRR receptor-like serine/ threonine-protein kinase, RLP | |
| Abiotic stress | Heat stress | Solyc06g011400.2.1 | 4.54 | 2.57 | 4.40 | 2.45 | ATP-dependent chaperone c1pB |
| Solyc06g11380.2.1 | 5.99 | 3.67 | 4.70 | 2.51 | Chaperone C1pB | ||
| Solyc06g011370.2.1 | 3.86 | 3.46 | 3.52 | 2.25 | Chaperone protein clpB 2 | ||
| Plant defence | Pathogen attack response | Solyc09g007020.1.1 | 1.70 | 1.24 | 1.07 | 1.14 | Pathogenesis-related protein |
| Pathogen resistance | Solyc07g009510.1.1 | 2.56 | 2.44 | 1.76 | 1.05 | Chitinase | |
| Gene to gene resistance/recognation | Solyc12g044190.1.1 | 1.07 | 1.33 | 2.37 | 2.18 | Nbs-lrr, resistance protein | |
| Redox state | Solyc01g081250.2.1 | 1.22 | 1.02 | 1.34 | 1.08 | Glutathione-S-transferase | |
| Solyc03g116120.1.1 | 2.80 | 1.87 | 2.18 | 1.99 | Glutathione S-transferase 12 | ||
| Solyc08g014330.2.1 | -2.24 | -1.19 | -1.06 | 1.08 | Primary amine oxidase | ||
| unknown | Solyc01g017600.2.1 | -2.17 | -5.86 | -10.57 | 1.48 | Plant viral-response family protein | |
| Transport | Amino acids | Solyc01g100390.2.1 | 2.12 | 2.04 | 2.63 | 1.21 | Pyrophosphate-energized proton pump |
| Peptides and oligopeptides | Solyc03g113430.2.1 | 1.63 | 1.24 | 3.70 | 1.28 | Peptide transporter | |
| unknown | Solyc01g73670.2.1 | -1,60 | -1.63 | -1.98 | 1.22 | Uncharacterized MFS-type transporter C19orf28 | |
| Protease inhibitor/seed storage /lipid transfer protein (LTP) family protein | Solyc03g083990.1.1 | -2.08 | 1.15 | 1.25 | -2.66 | Cortical cell-delineating protein | |
| Not assigned | unknown | Solyc04g015610.2.1 | -1.89 | -1.27 | -1.50 | -3.73 | Os01g0611000 protein (Fragment) |
| Solyc10g080380.1.1 | -1.53 | -3.73 | -3.61 | -1.55 | Unknown Protein | ||
| Solyc07g007770.1.1 | -6.36 | -5.19 | -5.23 | -4.54 | Unknown Protein | ||
| Solyc09g091810.1.1 | -2.80 | -1.29 | -1.62 | -2.78 | Unknown Protein | ||
| Solyc01g056370.2.1 | 12.79 | 12.99 | 13.67 | 13.64 | Unknown Protein | ||
| Solyc05g052880.2.1 | 1.93 | 1.37 | 1.89 | 3.33 | Unknown Protein |
Gene ID number and log2 values at each time point before and after inoculation are indicated. All genes were considered to be differentially expressed with a threshold q-value < 0.05.
Table 3.
Differentially expressed genes upon M. javanica inoculation
| Functional categories | Sub-category | Gene ID | OE2-KAN2 | OE3-KAN3 | OE4-KAN4 | Annotation |
|---|---|---|---|---|---|---|
| TCA/ org. transformation | Carbonic anhydrases | solyc02g067750.2.1 | 1.99565 | 2.704447 | 2.324205 | Carbonic anhydrase |
| Cell wall | Degradation pectate lyases and polygalacturonases | solyc08g068150.2.1 | -1.17686 | 1.198919 | 1.5876 | BURP domain-containing protein |
| Modification | solyc08g077900.2.1 | -1.5738 | -1.34033 | -1.07768 | Expansin-like protein | |
| solyc03g093390.2.1 | -1.23629 | -1.69975 | 1.122155 | Expansin protein | ||
| Lipid metabolism | FA synthesis and FA elongation | solyc03g005320.2.1 | -1.21734 | 1.118395 | 1.412513 | Fatty acid elongase 3-ketoacyl-CoA synthase |
| Lipid degradation | solyc01g100020.2.1 | -1.6392 | -1.60844 | -1.23022 | Phospholipase D | |
| Amino acid metabolism | Synthesis | solyc01g006620.2.1 | -1.39285 | 1.576919 | 1.629744 | Phosphoribosylanthranilate trnasferase |
| Secondary metabolism | Flavonoids | solyc05g053550.2.1 | 2.880173 | 1.665111 | -1.64308 | Chalcone synthase |
| Simple phenols | solyc06go76760.1.1 | -1.14152 | 1.327023 | 1.041531 | Laccase 1a | |
| Hormone metabolism | Gibberelin synthesis/degradation | solyc12g042980.1.1 | -2.48967 | -2.80308 | -1.41139 | 2-oxoglutarate- dependent dioxygenase |
| Tetrapyrrole synthesis | Unspecified | solyc12g005300.1.1 | -1.37617 | -1.44432 | 1.099574 | Chlorophyllase 2 |
| Plant Defense | Gen to resistance/recognition | solyc11g0066401.1 | -2.44176 | 1.276086- | 1.705295 | Cc-nbs-lrr, resistance protein |
| solyc07g006710.1.1 | -1.12202 | -2.25906 | -1.15715 | Pathogenesis-related protein PR-1 | ||
| Pathogen attack response | solyc01g106640.2.1 | -1.43339 | -2.26051 | -3.62562 | Pathogenesis-related protein 1 | |
| Redox state | solyc07g039410.2.1 | -2.20729 | -1.85636 | -1.61107 | Nbs-lrr, resistance protein | |
| solyc05g046030.2.1 | -1.44323 | -1.95476 | -1.75115 | Peroxidase | ||
| solyc01g006290.2.1 | -4.44176 | -2.66733 | 1.965054 | Peroxidase | ||
| solyc01g006310.2.1 | -2.19935 | -1.68906 | -1.71087 | Peroxidase | ||
| solyc05g006740.2.1 | -1.39462 | -1.67555 | -1.25058 | Glutathione S-transferase | ||
| Miscelaneous | CytochromeP450 | solyc07g052370.2.1 | 1.592891 | 1.144126 | 2.253552 | Cytochrome P450 |
| Transcription factor | C2c2(Zn) Co-like, Constans-like zinc finger family | solyc07g066510.2.1 | -3.16422 | -2.54704 | 2.541796 | Zinc finger protein CONSTANS-LIKE 2 |
| MYB domain transcription factor family | solyc06g005310.2.1 | -1.3004 | 1.639491 | 1.189045 | MYB transcription factor | |
| solyc10g008700.1.1 | -1.65232 | 2.339096 | 1.861414 | MYB transcription factor | ||
| bZIP transcription factor family | solyc02g072570.1.1 | -1.32419 | -1.23206 | -2.44191 | Transcription factor bZIP98 | |
| Aux/IAA family | solyc08g021820.2.1 | -10.0373 | -2.64797 | -4.15734 | Auxin responsive protein | |
| Putative transcription regulator | solyc01g081320.2.1 | -1.85679 | 1.327023 | 1.545638 | Pentatricopeptide repeat-containing protein | |
| Protein metabolism | Targeting | solyc07g017520.2.1 | -1.36494 | 1.651521 | 1.06459 | Conserved oligomeric Golgi complex subunit 3 |
| Posttranslational regulator | solyc04g15120.2.1 | -1.7365 | -1.3454 | -1.44333 | U-box domain containing protein expressed | |
| solyc09g083410.2.1 | 1.002774 | 1.290076 | 1.408902 | Amidase hydantoinase/carbamoylase family protein expressed | ||
| Degradation | solyc07g054370.2.1 | -1.74661 | 2.633149 | 1.463537 | F-box/LRR-repeat protein At3g59200 | |
| Signalling | Receptor kinases | solyc12g005620.1.1 | 1.813059 | 1.988114 | 1.028337 | LRR receptor-like serine/threonine-protein kinase,RLP |
| solyc06g069740.1. | 1.354104 | 1.251148 | -2.06553 | Calmodulin-like protein | ||
| Calcium | solyc03g083320.2.1 | -1.11983 | -1.18853 | 1.489756 | Calcineurin B-like calcium binding protein | |
| solyc01g097420.1.1 | 1.390368 | 1.531115 | -1.73009 | Calcuim ATPase | ||
| G-proteins | solyc03g078570.2.1 | -1.35177 | 1.730895 | 1.078512 | Ras-related protein Rab-6A | |
| Transport | Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | solyc06g054070.2.1 | -1.12655 | -1.38876 | -1.27683 | Non-specific lipid-transfer protein |
| Sugars | solyc03g113210.2.1 | -2.97781 | -10.8452 | 1.643367 | Porin/voltage-dependent anion-selective channel protein | |
| Not assigned | Unknown | solyco1g104720.2.1 | -3.27322 | -3.93722 | -2.36617 | Unknown Protein |
| solyc07g009020.1.1 | -2.17045 | -4.1934 | -1.14893 | Unknown Protein | ||
| solyc08g078920.1.1 | -1.1221 | -1.43749 | -1.25381 | Proline-rich Protein | ||
| solyc12g049140.1.1 | -2.16278 | -3.50997 | -2.2489 | Extensin-like protein Ext1 | ||
| solyc06g051500.2.1 | -1.31051 | 3.08695 | 2.375988 | Unknown Protein | ||
| solyc07g008980.2.1 | -3.19664 | -3.89496 | -1.52861 | Unknown Protein | ||
| solyc07g009030.2.1 | -2.28689 | -3.72919 | -1.38264 | Unknown Protein | ||
| solyc07g032170.2.1 | -1.37989 | 2.61237 | 1.682212 | Abhydrolase domain- containing protein 5 | ||
| solyc04g015700.1.1 | -1.09406 | -2.77071 | -1.14691 | Unknown Protein | ||
| solyc05g009580.2.1 | -1.41924 | -1.0847 | -1.80119 | Aluminum-activated malate transporter-like | ||
| solyc12g014120.1.1 | -1.51214 | 1.122747 | 1.442261 | Unknown Protein | ||
| solyc09g097770.2.1 | 1.242743 | 1.587252 | 1.011286 | Cell wall protein | ||
| solyc03g078580.2.1 | -1.2353 | 1.865868 | 1.701912 | Unknown Protein | ||
| solyc06g005210.1.1 | 2.124099 | 1.102057 | 1.169745 | Cytochrome P450 like_TBP | ||
| solyc01g097690.2.1 | -1.95437 | -3.72554 | -1.10791 | Extensin-like protien Dif54 |
Gene ID numbers along with log2 values at each time point before and after inoculation are indicated. All genes were considered to be differentially expressed with a threshold q-value < 0.05.
Gene ID number along with log2 values at each time point before and after inoculation are indicated. All genes were considered DEGs with a cutoff q-value < 0.05.
Principal component analysis and distribution of differentially expressed genes
The Pearson correlation coefficient was used to determine the significance of the correlation between mRNA datasets of all eight samples using R (version 3.0.0) (http://www.R-project.org) in the FactoMineR package [30]. As shown by the principal component analysis (PCA), root profiles of noninoculated and 2 DAI samples clustered with respect to temporal dynamics associated with nematode infection, whereas the effect of mj-far-1 overexpression was less important at these stages. The resulting dendrogram revealed small differences in the expression levels of DEGs between noninoculated OE and Kan roots as well as between 2 DAI OE and Kan roots (Figure 3A). At 5 and 15 DAI, i.e., once nematode infection had progressed, root samples clustered predominantly in accordance with mj-far-1 overexpression, revealing broad, global differences in the expression levels of DEGs between OE and Kan root lines (Figure 3A). These results indicate that mj-far-1 is more important in defining a common transcriptome at later time points (Figure 3A). In this analysis, the most variability in the data was accounted for by dimension 1 (34.76%), while dimension 2 accounted for 21.86% of the variability in the data. Analyzing the distribution of DEGs and measuring the transcriptional changes detected in the OE vs. Kan root lines demonstrated that most of the changes between the lines occurred at 5 DAI when 1959 genes were differentially expressed (Figure 3B).
Figure 3.
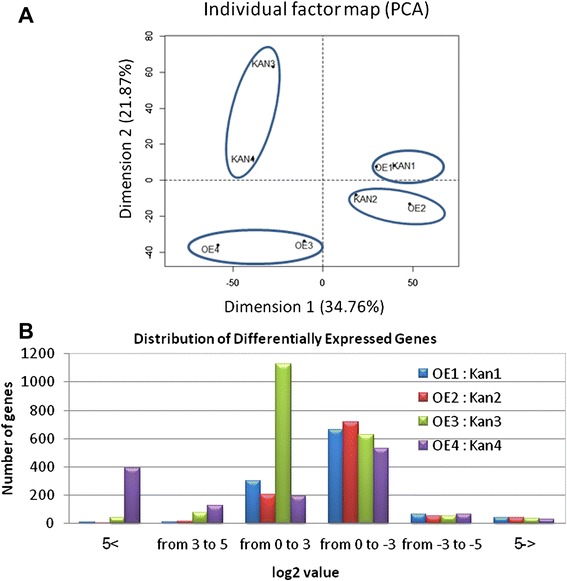
Distribution of differentially expressed genes. (A) Three-dimensional representation according to principle component analysis (PCA) of the differential gene expression data of eight treatments used in the RNA-Seq analysis (as implemented in JMP Genomics 5.1). Kan roots are root samples of vector 11.5 carrying the kanamycin-resistance gene (Kan control roots) and OE roots are mj-far-1.1 lines overexpressing mj-far-1 (OE roots). In this analysis, samples with similar expression profiles lie closer to each other than those with dissimilar profiles. Axes 1 and 2 show robust class separation into four major groups: Kan1 and OE1; Kan2 and OE2; Kan3 and Kan4; and OE3 and OE4. At the early time points (noninoculated and 2 DAI) the infection itself is responsible for most of the transcriptional variance. However, at 5 and 15 DAI, mj-far-1 is the variable responsible for most of the transcriptional variance among treatments with infection playing a lesser role. (B) Distribution of up- and downregulated differentially expressed genes and their fold change over all comparisons made between OE and Kan root lines.
Functional categorization of differentially expressed genes
To obtain an overview of the processes that are altered during the early and late stages of the plant’s response to nematode infection as a consequence of mj-far-1 overexpression, DEGs were classified using MapMan 2.0.0 [31] (Figure 4). Of the 3970 probe sets, 1144 corresponded to unassigned proteins, i.e., those with no known homolog in Arabidopsis. All other probe sets were grouped into functional categories, among them transcripts associated with secondary metabolism (104 probe sets), lipid metabolism (79 probe sets), cell wall (136 probe sets), transport (232 probe sets), hormone (155 probe sets), stress (180 probe sets), development (133 probe sets), and signaling (245 probe sets) (Figure 4). Probe sets that did not fit into any of these categories or fell into multiple categories were grouped as ‘miscellaneous’ (420 probe sets, Figure 4). In this study, we specifically focused on groups that were associated with stress and defense, fatty acids, phenylpropanoids (secondary metabolism), and cell walls. These groups of DEGs were further studied in relation to the increased susceptibility observed in roots overexpressing mj-far-1.
Figure 4.
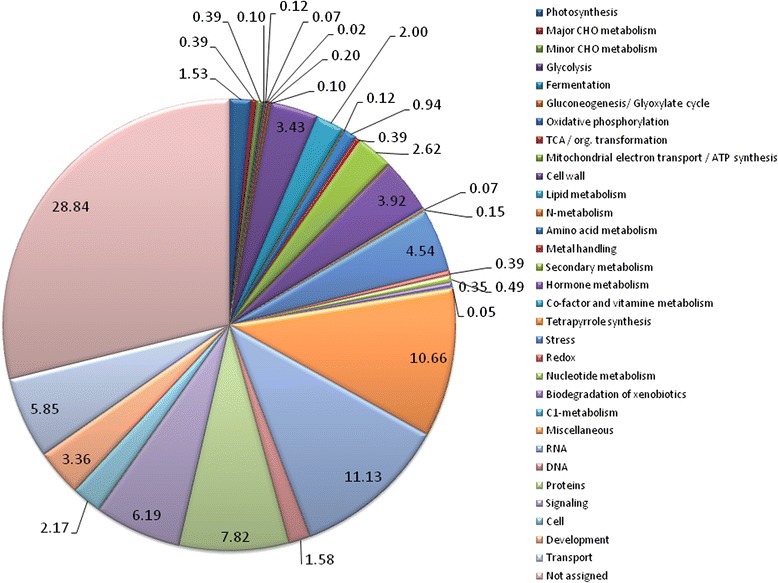
Functional classification of differentially expressed genes in OE root line vs. Kan root line ( q -value < 0.05) among all treatments as illustrated by MapMan categories.
Hormone metabolism- and fatty acid metabolism-related transcripts associated with mj-far-1 overexpression
Analysis of the ‘hormone metabolism’ category across all time points revealed differential expression of genes related to ET, auxin, methyl jasmonate and SA pathways in OE compared with Kan roots (as shown for 2 DAI in Additional file file 1: Table A1). A detailed analysis demonstrated a remarkable decrease in the expression of ET-related transcripts at 2 DAI in OE compared with Kan roots (Additional file 1: Table A1). These included transcripts encoding 2-oxoglutarate-dependent dioxygenase (Solyc09g089710.2.1), gibberellin 2-beta-dioxygenase (Solyc02g080120.1.1), and gibberellin 20-oxidase 4 (solyc01g093980.2.1), as well as aminocyclopropane-1-carboxylic acid synthase; expression of these transcripts decreased by 1072.16-, 13.03-, 693.7- and 1072.17-fold in OE vs. Kan roots at 2 DAI. Similarly, downregulation of auxin-related genes, such as the gene encoding the PIN6 auxin:hydrogen symporter/transporter (Solyc06g059730.1.1) by 2.83-fold and auxin-responsive family protein (Solyc12g017880.1.1) by 5.26-fold, was observed, together with additional auxin-related transcripts that demonstrated strong downregulation in expression at 2 DAI (Additional file 1: Table A1). With regard to SA-related transcripts, the 6.34-fold increase in expression of salicylic acid carboxyl methyltransferase (Solyc09g091550.2.1) in OE compared with Kan roots at 2 DAI might contribute to the observed root susceptibility [32] (Additional file 1: Table A1).
Given that FAR is implicated in fatty acid metabolism, we next focused on fatty acid-related signaling in OE vs. Kan roots before and throughout the time course of inoculation. Transcript analysis by MapMan showed that the gene encoding the repressor jasmonate ZIM-domain protein 1 (JAZ1) (Solyc12g009220.1.1) was downregulated in noninoculated OE roots. JAZ1 is a nucleus-localized protein belonging to the larger family of TIFY proteins [32] that act as repressors of JA signaling [33,34]. (Similarly, upregulation of the gene encoding allene oxide synthase (AOS) (Solyc01g109150.2.1) was observed in noninoculated OE roots; this protein might induce the JA pathway before inoculation, providing an advantage for RKN infection. At 2 DAI downregulation of a 9-LOX member, the LOXB transcript (Solyc01g099180.2.1; similar to Arabidopsis LOX1), by 360-fold and AOS (Solyc04g079730.1.1; similar to Arabidopsis AOS) by 4.43-fold was observed, indicating that changes in lipid metabolism are an early response to nematode inoculation. At 5 DAI additional downregulation of 9-LOX (Solyc09g075870.1.1; similar to Arabidopsis LOX5) in OE roots compared with Kan roots was observed; although these mentioned isoforms are not known to be involved in JA biosynthesis, their product might be active in local and systemic defense mechanisms against pathogens [35,36] (Table 4). At 15 DAI significant upregulation of 9-LOX (Solyc08g014000.2.1; highly similar to Arabidopsis LOX1) and AOS (Solyc10g0079601.1) transcripts was observed in inoculated OE roots compared with the noninoculated control (Table 4). Differential expression of JA- and fatty acid-related transcripts solely as a consequence of nematode infection was studied by analyzing the Kan control roots. Inoculated Kan roots at 5 and 15 DAI showed downregulation of the 9-LOX gene family as LOXB (Solyc01g099200.2.1 and Solyc01g099180.2.1; highly similar to Arabidopsis LOX1), and of the 13-LOX family (Solyc05g014790.2.1; similar to Arabidopsis LOX6) and as LOXD (Solyc03g122340.2.1; highly similar to Arabidopsis LOX3) (Table 4). Similarly, the genes encoding AOS (Solyc11g069800.1.1 and Solyc04g079730.1.1) and 12-oxophytodienoate reductase 3 (Solyc07g007870.2.1) were downregulated. Upregulation of genes encoding hydroperoxide lyase (HPL) (Solyc07g049690.2.1; similar to Arabidopsis HPL1), 9-LOX transcripts similar to Arabidopsis LOX5 (Solyc09g075870.1.1 and Solyc09g075860.2.1), and LOX1 (Solyc08g014000.2.1 and Solyc01g099210.2.1), AOS (Solyc01g109150.2.1) and OPR2 (12-oxophytodienoate reductase 2) (Solyc01g103390.2.1) was observed. Similar to noninoculated OE samples, in Kan control roots at 5 and 15 DAI transcripts similar to those of the negative regulators of JA signaling JAZ1 (Solyc12g009220.1.1, Solyc07g042170.2.1, Solyc12g049400.1.1) and JAZ2 (Solyc03g122190.2.1) were downregulated as infection proceeded.
Table 4.
Dynamics of the gene-expression profile related to the fatty acid pathway
| Number | Function (ITAG) | Tomato (NCBI) | Arabidopsis thaliana (TAIR) | KAN1/ KAN3 | KAN1/ KAN4 | OE1/ KAN1 | OE2/ KAN2 | OE3/ KAN3 | OE4/ KAN4 |
|---|---|---|---|---|---|---|---|---|---|
| solyc05g014790.2.1 | Lipoxygenase | XM_004239145.1 lipoxygenase 6 | AT4G15440 - HPL1 (HYDROPEROXIDE LYASE 1) | 1.20 | 1.03 | - | - | - | - |
| solyc09g075870.1.1 | Lipoxygenase | XM_004247319.1 lipoxygenase 5 | AT3G22400 - LOX5 (LIPOXYGENASE 5) | -1.82 | -2.15 | - | - | -2.06 | - |
| solyc08g014000.2.1 | Lipoxygenase | NM_001247927 lipoxygenase (LOX1.1) | AT1G55020 - LOX1 (LIPOXYGENASE 1) | -1.32 | - | - | - | - | 5.48 |
| solyc01g099210.2.1 | Lipoxygenase | XM_004230159.1 lipoxygenase 5 | AT1G55020 - LOX1 (LIPOXYGENASE 1) | -1.75 | -2.71 | - | - | - | - |
| solyc01g099200.2.1 | Lipoxygenase | XM_004231226 lipoxygenase 5 | AT1G55020 - LOX1 | - | 2.34 | - | - | - | - |
| solyc01g099180.2.1 | Lipoxygenase | XM_004230158 lipoxygenase B | AT1G55020 - LOX1 | - | 4.20 | - | -360.05 | - | - |
| solyc09g075860.2.1 | Lipoxygenase | XR_183132 lipoxygenase 5 | AT3G22400 - LOX5 | - | -1.66 | - | - | - | - |
| solyc03g122340.2.1 | Lipoxygenase | XM_004235501 lipoxygenase (loxD) | AT1G17420 - LOX3 (LIPOXYGENASE 3) | - | 1.37 | - | - | - | - |
| solyc07g049690.2.1 | Cytochrome P450 | NM_001247491.1 fatty acid hydroperoxide lyase (HPL) | AT4G15440 - HPL1 | -1.18 | - | - | - | - | - |
| solyc01g109150.2.1 | Cytochrome P450 | NM_001247573.1 cytochrome P450 CYP74C4 | AT5G42650 - AOS (ALLENE OXIDE SYNTHASE) | -1.79 | -3.06 | 2.98 | - | - | - |
| solyc11g069800.1.1 | Cytochrome P450 | NM_001247904.1 allene oxide synthase (AOS) | AT5G42650 - AOS | - | 1.02 | - | - | - | - |
| solyc04g079730.1.1 | Cytochrome P450 | DQ174273.1 allene oxide syntase | AT5G42650 - AOS | - | 1.40 | - | -4.43 | 2.74 | - |
| solyc10g007960.1.1 | Allene oxide synthase | DQ174273.1 allene oxide syntase AJ278331 | AT5G42650 - AOS | - | - | - | - | - | 7.29 |
| solyc01g103390.2.1 | Flavin oxidoreductase/NADH oxidase | 12-oxophytodienoate reductase 2 (OPR2) | AT1G76690 - OPR2 (12-OXOPHYTODIENOATE REDUCTASE 2) | -1.04 | -1.72 | - | - | - | - |
| solyc11g032130.1.1 | NADPH dehydrogenase 3 | XM_004250605.1 12-oxophytodienoate reductase 1-like | AT1G76690 - OPR2 | - | - | - | - | 2.59 | - |
| solyc07g007870.2.1 | NADH flavin oxidoreductase/12-oxophytodienoate reductase | NM_001246944 12-oxophytodienoate reductase 3 (opr3) | AT2G06050 - OPR3 (OPDA-REDUCTASE 3) | - | 1.06 | - | - | - | - |
| solyc03g122190.2.1 | Jasmonate ZIM domain 2 | NM_001247294.1 salt responsive protein 1 (SRG1) | AT1G74950 - JAZ2, TIFY10B (JASMONATE-ZIM-DOMAIN 2) | 1.23 | 2.07 | - | - | - | - |
| solyc12g009220.1.1 | Jasmonate ZIM-domain protein 1 | NM_001247954 jasmonate ZIM-domain protein 1 | AT1G19180 - JAZ1 (JASMONATE-ZIM-DOMAIN PROTEIN 1) | 1.97 | 1.92 | -2.39 | - | 2.42 | - |
| solyc12g049400.1.1 | Protein TIFY 3B | XM_004252359 TIFY 10A-like | AT1G19180 - JAZ1 | 2.63 | 2.66 | - | - | - | - |
| solyc07g042170.2.1 | Jasmonate ZIM-domain protein 3 | XM_004243648.1 TIFY 10A-like | AT1G19180 - JAZ1 | - | 1.33 | - | - | - | - |
Given that the LOXD isoform is involved in JA biosynthesis, we next studied the dynamic expression of LOXD by means of a LOXD promoter–GUS construct. For these experiments, primers corresponding to the 5′ upstream sequences of LOXD (Solyc03g122340.2.1; similar to Arabidopsis LOX3) were designed with reference to the recently released genome (ITAG Release 2 [2010-11-28] official annotations on the SL2.31 genome built by ITAG). Promoter fragments were amplified by polymerase chain reaction (PCR) using tomato line 870 genomic DNA as a template, cloned upstream of the GUS reporter gene in the vector pUC19_Y [37], and subsequently cloned in the binary vector pCAMBIA2300 [38]. Transgenic hairy roots were generated in the background of tomato line 870 using the LOXD promoter–GUS construct transformed into Agrobacterium rhizogenes. Five positive transgenic hairy roots events were then infected with the avirulent M. javanica population. For the LOXD promoter–GUS line (pLOXD–GUS), LOXD expression was conspicuous, but restricted to the vascular cylinder, in noninoculated roots (Figure 5A,C,E). Following infection, weak signal corresponding to LOXD expression was observed within the vascular tissue at 2 and 5 DAI (Figure 5B,D), which corresponded well with the transcriptome results (Table 4). At 15 DAI, by which time galls had developed, extremely intense signal was detected within the gall, restricted to the vascular tissue associated with GCs (Figure 5F). A similar phenotype was observed for all pLOXD–GUS transformed root events.
Figure 5.
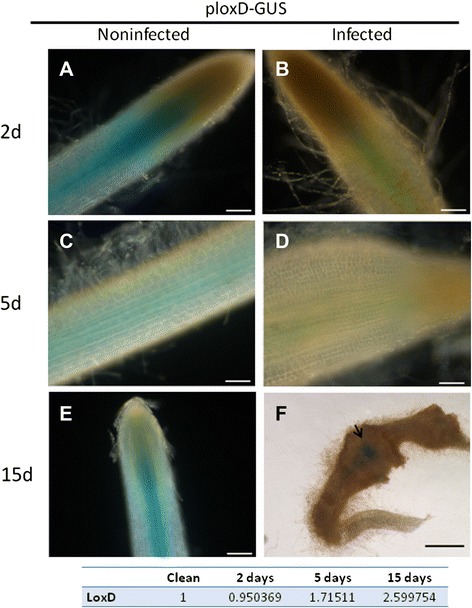
LOXD promoter (pLOXD)–GUS expression in transgenic tomato hairy root line infected with M. javanica second-stage juveniles (J2s). Noninfected control roots harboring the pLOXD–GUS fusion construct (A, C, E) show GUS staining of the root tip and vascular cylinder. Infected roots harboring pLOXD–GUS (B, D) show a decrease in GUS signal at 2 and 5 DAI. However, at 15 DAI (E), GUS signal is observed in the center of the developing gall in the giant cell area induced by the invading nematodes. (A–E) Light micrographs as viewed under a light microscope. (F) Bright-field image of galls photographed using a stereomicroscope. Bars: A–E = 100 μm, F = 1000 μm. Differential expression of LOXD in OE roots compared with Kan roots obtained in the RNA-Seq data is shown at the bottom of the table.
Differential expression of cell wall biosynthesis-, modification-, and remodeling-related genes associated with mj-far-1 overexpression
A distinct difference in the expression of genes showing strong or moderate association with cell wall-related activities was detected in OE vs. Kan roots. This pattern was demonstrated by the high representation of genes belonging to the different cell wall subcategories, as illustrated in Figure 6, whereby transcripts belonging to certain subcategories are overrepresented among the DEGs at a specific time point compared with their frequency in the tomato genome. Among these subcategories, a high representation of cell wall modification- and remodeling-related genes (e.g., those encoding pectin esterases) and expansin-encoding transcripts was observed (Figure 7, Additional file 1: Table A1 and Additional file 2: Table A2). The group of genes associated with cell wall-synthesis activity, such as the gene encoding cellulose synthase (Figure 7, Additional file 1: Table A1 and Additional file 2: Table A2), and cell wall degradation-related genes, such as genes encoding pectate lyases and polygalacturonases, were also overrepresented (Figure 7, Additional file 1: Table A1 and Additional file 2: Table A2). As noted already, at the early time points most genes belonging to these subcategories showed downregulation of the corresponding transcripts in OE roots compared with Kan roots. However, several transcripts belonging to the different subcategories showed remarkable upregulation, in particular at later time points (Figure 7).
Figure 6.
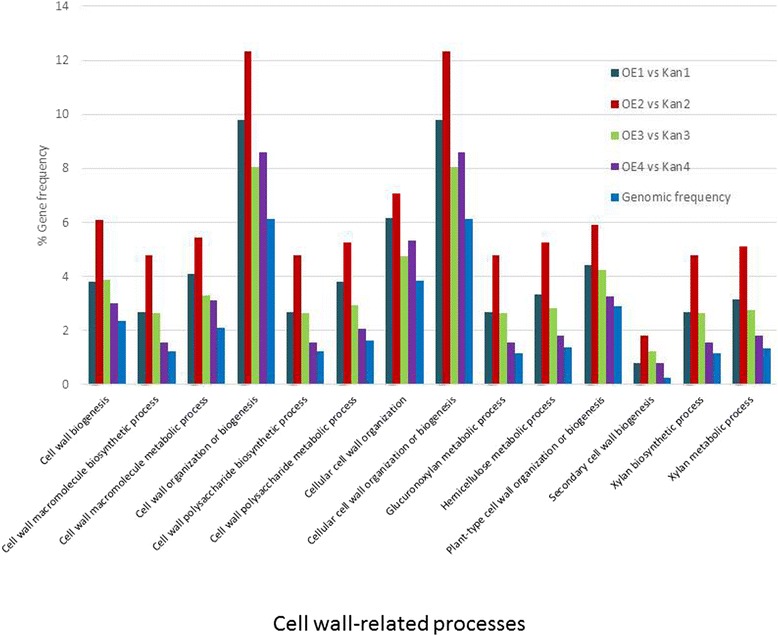
Frequency distribution of reads putatively associated with cell wall processes identified among differentially expressed genes (DEGs) from the root transcriptomes of all noninfected and infected samples. All Kan roots are root samples of vector 11.5 carrying the kanamycin-resistance gene (Kan1, Kan2, Kan3, and Kan4 refer to Kan control roots that were noninoculated and at 2, 5, and 15 DAI, respectively) and OE roots are mj-far-1.1 lines overexpressing mj-far-1 (OE1, OE2, OE3, and OE4 refer to OE roots that were noninoculated and at 2, 5, and 15 DAI, respectively). Each category of cell wall-related genes is indicated on the x-axis and the percentage of genes in each category relative to the ITAG2.3 reference tomato genome and the DEGs is indicated on the y-axis.
Figure 7.
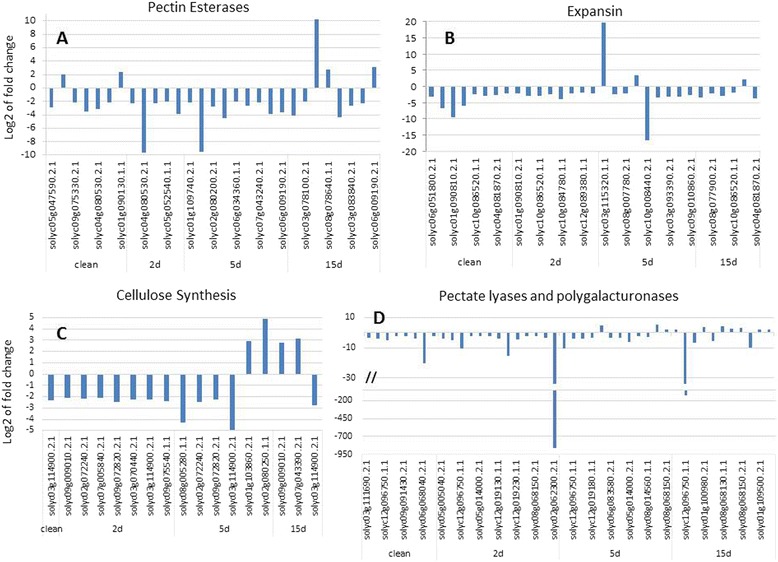
Regulation of cell wall protein related transcripts. Fold change in expression of (A) pectin esterase-related genes, (B) expansin-related genes, (C) cellulose synthesis-related genes, (D) pectate lyase and polygalacturonase-related genes in OE vs. Kan root lines that were noninoculated and at 2, 5, and 15 DAI.
To further validate the spatial and temporal expression patterns of cell wall-related genes, we used the gene encoding CWP (Solyc09g097770.2.1) in the aforementioned promoter–GUS construct assay. In noninoculated roots of the CWP promoter–GUS hairy root line (pCWP–GUS), no CWP was detected at any of the tested time points (Figure 8A,C,E). Interestingly, after inoculation, the lateral roots adjacent to the galls showed strong signal, which indicated induction of CWP by nematode infection (Figure 8F). A similar phenotype was observed for all pCWP–GUS transformed root events.
Figure 8.
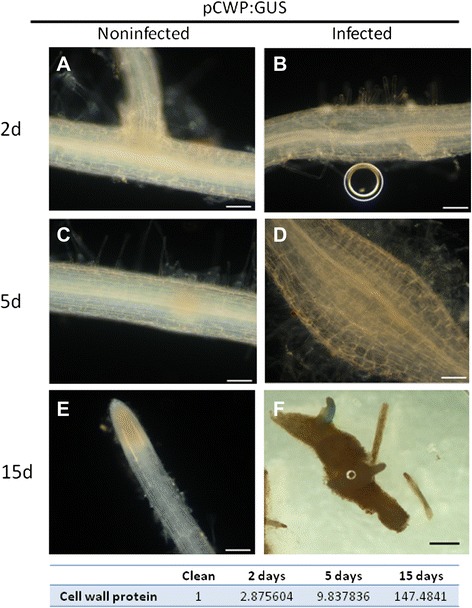
Cell wall protein (CWP) promoter (pCWP)–GUS expression in transgenic tomato hairy root line infected with M. javanica second-stage juveniles (J2s). Noninfected control (A, C) and infected (B, D) roots harboring the pCWP–GUS fusion construct show no GUS staining of the root or lateral root at 2 and 5 DAI. At 15 DAI, no signal is observed in noninfected roots (A, C, E). Although infected roots at 2 and 5 DAI (B, D) showed no signal, GUS signal is observed in lateral roots associated with galls induced by the invading nematodes at 15 DAI (F). (A–E) Light micrographs as viewed under a light microscope. (F) Bright-field image of galls photographed using a stereomicroscope. Bars: A–E = 100 μm, F = 1000 μm. Differential expression of CWP in OE roots compared with Kan roots obtained in the RNA-Seq data is shown at the bottom of the table.
Transcriptome changes in the phenylpropanoid and phenylalanine pathways associated with mj-far-1 overexpression
All 3970 DEGs were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [39] for pathway-enrichment analysis relative to the available whole transcriptome annotation for tomato (ITAG2.3), using the hypergeometric test. Enrichment of the different pathways at each time point is summarized in Figure 9. Pathway-enrichment analysis revealed the predominance of both phenylalanine and phenylpropanoid pathways in roots overexpressing mj-far-1 compared with Kan roots at all tested time points (Figure 9). This pattern was demonstrated by a high representation of genes belonging to the secondary metabolism subcategories (Figure 10), whereby transcripts belonging to certain subcategories were overrepresented in the DEGs at specific time points compared with their frequency in the tomato genome.
Figure 9.
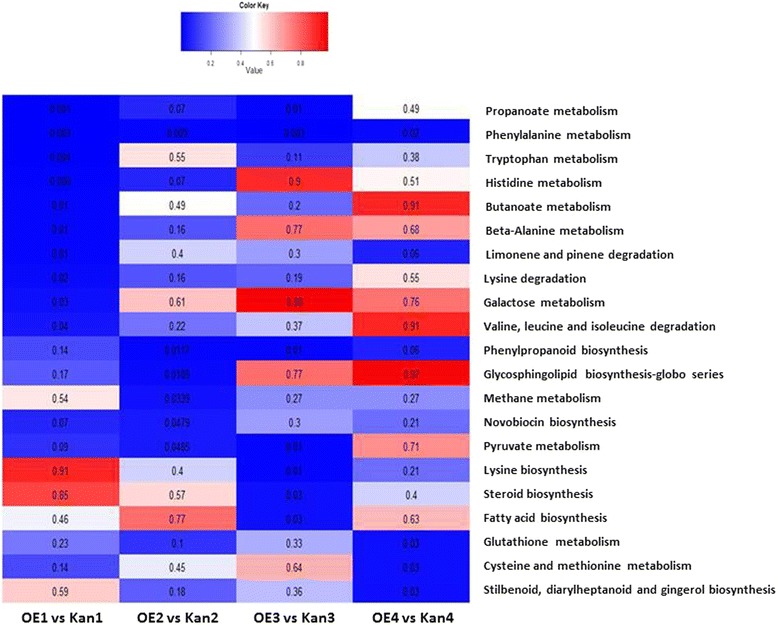
Enriched KEGG pathway at different time points. All 3970 differentially expressed genes were assigned to a pathway according to the KEGG database. All Kan roots are root samples of vector 11.5 carrying the kanamycin-resistance gene (Kan1, Kan2, Kan3, and Kan4 refer to Kan control roots that were noninoculated and at 2, 5, and 15 DAI, respectively) and OE roots are mj-far-1.1 lines overexpressing mj-far-1 (OE1, OE2, OE3, and OE4 refer to OE roots that were noninoculated and at 2, 5, and 15 DAI, respectively). KEGG pathway enrichment at each time point was calculated using the hypergeometric test. The P-values are presented on the heat map: the color gradient from dark blue to red represents strongly and significantly enriched pathways to nonsignificantly enriched pathways, respectively.
Figure 10.
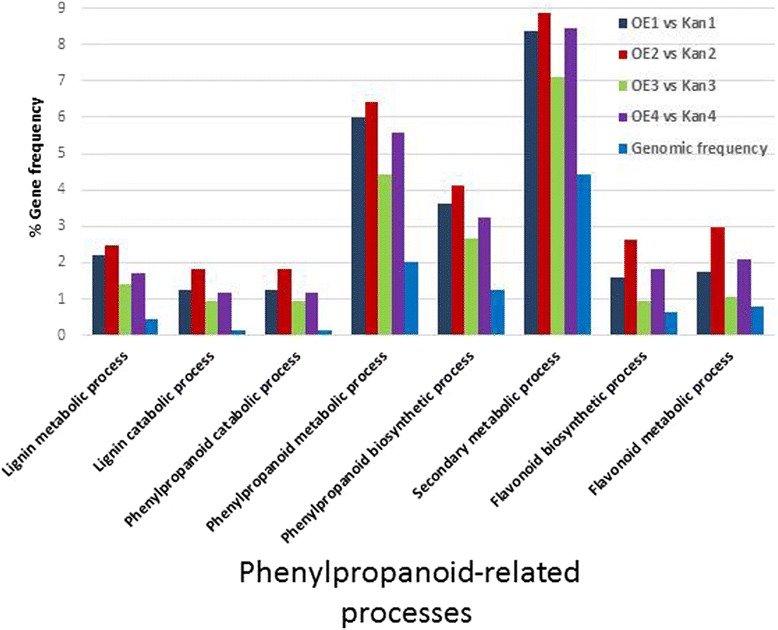
Frequency distribution of reads putatively associated with phenylpropanoid processes identified among differentially expressed genes (DEGs) from root transcriptomes of all noninfected and infected samples. All Kan roots are root samples of vector 11.5 carrying the kanamycin-resistance gene (Kan1, Kan2, Kan3, and Kan4 refer to Kan control roots that were noninoculated and at 2, 5, and 15 DAI, respectively) and OE roots are mj-far-1.1 lines overexpressing mj-far-1 (OE1, OE2, OE3, and OE4 refer to OE roots that were noninoculated and at 2, 5, and 15 DAI, respectively). Each category of secondary metabolite-related genes is indicated on the x-axis and the percentage of genes from each category relative to the ITAG2.3 reference tomato genome and to the DEGs is indicated on the y-axis.
The phenylpropanoid pathway leads to the synthesis of coumarins, flavonoids, phytoalexins, lignins, and lignans, all of which can contribute to plant defense. In roots overexpressing mj-far-1, decreased levels of a transcript similar to phenylalanine ammonia-lyase (PAL) 2 (Solyc03g042560.1.1) and a transcript similar to 4-coumarate-CoA ligase (4CL3) (Solyc03g097030.2.1) were observed before nematode inoculation. The latter protein has a pivotal role in the biosynthesis of plant secondary compounds at the divergence point from general phenylpropanoid metabolism to several major branch pathways [40]. At 15 DAI, upregulation of two transcripts similar to PAL1 (Solyc00g282510.1.1 and Solyc10g011930.1.1) and a transcript similar to PAL4 (Solyc03g036480.1.1) by 3.3-, 4.18- and 2.82-fold, respectively, was observed. Transcripts similar to cinnamyl alcohol dehydrogenase 9 (CAD9) (Solyc08g014360.1.1 and Solyc02g069250.2.1), a key enzyme in lignin biosynthesis [41], were strongly upregulated (553.4- and 2.19-fold, respectively) in roots overexpressing mj-far-1 at 2 DAI. Similarly, expression of the gene encoding CHS (Solyc05g053550.2.1), a key enzyme in flavonoids biosynthesis, was differentially regulated in all inoculated OE vs. Kan roots (Table 3), showing increased transcript levels at 2 and 5 DAI followed by a decreased transcript level at 15 DAI.
Quantitative reverse-transcription (qRT)-PCR validation of RNA-Seq data
To confirm the expression profiles obtained from the RNA-Seq data, qRT-PCR analysis was carried out for 22 genes selected from among the 61 and 52 common DEGs in OE roots compared with Kan roots for overall noninoculated and inoculated samples, and for inoculated-only samples, respectively (Tables 2 and 3). Quantitative RT-PCR was performed using RNA isolated from infected and noninfected root samples of both root lines from the same batch used for preparation of the whole root transcriptome (Additional file 3: Table A3). The genes were selected to represent both up- and downregulated genes with log2 changes ranging from 3.11-fold upregulation to 12.08-fold downregulation in the transcriptome analysis. Of the 22 genes tested, 19 (86.3%) showed differential expression in the direction observed in the transcriptome profiling (Additional file 3: Table A3). For example, we confirmed the constant downregulation of the genes encoding long-chain fatty acid-CoA ligase (Solyc08g008310.2.1) and indole-3-acetic acid-amido synthetase (Solyc02g092820.2.1), together with constant upregulation of the genes encoding nodulin family protein (Solyc05g055540.1.1) and chitinase (Solyc07g009510.1.1). Only two probe sets that were shown to be downregulated in the RNA-Seq analysis, namely esterase/lipase/thioesterase (Solyc05g018770.1.1) and auxin-responsive protein (Solyc08g021820.2.1), were slightly upregulated in the qRT-PCR analysis at 5 and 15 DAI. Similarly, the gene encoding the fatty acid elongase 3-ketoacyl-CoA synthase (Solyc03g005320.2.1), which was shown to be upregulated in the RNA-Seq analysis, was downregulated in the qRT-PCR analysis at 5 and 15 DAI (Additional file 3: Table A3). Thus, overall, the qRT-PCR and RNA-Seq results were in agreement.
Discussion
Following deposition of effectors by the nematode through the stylet [5-9] or other organs in contact with the external environment, such as the amphids or cuticle [4,12,13], the question of how these proteins ultimately perform their function in the host cells remains to be answered. This is a complex aspect of plant–nematode interactions that are being dissected by several research groups [42]. A range of functions are attributed to nematode effectors, apart from reprogramming cell metabolism for the generation and maintenance of the nematodes’ feeding sites. A relatively large subset of effectors deals with the suppression of defense responses triggered by parasitism [42,43]. Data to date indicate that, as with other pathogens, active suppression of host defense responses is a critical component of successful parasitism by nematodes [42].
The involvement of nematode effectors in promoting host susceptibility to nematode infection has been reported for the cyst nematode Heterodera effectors CBP, 10A06, 4FO1, and 30CO2, and for the RKN Meloidogyne incognita effector CRT, whose overexpression in Arabidopsis increases susceptibility to nematodes [27,28,42-45]. Similarly, studies of the FAR protein from M. javanica (Mj-FAR-1) have shown that tomato hairy roots overexpressing mj-far-1 are remarkably more susceptible to RKNs [26]. The observed increase in host susceptibility conferred by successful pathogens could be the result of their overcoming pattern-triggered immunity, via suppression of the pattern-triggered immunity response by secreted effectors, leading to effector-triggered susceptibility (ETS) [42]. Even though the function of nematode infection in the manipulation of plant defense has been extensively studied, it is clear that nematodes target different levels of the plant’s immune system.
In this study, we aimed to elucidate the contribution of the protein Mj-FAR-1, which is secreted to suppress plant defense responses and to promote ETS, by exploring the broad transcriptional events underlying the increased susceptibility observed in roots overexpressing mj-far-1 relative to control roots. Using the RNA-Seq approach, we observed that mj-far-1 overexpression accounts for the differential expression of 3970 transcripts before and after inoculation. Of these transcripts, 2069 were upregulated and 2205 were downregulated in the OE vs. Kan roots for all inoculated and noninoculated samples. This finding is in agreement with previous results in which nematode infection induced not only upregulation of the transcription of specific enzymes, but also downregulation of transcriptional, translational, and catalytic events [14]. Collectively, these results suggest that the nematode might manipulate multiple pathways to suppress the host defense response. Pearson correlation tests indicated that samples of noninoculated roots and roots in the first phase of parasitism, i.e., prior to feeding (2 DAI) when the nematode is initiating attempts to establish itself in the host, clustered predominantly according to the infection step. However, at 5 and 15 DAI, during feeding-site formation and execution of the compatibility response, mj-far-1 overexpression had a greater impact on defining the root transcriptome of OE and Kan lines (Figure 3A). These results support our transcriptomic data, as global changes occurring in response to nematode infection are predicted to be similar in both lines, but the number and response level of modulated genes should provide a global overview of the changes that are attributable to Mj-FAR-1.
Regulation of hormone signaling-related genes by mj-far-1
Examination of genes encoding host biochemical pathways that have been implicated in the response to RKNs identified several hormone pathways that might be subject to mj-far-1 manipulation. The observed downregulation of JAZ1 together with upregulation of AOS in noninoculated roots expressing mj-far-1 might induce the JA pathway and support nematode invasion and establishment in the first stages of infection. Furthermore, the observed upregulation of Ethylene Response Factor 1 might indicate that JA levels are increased because this gene is known to be activated by both JA and ET [46,47]. Coincident with the suggested upregulation of the JA pathway in noninoculated OE roots, a reduction in PAL levels was indicated. These observations might support an antagonistic interaction between SA and JA in OE roots [48-50]. Moreover, upregulation of salicylic acid carboxyl methyltransferase (solyc09g091550.2.1) at 2 DAI might indicate a decrease in SA accumulation. This suggestion is supported by previous findings in which overexpression of salicylic acid carboxyl methyltransferase reduces SA-mediated pathogen resistance in Arabidopsis thaliana [51]. Evidence for the possible role of JA in promoting nematode development has been reported by Bhattarai et al. [52], who analyzed tomato mutants altered in JA signaling and concluded that an intact JA-signaling pathway is required for tomato susceptibility to RKNs. Similarly, in maize, Mu-insertional lox3-4 mutants displayed increased attractiveness to RKNs, and an increased number of juveniles and eggs were accompanied by elevated levels of JA [53]. More recently, in Arabidopsis, the 13-LOX member, lox4-1 mutant, characterized by increased levels of JA, demonstrated increased susceptibility to RKNs [54]. Overall, these results support our findings of increased JA signal promoting root susceptibility to RKNs. Following inoculation, several transcripts similar to Arabidopsis 9- and 13-LOX were differentially regulated in roots overexpressing mj-far-1 relative to control roots. LOXs are widely present in higher plants; they are important enzymes in the biosynthesis of oxylipins and in the plant response to wounding and pathogen attack [55]. Although only the 13-LOX pathway has been implicated in JA biosynthesis, other studies have suggested that there may be another as-yet-unknown pathway leading to LOX-mediated defense responses [56]. The observed fluctuation in the expression patterns of LOX-encoding transcripts suggests that nematode development requires the dynamic coordinated expression of LOX genes in the proper order for successful establishment in a susceptible root. The altered expression of LOX genes as a result of Mj-FAR-1 in OE compared with Kan roots is supported by results from an in vitro study indicating that LOX activity was inhibited by FAR-1 of the potato nematode G. pallida [11]. Thus, lipid-binding activity of Mj-FAR-1 toward free fatty acids which, among others, are LOX substrates might be involved in manipulating the plant defense response mediated by fatty acid signaling.
Downregulation of several auxin-related genes was observed in OE vs. Kan control roots, including those genes encoding indole-3-acetic acid-amido synthetase (GH3.8) and auxin-responsive GH3-like, which were downregulated in all OE root samples (Additional file 1: Table A1). These results are consistent with the hypothesis that, in general, global alterations of auxin balance accompany RKN infection [57]. Moreover, the finding that all transcripts similar to JAZ1 were downregulated at later time points after infection suggests that strict regulation of JAZ1 is an infection strategy that enables nematode development. Recent evidence indicating that jaz1 is not only a JA-responsive but also an auxin-responsive gene further illustrates the intimate molecular interplay between auxin and JA signaling [58]. Collectively, these results suggest that Mj-FAR-1 is involved in manipulating multiple pathways to coordinate feeding-site formation, protection from host defense responses, and maintenance of GCs.
Regulation of cell wall organization-related genes by mj-far-1
In the present study, strong representation of DEGs associated with cell wall-related activities was detected. The extensive modifications in cell wall architecture (i.e., thickening, ingrowth, disassembly, and dissolution) that occur in cyst nematode and RKN feeding cells are likely mediated by the activity of both cell wall-biosynthetic and cell wall-degrading enzymes. Interestingly, most of the cell wall biosynthesis-, organization-, and modification-related genes were downregulated (more than 3-fold) in OE vs. Kan roots, and particularly in noninoculated roots at 2 and 5 DAI. These data further confirm the hypothesis that cell wall biosynthesis, modification or fortification is essential to the plant’s response to nematode infection. Similar to other transcriptomic studies, RNA-Seq data indicated that the expression of many genes involved in cell wall extension and remodeling is altered following nematode inoculation [59-61]. For example, a group of genes involved in cellulose synthesis was downregulated at an early time point, whereas several transcripts were upregulated at 15 DAI in OE vs. Kan control roots (Figure 7C). Cellulose synthesis is expressed in the initial expansion phase of GC development. Concomitant hyperplasia of root cells surrounding the GCs to form the visible gall also likely requires synthesis of new (primary) cell wall [62-64]. It might be that upregulation of this group of genes at 5 and 15 DAI reflects the accelerated disease development observed on the OE root line. Similarly, there was impressive representation of expansin-encoding genes (Figure 7B). Expansins are encoded by a large multigene family. They are identified as wall-loosening factors and facilitators of cell expansion [64,65]. A previous study indicated that a decrease in tomato EXPA5 expression by means of RNAi-transgenic root generation reduces the nematode’s ability to complete its life cycle in transgenic roots [65]. Following GC formation, and possibly as a secondary response, division and expansion of cortical and pericycle cells around the GCs occur, causing the formation of galls. Similarly, specific transcripts among the group of genes encoding pectin esterases, pectate lyase and polygalacturonase were upregulated in OE vs. Kan roots at the later time points (Figure 7).
Cell expansion, cell elongation, cell wall biosynthesis, and cell wall dissolution are all physiological processes that have been observed indirectly within nematode-induced feeding cells [66,67]. The differential regulation of cell wall biosynthesis- and modification-related gene expression at the early time point might facilitate nematode establishment in root tissues.
Regulation of phenylpropanoid-related genes by mj-far-1
Notable differences in the expression of genes encoding enzymes in the phenylpropanoid pathway were observed in OE vs. Kan control roots at all time points (Figure 9). Many secondary metabolites derived from multiple branches of the phenylpropanoid pathway, including lignins, isoflavonoid-phytoalexins, other phenolic compounds, and SA, are instrumental in the plant’s ability to mount a successful defense against invading pathogens [68]. A remarkable decrease in the expression of genes encoding enzymes at the initiation of the phenylpropanoid pathway, e.g., genes encoding PAL and 4CL3, was observed in noninoculated OE vs. Kan control roots. PAL (EC 4.3.1.34) can be considered a control point for entry into the phenylpropanoid pathway [69], whereas 4CL3 has a pivotal role in the biosynthesis of plant secondary compounds at the divergence point from general phenylpropanoid metabolism to several major branch pathways [38]. At 5 and 15 DAI, four genes encoding different PAL isoforms increased in expression, thereby suggesting increased metabolic flow into the phenylpropanoid pathway. Increased PAL enzyme activity has been noted in resistant tomato roots infected with RKNs, whereas PAL activity is depressed in susceptible tomato roots [70]. Similarly, in potato, PAL activity is higher in resistant plants [71]. It may be hypothesized that the decrease in PAL expression before inoculation facilitates nematode infection, whereas the high level of PAL at 5 and 15 DAI reflects acceleration of the infection progress promoted in the OE root. Two transcripts encoding CAD9 showed increased expression at 2 DAI; CAD9 is a key enzyme in lignin biosynthesis as it catalyzes the final step in the synthesis of monolignols. Its expression may be the result of increased penetration and accelerated disease progression in the OE line (Solyc02g069250.2.1 and Solyc08g014360.1.1, with 2.19- and 553.4-fold increases in expression, respectively). An additional important gene is CHS, which was upregulated at 2 and 5 DAI and downregulated at 15 DAI. CHS is involved in glyceollin synthesis, which is known to inhibit oxygen uptake by Meloidogyne [72]. The decrease in CHS expression at 15 DAI might support nematode infection. Invasion of roots with RKNs and cyst nematodes induces the flavonoid pathway in infection structures [57,73], and flavonoids are hypothesized to act as regulators of auxin transport and accumulation during gall formation [57,58]. In flavonoid-deficient Medicago truncatula plants, gall formation still occurred, although galls were smaller and showed fewer cell divisions [74]. In flavonoid-deficient Arabidopsis and tobacco mutants, reproduction of several species of nematodes was not affected [73,75]. However, flavonoids did affect nematode behavior; for example, certain flavonoids acted as repellents for specific nematode species and inhibited their motility and hatching at millimolar concentrations [76]. Although synthesis of flavonoid or isoflavonoid phytoalexins, deposition of lignin or cell wall-bound phenolics, and synthesis of other defense chemicals via the phenylpropanoid pathway are often characteristic of both the localized hypersensitive response and systemic acquired resistance [77], we suggest that in OE roots, decreased abundance of transcripts associated with synthesis and regulation of defense chemicals derived from the phenylpropanoid pathway might facilitate nematode infection.
Conclusions
The present study provides evidence for the potential mediation by Mj-FAR-1 of a complex defense-related response, including differential regulation of cell wall-, hormone- and fatty acid-related genes, as well as changes in the phenylpropanoid pathway. Our results indicate that roots overexpressing mj-far-1 still mount a defense response against nematode infection; however, this rapid response might reflect the accelerated disease progress in OE roots upon nematode infection. While the direct effects of mj-far-1 might be related only to fatty acid metabolism, the indirect effect mediated by lipid signaling may drive other pathways that affect plant responses to nematodes. This study adds to our understanding of the role of mj-far-1 and may ultimately indicate novel pathways that are required for nematode establishment and parasitism.
Methods
Plant materials and growth conditions
Tomato ‘Avigail’ )870) was used as the background line for both transgenic root lines: mj-far-1 OE and the control Kan, as described previously [26]. Both root lines were subcultured on standard-strength Gamborg’s B5 salt medium (Duchefa, Haarlem, The Netherlands), supplemented with 2% (w/v) sucrose and solidified with 0.8% (w/v) Gelrite agar (Duchefa). Roots were subcultured on B5 medium, with one root section per petri dish (Miniplast, Ein Shemer, Israel), and incubated horizontally in a growth chamber at 26°C in the dark for 1 week to allow root branching before nematode inoculation.
Nematode culture and infection assays
Meloidogyne javanica was propagated on greenhouse-grown tomato ‘Avigail’ (870) plants. Nematode egg masses were extracted from roots with 0.05% (v/v) sodium hypochlorite followed by sucrose flotation [78]. For sterilization, eggs were placed on a cellulose–acetate filter membrane (Sartorius Stedim Biotech GmbH, Goettingen, Germany, pore size 5 μm) in a sterile Whatman® filter holder (Whatman International Ltd., Dassel, Germany). Eggs on the filter were exposed for 10 min to 0.01% (w/v) mercuric chloride (Sigma-Aldrich, St Louis, MO, USA), followed by 0.7% (v/v) streptomycin solution (Sigma-Aldrich), and three washing steps with 50 ml sterilized distilled water [79]. The sterilized eggs were collected from the membrane and placed on 25-μm-pore sieves in 0.01 M 2-morpholinoethanesulfonic acid buffer (Sigma-Aldrich) under aseptic dark conditions for 3 days, allowing J2s to hatch. Freshly hatched preparasitic J2s were collected in a 50 ml falcon tube. For nematode infection, 1-week-old transgenic tomato root lines, growing on standard-strength Gamborg’s B5 salt medium, were inoculated with 200 sterile freshly hatched M. javanica preparasitic J2s. Plates were left uncovered in a laminar flow hood until water had completely soaked into the medium [80]. The inoculated and noninoculated roots were incubated horizontally in the dark, and root samples were taken for either RNA extraction or GUS bioassay at the designated time points after inoculation.
cDNA library preparation and high-throughput sequencing
Total RNA was extracted using TRI reagent (Sigma-Aldrich) from Kan and OE tomato root lines at different time points postinoculation. Beads containing oligo (dT) were used to isolate poly(A) mRNA from 500 μg total RNA for each sample. Purified mRNA was then fragmented in fragmentation buffer. Using these short fragments as templates, random hexamer primers were used to synthesize the first-strand cDNA. The second-strand cDNA was synthesized using buffer, dNTPs, RNase H and DNA polymerase I. Short double-stranded cDNA fragments were purified with the QIAquick PCR Purification Kit (QIAGEN Inc., Valencia, CA) and eluted with elution buffer (EB) for end repair and the addition of an ‘A’ base. The short fragments were ligated to Illumina sequencing adaptors. DNA fragments of a selected size were gel-purified and amplified by PCR. The amplified library was sequenced on an Illumina HiSeq™ 2000 platform. The details of the experiment were as follows: expected library size, 200 bp; read length, 90 nucleotides; sequencing strategy, paired-end sequencing. The library size and read length are provided in the Additional file 1: Table A1.
Read alignment to the reference tomato genome
In total, 212,975,340 2 × 100 bp reads were sequenced. Read mapping to the ITAG Solanum lycopersicum protein reference version 2.3 (ITAG2.3; http://solgenomics.net) was performed with SoapAligner/SOAP2 [29]. An average of 20.1 million reads from each library paired-end sequencing were uniquely aligned to the reference sample, and overall made up ca. 75.6% of the total reads (Table 1) used in the bioinformatics analysis. Gene-expression level was normalized using the RPKM (reads per kilobase transcriptome per million mapped reads) method [81].
Differences in gene expression between two samples were calculated based on Poisson distribution for gene expression. FDR was calculated using the Benjamini and Yekutieli (2001) FDR method [82]. We used FDR ≤ 0.001 and the absolute value of log2 ratio ≥ 1 as the thresholds to judge the significance of the differences in gene expression. All sequences were uploaded to the NCBI SRA database under accession no. SRX504894.
Differences in gene expression were visualized using MapMan [31,83]. The MapMan mapping file was obtained from http://www.gomapman.org/; 27,212 of the 29,549 genes on the microarray were present in the mapping file. Enrichments of functional categories of the MapMan annotation in the significantly DEGs were tested for significance by applying Fisher’s test with Bonferroni correction for multiple tests using Mefisto Version 0.23beta (http://www.usadellab.org). Enrichment of Gene Ontology (GO) terms in significantly DEGs was evaluated using the agriGO GO analysis toolkit (http://bioinfo.cau.edu.cn/agriGO) [84] with Fisher’s test and Bonferroni multiple testing correction (P < 0.05). Pathway analysis was done using the KEGG database [38] and enrichment was calculated using the hypergeometric test followed by the FDR test. PCA analysis was performed using the FactoMineR package [29].
Real-time qPCR analysis
For qPCR experiments, contaminant genomic DNA was removed from the RNA with the Turbo DNA-free Kit from Ambion (Applied Biosystems, Foster City, CA, USA). DNA-free RNA (1 μg) was converted into first-strand cDNA using the Verso™ cDNA Synthesis kit (ABgene, Epsom, UK), and reactions were performed using the ABsolute SYBR Green ROX mix (ABgene). Primers for qRT-PCR experiments were designed with Primer Express software (Applied Biosystems; see Additional file 4: Table A4). The real-time PCR contained 3.4 μl cDNA in a total volume of 10 μl, consisting of 1× SYBR-Green Amplification Kit (ABgene), 150 nM forward primer and 150 nM reverse primer, and was run in real-time PCR plasticware (Axygen, Union City, CA, USA). All PCR cycles began with 2 min at 50°C, then 10 min at 95°C, followed by 40 cycles of 10 s at 95°C and 1 min at 60°C. After the PCR, a melting curve was generated by gradually increasing the temperature to 95°C to test for amplicon specificity. For qPCR, a mixture of all cDNAs was used for all treatments, as a template for calibration curves designed for each pair of primers. Each reaction was performed in triplicate and the results represent the mean of two independent biological experiments. Three constitutively expressed genes, namely actin (ACT; GenBank accession no. U60482.1), β-tubulin (TUB; GenBank accession no. NM_001247878.1) and 18S (GenBank accession no. BH012957.1), were used as endogenous controls for gene expression analysis (Additional file 4: Table A4). Transcript levels were normalized for each sample with the geometric mean of the corresponding selected housekeeping genes. All of the housekeeping genes were confirmed to display minimal variation across the treatment and were the most stable housekeeping genes from a set of tested genes in a given cDNA sample. Values were expressed as the increase or decrease in level relative to a calibration sample. The following control reactions were included: PCR negative control without cDNA template to confirm the absence of nonspecific PCR products (NTC), and a second reaction containing mRNA that had not been subjected to reverse transcription (NRT control). To confirm the expression profiles obtained from the RNA-Seq expression data, RT-qPCR analysis was carried out for 22 genes selected on the basis of their biological significance: genes involved in fatty acid metabolism, such as long-chain fatty acid-CoA ligase and fatty acid elongase, cell wall-related transcripts such as chitinase, expansin-1 and CWP, and hormone-related transcripts such as auxin-responsive protein and indole-3-acetic acid-amido synthetase.
Plasmid construction and generation of transgenic tomato roots
All PCR amplifications used for plasmid construction were performed using the Dream Taq Green Master Mix (Thermo Fisher Scientific, Pittsburgh, PA, USA) in accordance with the manufacturer’s instructions and using tomato genomic DNA as the template. To clone the different promoter sequences (pLOXD and pCWP), specific primers designed to amplify a 2000 bp fragment and to create the SacI and SmaI restriction sites at the 5′ and 3′ ends of the promoter, respectively, were used (Additional file 5: Table A5). The SmaI restriction site was placed before the ATG sequence of the respective genes to guarantee the correct reading frame when the promoter was fused to the β-glucuronidase (GUS) gene. The 2000 bp fragment was then cloned into the pUC19_Y vector [37] at the SacI and SmaI restriction sites. The 4010 bp cassette containing the specific gene promoter and the GUS reporter gene was then isolated by restriction digestion with SacI and SalI and subsequently cloned into the pCAMBIA2300 binary vector [38]. The identity, orientation, and junctions of the resulting pCAM-LOXD and pCAM-CWP constructs were confirmed by digestion patterns and sequencing. Five different events of transformed roots with pCAM-LOXD or pCAM-CWP were subjected to the GUS assay, with 10 specimens sampled for each root line. A supplementary plasmid pME-524, expressing GUS under the control of the 35S promoter, was used as a positive control. The pCAMBIA2300 empty-vector control and the construct plasmids were subsequently used for A. rhizogenes-mediated root transformation.
Agrobacterium-mediated root transformation and production of hairy root cultures
The binary vector pCAM-LOXD, pCAM-CWP and the empty-vector control pCAMBIA2300 were electrotransformed into A. rhizogenes ATCC 15834 [85]. Individual cotyledons were excised from 8–10-day-old tomato seedlings and immersed in an A. rhizogenes suspension (OD600 1.0) for 15 min. The excised cotyledons were blot-dried on sterile filter paper, then co-cultivated on standard-strength Gamborg’s B5 salt medium for 3 days. Explants were then washed with liquid B5 medium supplemented with the antibiotics kanamycin (50 μg ml−1) (Duchefa Biochemie) and Timentin (300 μg ml−1; ticarcillin disodium:potassium clavulanate, 15:1) (Duchefa Biochemie) and incubated at room temperature for 1 h with mild shaking. The explants were blot-dried on sterile filter paper and placed on B5 agar medium supplemented with the same antibiotics. Within 7 to 10 days of incubation at 25°C in the light, roots emerged on the surface of the cotyledons. Hairy roots were transferred to Gamborg’s B5 medium supplemented with 0.8% (w/v) Gelrite and kanamycin (50 μg ml−1).
Histochemical localization of GUS activity and microscopic analysis
One-week-old control and promoter-GUS tomato roots were inoculated as described above, and assayed histochemically for GUS activity at 2, 5 and 15 DAI. A set of noninfected plates served as the control group. For GUS assays, infected and noninfected transgenic root tissues were removed from the petri dishes at specific time points after inoculation and infiltrated with GUS-staining buffer containing 50 mM sodium phosphate (pH 7.0), 10 mM EDTA, 5 mM K4[Fe2(CN)6], 5 mM K3[Fe2(CN)6], 0.2% (v/v) Triton X-100 and 2 mM 5-bromo-4-chloro-3-indolyl ß-D-glucuronide (X-Gluc). GUS staining was performed for 12 h at 37°C. For observation and documentation, GUS-stained roots were mounted on microscope slides or in small wells, and photographed with either a Leica DMLB light microscope and a Nikon Eclipse 90i (Leica Microsystems GmbH, Wetzlar, Germany; Nikon Corporation, Tokyo, Japan), or a stereomicroscope (Leica MZFLIII, Leica Microsystems GmbH) equipped with a Nikon DS-Fi1 camera.
Availability of supporting data
All the referred supporting data are included as additional files.
Acknowledgments
We appreciate the financial support of a grant from the Israeli Chief Scientist’s Office, Ministry of Agriculture and Rural Development (no. 020-0687-02).
Additional files
Differentially expressed genes related to biotic stress as represented in the MapMan illustration.
Differentially expressed genes involved in cell wall metabolism.
Validation of RNA-Seq results by qRT-PCR.
Forward and reverse primer sequences used for qRT-PCR confirmation of expression levels of differentially expressed genes from the transcriptomic data, and primers sets for internal control genes used to normalize gene expression in the qRT-PCR analysis.
Primer pairs used for amplification of promoter fragments.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SBM and II designed the experiments and wrote the manuscript. II performed the research and data analyses. II provided all infected and noninfected materials for RNA-Seq and constructed all plasmids for promoter-GUS assays. NS performed all of the bioinformatics analyses. NS made critical comments on the manuscript and performed detailed analysis of the RNA-Seq data. All of the authors have read and approved the final manuscript.
Authors’ information
The author Sigal Brown Miyara is a Nematologist researcher in the Entomology Department and the Units of Nematology and Chemistry. Ionit Iberkleid is now doing her Postdoctoral studies at San–Diego State University.
Contributor Information
Ionit Iberkleid, Email: sho132@gmail.com.
Noa Sela, Email: noa@volcani.agri.gov.il.
Sigal Brown Miyara, Email: sigalhor@volcani.agri.gov.il.
References
- 1.Bird DM. Signaling between nematodes and plants. Curr Opin Plant Biol. 2004;7(4):372–376. doi: 10.1016/j.pbi.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Dubreuil G, Magliano M, Deleury E, Abad P, Rosso MN. Transcriptome analysis of root-knot nematode functions induced in the early stages of parasitism. New Phytol. 2007;176(2):426–436. doi: 10.1111/j.1469-8137.2007.02181.x. [DOI] [PubMed] [Google Scholar]
- 3.Gheysen G, Fenoll C. Gene expression in nematode feeding sites. Annu Rev Phytopathol. 2002;40:191–219. doi: 10.1146/annurev.phyto.40.121201.093719. [DOI] [PubMed] [Google Scholar]
- 4.Jones JT, Reavy B, Smant G, Prior AE. Glutathione peroxidases of the potato cyst nematode Globodera rostochiensis. Gene. 2004;324:47–54. doi: 10.1016/j.gene.2003.09.051. [DOI] [PubMed] [Google Scholar]
- 5.Baum TJ, Hussey RS, Davis EL. Root-knot and cyst nematode parasitism genes: the molecular basis of plant parasitism. Genet Eng (N Y) 2007;28:17–43. doi: 10.1007/978-0-387-34504-8_2. [DOI] [PubMed] [Google Scholar]
- 6.Davis EL, Hussey RS, Baum TJ, Bakker J, Schots A, Rosso MN, et al. Nematode parasitism genes. Annu Rev Phytopathol. 2000;38:365–396. doi: 10.1146/annurev.phyto.38.1.365. [DOI] [PubMed] [Google Scholar]
- 7.Niebel A, De Almeida EJ, Tire C, Engler G, Van Montagu M, Gheysen G. Induction patterns of an extensin gene in tobacco upon nematode infection. Plant Cell. 1993;5(12):1697–1710. doi: 10.1105/tpc.5.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sijmons PC. Plant-nematode interactions. Plant Mol Biol. 1993;23(5):917–931. doi: 10.1007/BF00021809. [DOI] [PubMed] [Google Scholar]
- 9.Vanholme B, De Meutter J, Tytgat T, Van Montagu M, Coomans A, Gheysen G. Secretions of plant-parasitic nematodes: a molecular update. Gene. 2004;332:13–27. doi: 10.1016/j.gene.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Jones D, Candido EP. The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev Biol. 2000;226(1):152–165. doi: 10.1006/dbio.2000.9847. [DOI] [PubMed] [Google Scholar]
- 11.Prior A, Jones JT, Blok VC, Beauchamp J, McDermott L, Cooper A, et al. A surface-associated retinol- and fatty acid-binding protein (Gp-FAR-1) from the potato cyst nematode Globodera pallida: lipid binding activities, structural analysis and expression pattern. Biochem J. 2001;356(Pt 2):387–394. doi: 10.1042/0264-6021:3560387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson L, Robertson WM, Sobczak M, Helder J, Tetaud E, Ariyanayagam MR, et al. Cloning, expression and functional characterisation of a peroxiredoxin from the potato cyst nematode Globodera rostochiensis. Mol Biochem Parasitol. 2000;111(1):41–49. doi: 10.1016/S0166-6851(00)00295-4. [DOI] [PubMed] [Google Scholar]
- 13.Spiegel Y, Kahane I, Cohen L, Sharon E. Meloidogyne javanica surface proteins: characterization and lability. Parasitology. 1997;115(Pt 5):513–519. doi: 10.1017/S0031182097001637. [DOI] [PubMed] [Google Scholar]
- 14.Jammes F, Lecomte P, de Almeida-Engler J, Bitton F, Martin-Magniette ML, Renou JP, et al. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 2005;44(3):447–458. doi: 10.1111/j.1365-313X.2005.02532.x. [DOI] [PubMed] [Google Scholar]
- 15.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5(4):325–331. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 17.Basavaraju SV, Zhan B, Kennedy MW, Liu Y, Hawdon J, Hotez PJ. Ac-FAR-1, a 20 kDa fatty acid- and retinol-binding protein secreted by adult Ancylostoma caninum hookworms: gene transcription pattern, ligand binding properties and structural characterisation. Mol Biochem Parasitol. 2003;126(1):63–71. doi: 10.1016/S0166-6851(02)00253-0. [DOI] [PubMed] [Google Scholar]
- 18.Bath JL, Robinson M, Kennedy MW, Agbasi C, Linz L, Maetzold E, et al. Identification of a secreted fatty acid and Retinol-Binding Protein (Hp-FAR-1) from Heligmosomoides polygyrus. J Nematol. 2009;41(3):228–233. [PMC free article] [PubMed] [Google Scholar]
- 19.Fairfax KC, Vermeire JJ, Harrison LM, Bungiro RD, Grant W, Husain SZ, et al. Characterization of a fatty acid and retinol binding protein orthologue from the hookworm Ancylostoma ceylanicum. Int J Parasitol. 2009;39(14):1561–1571. doi: 10.1016/j.ijpara.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garofalo A, Kennedy MW, Bradley JE. The FAR proteins of parasitic nematodes: their possible involvement in the pathogenesis of infection and the use of Caenorhabditis elegans as a model system to evaluate their function. Med Microbiol Immunol. 2003;192(1):47–52. doi: 10.1007/s00430-002-0158-6. [DOI] [PubMed] [Google Scholar]
- 21.Garofalo A, Klager SL, Rowlinson MC, Nirmalan N, Klion A, Allen JE, et al. The FAR proteins of filarial nematodes: secretion, glycosylation and lipid binding characteristics. Mol Biochem Parasitol. 2002;122(2):161–170. doi: 10.1016/S0166-6851(02)00097-X. [DOI] [PubMed] [Google Scholar]
- 22.Garofalo A, Rowlinson MC, Amambua NA, Hughes JM, Kelly SM, Price NC, et al. The FAR protein family of the nematode Caenorhabditis elegans. Differential lipid binding properties, structural characteristics, and developmental regulation. J Biol Chem. 2003;278(10):8065–8074. doi: 10.1074/jbc.M206278200. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy MW, Garside LH, Goodrick LE, McDermott L, Brass A, Price NC, et al. The Ov20 protein of the parasitic nematode Onchocerca volvulus. A structurally novel class of small helix-rich retinol-binding proteins. J Biol Chem. 1997;272(47):29442–29448. doi: 10.1074/jbc.272.47.29442. [DOI] [PubMed] [Google Scholar]
- 24.Jordanova R, Groves MR, Kostova E, Woltersdorf C, Liebau E, Tucker PA. Fatty acid- and retinoid-binding proteins have distinct binding pockets for the two types of cargo. J Biol Chem. 2009;284(51):35818–35826. doi: 10.1074/jbc.M109.022731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michalski ML, Monsey JD, Cistola DP, Weil GJ. An embryo-associated fatty acid-binding protein in the filarial nematode Brugia malayi. Mol Biochem Parasitol. 2002;124(1–2):1–10. doi: 10.1016/S0166-6851(02)00081-6. [DOI] [PubMed] [Google Scholar]
- 26.Iberkleid I, Vieira P, de Almeida EJ, Firester K, Spiegel Y, Horowitz SB. Fatty acid-and retinol-binding protein, Mj-FAR-1 induces tomato host susceptibility to root-knot nematodes. PLoS One. 2013;8(5):e64586. doi: 10.1371/journal.pone.0064586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewezi T, Howe P, Maier TR, Hussey RS, Mitchum MG, Davis EL, et al. Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. Plant Cell. 2008;20(11):3080–3093. doi: 10.1105/tpc.108.063065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel N, Hamamouch N, Li C, Hewezi T, Hussey RS, Baum TJ, et al. A nematode effector protein similar to annexins in host plants. J Exp Bot. 2010;61(1):235–248. doi: 10.1093/jxb/erp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Le S, Josse J, Husson F. FactoMineR: An R package for multivariate analysis. JSS J Stat Softw. 2008;25(1):1–18. [Google Scholar]
- 31.Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, et al. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol. 2005;138(3):1195–1204. doi: 10.1104/pp.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. The tify family previously known as ZIM. Trends Plant Sci. 2007;12:239–244. doi: 10.1016/j.tplants.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 33.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu GH, et al. JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. J Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 34.Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 35.Vicente J, Cascon T, Vicedo B, Garcia-Agustin P, Hamberg M, Castresana C. Role of 9-lipoxygenase and alpha-dioxygenase oxylipin pathways as modulators of local and systemic defense. Mol Plant. 2012;5(4):914–928. doi: 10.1093/mp/ssr105. [DOI] [PubMed] [Google Scholar]
- 36.Yan L, Zhai Q, Wei J, Li S, Wang B, Huang T, et al. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 2013;9(12):e1003964. doi: 10.1371/journal.pgen.1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vancanneyt G, Schmidt R, O’Connor-Sanchez A, Willmitzer L, Rocha-Sosa M. Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet. 1990;220(2):245–250. doi: 10.1007/BF00260489. [DOI] [PubMed] [Google Scholar]
- 38.Remy S, Thiry E, Coemans B, Windelinckx S, Swennen R, Sagi L. Improved T-DNA vector for tagging plant promoters via high-throughput luciferase screening. Biotechniques. 2005;38(5):763–770. doi: 10.2144/05385RR01. [DOI] [PubMed] [Google Scholar]
- 39.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamberger B, Hahlbrock K. The 4-coumarate: CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci U S A. 2004;101(7):2209–2214. doi: 10.1073/pnas.0307307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barakat A, Bagniewska-Zadworna A, Choi A, Plakkat U, DiLoreto DS, Yellanki P, et al. The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biol. 2009;9:26. doi: 10.1186/1471-2229-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hewezi T, Baum TJ. Manipulation of plant cells by cyst and root-knot nematode effectors. Mol Plant Microbe Interact. 2013;26(1):9–16. doi: 10.1094/MPMI-05-12-0106-FI. [DOI] [PubMed] [Google Scholar]
- 43.Hamamouch N, Li C, Hewezi T, Baum TJ, Mitchum MG, Hussey RS, et al. The interaction of the novel 30C02 cyst nematode effector protein with a plant beta-1,3-endoglucanase may suppress host defence to promote parasitism. J Exp Bot. 2012;63(10):3683–3695. doi: 10.1093/jxb/ers058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaouannet M, Magliano M, Arguel MJ, Gourgues M, Evangelisti E, Abad P, et al. The root-knot nematode calreticulin Mi-CRT is a key effector in plant defense suppression. Mol Plant Microbe Interact. 2013;26(1):97–105. doi: 10.1094/MPMI-05-12-0130-R. [DOI] [PubMed] [Google Scholar]
- 45.Hewezi T, Howe PJ, Maier TR, Hussey RS, Mitchum MG, Davis EL, et al. Arabidopsis spermidine synthase is targeted by an effector protein of the cyst nematode Heterodera schachtii. Plant Physiol. 2010;152:968–984. doi: 10.1104/pp.109.150557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004;16(7):1938–1950. doi: 10.1105/tpc.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15(1):165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- 49.Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, Van Loon LC, et al. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 2008;147(3):1358–1368. doi: 10.1104/pp.108.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 51.Koo YJ, Kim MA, Kim EH, Song JT, Jung C, Moon JK, et al. Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol Biol. 2007;64(1–2):1–15. doi: 10.1007/s11103-006-9123-x. [DOI] [PubMed] [Google Scholar]
- 52.Bhattarai KK, Xie QG, Mantelin S, Bishnoi U, Girke T, Navarre DA, et al. Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Mol Plant Microbe Interact. 2008;21(9):1205–1214. doi: 10.1094/MPMI-21-9-1205. [DOI] [PubMed] [Google Scholar]
- 53.Gao X, Starr J, Gobel C, Engelberth J, Feussner I, Tumlinson J, et al. Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol Plant-Microbe Interact. 2008;21(1):98–109. doi: 10.1094/MPMI-21-1-0098. [DOI] [PubMed] [Google Scholar]
- 54.Ozalvo R, Cabrera J, Escobar C, Christensen SA, Borrego EJ, Kolomiets MV, et al. Two closely related members of Arabidopsis 13-lipoxygenases (13-LOXs), LOX3 and LOX4, reveal distinct functions in response to plant-parasitic nematode infection. Mol Plant Pathol. 2014;15:319–332. doi: 10.1111/mpp.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gobel C, Feussner I, Schmidt A, Scheel D, Sanchez-Serrano J, Hamberg M, et al. Oxylipin profiling reveals the preferential stimulation of the 9-lipoxygenase pathway in elicitor-treated potato cells. J Biol Chem. 2001;276(9):6267–6273. doi: 10.1074/jbc.M008606200. [DOI] [PubMed] [Google Scholar]
- 56.Reddy PS, Kumar TC, Reddy MN, Sarada C, Reddanna P. Differential formation of octadecadienoicacid and octadecatrienoic acid products in control and injured/infected potato tubers. Biochim Biophys Acta. 2000;1483:294–300. doi: 10.1016/S1388-1981(99)00191-2. [DOI] [PubMed] [Google Scholar]
- 57.Hutangura P, Mathesius U, Jones MGK, Rolfe BG. Auxin induction is a trigger for root gall formation caused by root-knot nematodes in white clover and is associated with the activation of the flavonoid pathway. Aust J Plant Physiol. 1999;26(3):221–231. doi: 10.1071/PP98157. [DOI] [Google Scholar]
- 58.Grunewald W, Cannoot B, Friml J, Gheysen G. Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog. 2009;5(1):e1000266. doi: 10.1371/journal.ppat.1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ibrahim HM, Hosseini P, Alkharouf NW, Hussein EH, Gamal El-Din Ael K, Aly MA, et al. Analysis of gene expression in soybean (Glycine max) roots in response to the root knot nematode Meloidogyne incognita using microarrays and KEGG pathways. BMC Genomics. 2011;12:220. doi: 10.1186/1471-2164-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ithal N, Recknor J, Nettleton D, Maier T, Baum TJ, Mitchum MG. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol Plant Microbe Interact. 2007;20(5):510–525. doi: 10.1094/MPMI-20-5-0510. [DOI] [PubMed] [Google Scholar]
- 61.Klink VP, Hosseini P, Matsye P, Alkharouf NW, Matthews BF. A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode) Plant Mol Biol. 2009;71(6):525–567. doi: 10.1007/s11103-009-9539-1. [DOI] [PubMed] [Google Scholar]
- 62.Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279(5351):717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 63.Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, et al. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000;12(12):2409–2424. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall extension in plants. Plant Cell. 1992;4(11):1425–1433. doi: 10.1105/tpc.4.11.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gal TZ, Aussenberg ER, Burdman S, Kapulnik Y, Koltai H. Expression of a plant expansin is involved in the establishment of root knot nematode parasitism in tomato. Planta. 2006;224(1):155–162. doi: 10.1007/s00425-005-0204-x. [DOI] [PubMed] [Google Scholar]
- 66.Endo BY. Histology and ultrastructural modification induced by cyst nematodes. In: Taylor LaCE, editor. Cyst nematodes. New York: Plenum Press; 1986. pp. 133–146. [Google Scholar]
- 67.Jones MGK. Host cell responses to endoparasitic nematode attacks: Structure and function of giant cells and syncytia. Ann Appl Biol. 1981;97:353–372. doi: 10.1111/j.1744-7348.1981.tb05122.x. [DOI] [Google Scholar]
- 68.Zabala G, Zou J, Tuteja J, Gonzalez DO, Clough SJ, Vodkin LO. Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol. 2006;6:26. doi: 10.1186/1471-2229-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matthews BF, Ibrahim HMM, Klink VP. Changes in the expression of genes in soybean roots infected by Nematodes. In: Krezhova D, ed. Soybean - Genetics and Novel Techniques for Yield Enhancement. 2011. ISBN: 978-953-307-721-5, InTech, Available from: http://www.intechopen.com/books/soybean-genetics-and-novel-techniques-for-yield-enhancement/changes-inthe-expression-of-genes-in-soybean-roots-infected-by-nematodes.
- 70.Bruske CH. Phenylalanine ammonia lyase activity in tomato roots infected and resistant to the root-knot nematode Meloidogyne incognita. Physiol Pl Path. 1980;16:409–414. doi: 10.1016/S0048-4059(80)80012-9. [DOI] [Google Scholar]
- 71.Giebel J. Phenylalanine and tyrosine ammonia-lyase activities in potato roots and their significance in potato resistance to Heterodera rostochiensis. Nematologica. 1973;19(1):1–6. doi: 10.1163/187529273X00024. [DOI] [Google Scholar]
- 72.Kaplan DT, Keen NT, Thomason IJ. Studies on the mode of action of glyceollin in soybean incompatibility to the root-knot nematode, Melidogyne incognita. Physiol Pl Path. 1980;16:319–325. doi: 10.1016/S0048-4059(80)80003-8. [DOI] [Google Scholar]
- 73.Jones JT, Furlanetto C, Phillips MS. The role of flavonoids produced in response to cyst nematode infection of Arabidopsis thaliana. Nematology. 2007;9:671–677. doi: 10.1163/156854107782024875. [DOI] [Google Scholar]
- 74.Wasson AP, Pellerone FI, Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wuyts N, Lognay G, Swennen R, De Waele D. Nematode infection and reproduction in transgenic and mutant Arabidopsis and tobacco with an altered phenylpropanoid metabolism. J Exp Bot. 2006;57(11):2825–2835. doi: 10.1093/jxb/erl044. [DOI] [PubMed] [Google Scholar]
- 76.Wuyts N, Swennen R, De Waele D. Effects of plant phenylpropanoid pathway products and selected terpenoids and alkaloids on the behaviour of the plant-parasitic nematodes Radopholus similis, Pratylenchus penetrans and Meloidogyne incognita. Nematology. 2006;8:89–101. doi: 10.1163/156854106776179953. [DOI] [Google Scholar]
- 77.Edens RM, Anand SC, Bolla RI. Enzymes of the Phenylpropanoid Pathway in Soybean Infected with Meloidogyne incognita or Heterodera glycines. J Nematol. 1995;27(3):292–303. [PMC free article] [PubMed] [Google Scholar]
- 78.Hussey RS, Baker KR. Comparison of methods of collecting inocula for Meloidogyne spp., including a new technique. Plant Dis Rep. 1973;57:1025–1028. [Google Scholar]
- 79.Jansen van Vuuren R, Woodward B. The response of cassava cultivars to root-knot nematode infestation: an in vitro method. Euphyrica. 2001;120(1):109–113. doi: 10.1023/A:1017524210671. [DOI] [Google Scholar]
- 80.Sijmons PC, Grundler FMW, Von Mende N, Burrows PR, Wyss U. Arabidopsis thaliana as a new model host for plant parasitic nematodes. Plant J. 1991;1:245–254. doi: 10.1111/j.1365-313X.1991.00245.x. [DOI] [Google Scholar]
- 81.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 82.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):919–1188. [Google Scholar]
- 83.Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37(6):914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 84.Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen WJ, Forde BG. Efflicient transformation of Agrobactenum spp. by high voltage electroporation. Nucleic Acid Res. 1989;17(20):8385. doi: 10.1093/nar/17.20.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]


