Genomic rearrangements of the cytokine receptor-like factor 2 (CRLF2) gene,1,2 which is part of the thymic stromal lymphopoietin receptor (TSLPR), result in overexpression of CRLF2 itself leading to JAK2-mediated activation of STAT5, which regulates cell proliferation, survival, and apoptosis.3,4,11–13 In this regard, childhood B-cell precursor acute lymphoblastic leukemias (BCP-ALLs), bearing a rearranged CRLF2, display a high rate of relapse.5–10 Furthermore, CRLF2 genomic rearrangements are strictly associated with its surface overexpression, rendering this marker suitable for detection by flow cytometry (FCM).14
To determine CRLF2 expression in childhood BCP-ALLs, we first assessed TSLPR surface expression. For this purpose, we carried out, at diagnosis, standard multiparametric FCM (Dworzak et al., manuscript in preparation) on 421 consecutive diagnostic bone marrow (BM) samples from BCP-ALL children (256 males and 165 females), enrolled in six centers of the AIEOP-BFM-ALL-2009 trial between December 2010 and June 2013. Our gating strategy used to measure TSLPR surface expression (Online Supplementary Figure S1) allowed us to distinguish three blast subpopulations according to the intensity of TSLPR staining: the first one was defined as negative (i.e. positivity <10%), the second one was moderately positive (i.e. positivity ≥ 10% to <50%), and the third one was strongly positive (i.e. positivity ≥ 50%). We found 383 (91.2%) negative samples, 8 (1.9%) moderately positive, and 29 (6.9%) strongly positive. Inter-center distribution of patients’ subgroups is shown in Table 1. Representative examples are reported in Figure 1A–C.
Table 1.
TSLPR reactivity in BCP-ALL blasts at diagnosis analyzed in six different centers.
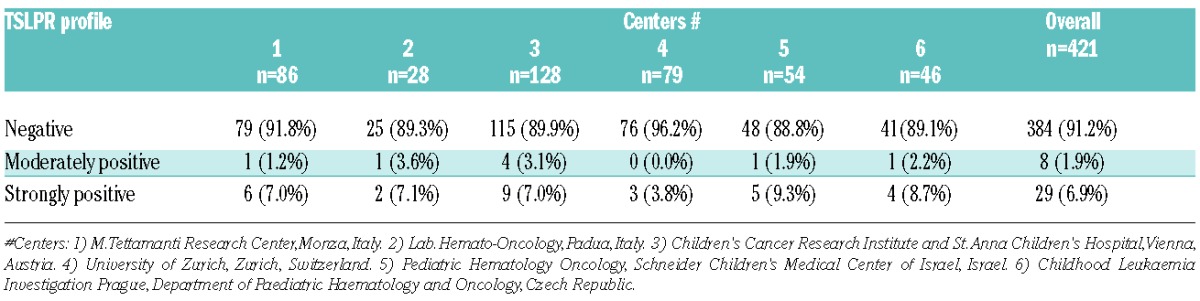
Figure 1.

Different patterns of TSLPR expression in representative BCP-ALL cases: strongly positive (positivity ≥50%, (A), moderately positive (positivity ≥10% – <50%), (B), and negative (positivity <10%), (C). Fine tuning of TSLPR negative cases revealed three possible patterns of TSLPR positivity below the threshold of 10%: TSLPR fully negative (D); moderately positive (E); and partially positive (F). The blue histograms represent the blast cells, the red ones represent the normal residual lymphocytes. Mean fluorescence intensity (MFI) of lymphocytes vs. blasts were measured in each representative case: (A): 174.0 vs. 3.899; (B): 149.0 vs. 333; (C) and panel (D) (same representative patient): 93.0 vs. 97.0; (E): 88.0 vs. 175.0; (F): 48.0 vs. 1001.0.
We then studied the immunophenotypic profile of TSLPR among the 86 patients enrolled in Center 1 during initial screening. Fine tuning of fluorescence distribution of 79 of 86 patients that had been previously found negative for TSLPR (i.e. positivity <10%) (Table 1) allowed us to further distinguish three different expression patterns: 1) TSLPR-stained blasts overlapping with control fluorescence (n. 72, mean positivity 0.52%±0.52%, range 0.0%–2.2%); 2) a second population of TSLPR-stained blasts clearly shifted to the right (n=5, mean % positivity 2.72%±0.16%, range 2.5%–2.9%), which was identical to the TSLPR moderate pattern we observed previously in the diagnostic screening apart from TSLPR positivity being less than 10%; 3) a third pattern showing two clearly distinct blast populations: a larger one, TSLPR fully negative, and a smaller one positive, shifted to the right (n=2, positivity was 1% and 3.5%, respectively). Hereinafter, we will refer to these three patterns as fully negative, moderately positive (<10%), and partially positive, respectively. Representative examples are shown in Figure 1D–F. Interestingly, one TSLPR moderately positive (<10%), and 2 TSLPR partially positive patients (UPNs 016, 013, and 039, respectively) showed low levels of P2RY8-CRLF2 expression (F.C. < 0.50), suggesting the presence of a minor CRLF2 sub clone (Online Supplementary Table S2).
Next, CRLF2 transcripts levels, CRLF2 aberrations (P2RY8-CRLF2, IGH@-CRLF2, CRLF2 F232C), and JAK2 and IL7R mutations were analyzed in 86 of our BCP-ALL samples collected in Center 1 as described previously.9 We detected CRLF2 overexpression in 9.3% of BCP-ALL patients. Seventy-nine of these patients (91.8%) were negative for surface TSLPR expression as assessed by both FCM (<10%) and RQ-PCR (<20 FC), while only 7 (8.1%) were concordantly positive. Intriguingly, one patient (UPN 084) showed overexpression of CRLF2 (FC 33.2), whereas TSLPR expression level was undetectable. However, this patient did not display P2RY8-CRLF2 gene fusion. Two of the 7 patients with CRLF2 overexpression (UPN 30 and UPN 62), as assessed by both techniques, were negative for P2RY8-CRLF2 fusion and IGH@-CRLF2 translocation. Conversely, 5 non-over-expressed cases showed barely detectable levels of P2RY8-CRLF2 gene fusion. Thus, while our results seem to indicate a lack of correlation between genomic rearrangement and CRLF2 overexpression, as assessed by PCR, they clearly show that CRLF2-over-expressing BCP-ALLs are characterized by a strong positivity for TSLPR when analyzed by FCM.
To determine a functional read out of CRLF2 genomic rearrangements, MUTZ5 cells (IGH@-CRLF2; JAK2 R683G), MHH-CALL4 cells (IGH@-CRLF2; JAK2 I682F), or primary thawed cells were subject to phospho flow cytometric assay (Online Supplementary Appendix). Likewise, a total of 41 cryopreserved BCP-ALL samples obtained according to their availability in cell banks [28 were obtained from the consecutive series of Center 1 (total 86) and 15 from a local cell bank] and viability after thawing (cut off ≥80%) were subject to phospho flow assay. Twenty-four BCP-ALL samples were TSLPR fully negative, 5 moderately positive (all of them <10%), and 12 strongly positive.
Next, we sought to determine basal and TSLP-induced pSTAT5 expression in CD45 intermediate/CD10+/CD7− blasts. The mean level of basal pSTAT5 detected in the three subgroups fully negative, moderately positive, and strongly positive for TSLPR was 0.71%±1.03% (range 0.0%–4.0%), 2.64%±3.64% (range 0.2%–9.0%), and 11.30%±18.31% (range 0.0%–65.6%). Statistical differences were calculated by one-way ANOVA analysis of variance (P=0.0200). As expected, we observed much higher phosphorylation of STAT5 in the TSLPR strongly positive samples than the fully negative ones, with a mean of pSTAT5+ cells of 60.79%±12.79% (range 37.0%–83.6%) and 2.95%±3.26% (range 0.2%–11.0%), respectively (P<0.001 by Bonferroni test) (Figure 2A). Furthermore, CRLF2-rearranged MUTZ5 and MHH-CALL4 cells showed aberrant TSLP-induced pSTAT5 compared with CRLF2 wild-type REH cells (data not shown). Interestingly, the group of 5 patients who were TSLPR moderately positive (<10%) showed enhanced pSTAT5 response with a mean of 22.36%±.63% (range 16.0%–34.3%), significantly higher than TSLPR fully negative patients (P<0.001 by Bonferroni test) (Figure 2A).
Figure 2.
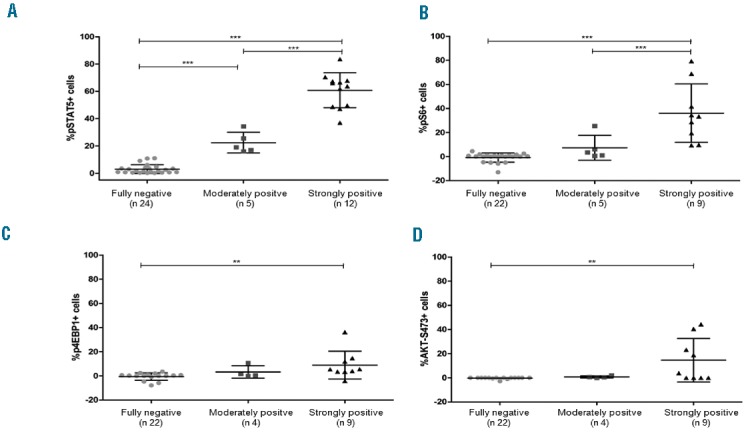
TSLP-induced phosphoprotein responses in BCP-ALL patients according to TSLPR expression (fully negative, moderately positive, or strongly positive). Distribution of positive cells is represented as whiskers plot of 5th and 95th percentile with means and standard deviations. Statistical significance among groups was determined by one-way ANOVA analysis of variance followed by post hoc Bonferroni’s multiple comparison test (*P<0.05, **P<0.01, ***P<0.001). (A) Shows pSTAT5 response (n 41); (B), (C), and (D) show TSLP-induced pS6, p4EBP1 and pAKT expression (n 36, 35, and 35, respectively). All groups were compared, but only those with statistical significance are indicated by stars and horizontal bars. Data were normalized to the basal phosphorylation level of each phosphoprotein.
We also studied TSLP-induced signaling through the PI3K/AKT/mTOR pathway (S6, 4EBP1 and AKT) in 36 out of 41 BCP-ALL patients [9 TSLPR strongly positive, 5 moderately positive (i.e. <10%), and 22 fully negative]. TSLP stimulation led to a significant increase in phosphorylation levels of S6, 4EBP1, and AKT in TSLPR strongly positive samples as compared to both the fully negative and moderately positive cases (one-way ANOVA P<0.0001, P=0.0119, and P=0.0065, respectively), in good agreement with Tasian et al.14 Differences between groups are detailed in Figure 2B–D. Contrary to reports by Tasian et al., in our samples, we observed no significant difference in basal phosphorylation of S6, 4EBP1, and AKT-S473 that could be ascribed to differences in TSLPR expression levels.
Strikingly, neither TSLPR fully negative nor TSLPR moderately positive cases showed mutations in JAK2, CRLF2, or IL7RA. However, the observation of enhanced level of basal pSTAT5 in TSLPR moderately positive as compared to the fully negative patients may indicate the presence of a CRLF2 rearranged sub clone below the level of detection in this latter subgroup of patients. In favor of this hypothesis, TSLPR strongly positive patients displayed an heterogeneous mutational profile: 10 of 12 carried P2RY8-CRLF2 rearrangement, one of these also carrying a mutation in JAK insertion L681-I682 insEA and another one carrying the IL7RA mutation S185C; 1 of 12 displayed IGH@-CRLF2 translocation and JAK2 point mutation R683G; 1 of 12 was wild type also for P2RY8-CRLF2 and IGH@-CRLF2 rearrangements. SNP at codon 244 (rs151218732) of CRLF2 as well as SNP at codon 244 (rs6897932) of IL7RA were randomly distributed independently of TSLPR over-expression. A summary of phenotypic, molecular and signaling features of the analyzed patients is provided in Online Supplementary Table S2.
To the best of our knowledge, this is the first report showing BCP-ALL patients moderately positive for TSLPR characterized by aberrant pSTAT5 and pS6 expression. We are currently investigating whether this signature refers to the presence of minor clones or is due to additional mechanisms driving aberrant JAK/STAT and PI3K/mTOR signal transduction.
In this regard, Tasian et al. has pointed to a potential diagnostic value of TSLP-mediated phosphosignaling in patients moderately positive for TSLPR staining (i.e. TSLPR-dim) as it would be a bona fide functional read out of the CRLF2 status. However, they did not provide any evidence of TSLPR-dim patients. In our study, we demonstrate the existence of CRLF2 moderately positive patients characterized by an activated phosphosignaling cascade. Thus, it is possible that Tasian et al. failed to identify TSLPR moderately positive patients because TSLPR expression was assessed after fixation and permeabilization, a procedure that is known to mask the presence of several surface antigens.
In summary, screening of TSLPR expression in BCP-ALL patients can be successfully achieved using standardized FCM protocols. FCM and PCR are highly concordant in detecting both CRLF2 over-expressed and non-over-expressed patients. However, patients characterized by a moderately or partially positive TSLPR expression associated with aberrant pSTAT5 and pS6 expression could only be detected by FCM analysis. Thus, our findings might prove useful in refining future diagnostic screening of ALL patients and help develop novel CRLF2 inhibitor-based therapies. In this regard, it is important to point out that approximately 50% of ALL patients with a Ph-like gene expression profile, which is associated with a poor outcome, have CRLF2 rearrangements.15
Acknowledgments
We would like to thank Dr. Chiara Buracchi for her technical support in preparing the manuscript.
Footnotes
Funding: this work was supported by Fondazione Tettamanti, Fondazione Benedetta è la Vita ONLUS, Comitato M.L. Verga, Fondazione Città della Speranza and Grant Ric. Corrente OBG 2006/02/R/001822, Associazione Italiana per la Ricerca sul Cancro (AIRC; to ABi and FL), Fondazione Cariplo (ABi), Ministero dell’Istruzione, Università e Ricerca (MIUR; ABi). We also thank the AIEOP centers for their support.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009; 41(11):1243–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell LJ, Capasso M, Vater I, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009; 114(13):2688–2698. [DOI] [PubMed] [Google Scholar]
- 3.Harvey RC, Mullighan CG, Chen IM, et al. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115(26):5312–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roll JD, Reuther GW. CRLF2 and JAK2 in B-progenitor acute lymphoblastic leukemia: a novel association in oncogenesis. Cancer Res. 2010;70(19):7347–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attarbaschi A, Morak M, Cario G, et al. Treatment outcome of CRLF2-rearranged childhood acute lymphoblastic leukaemia: a comparative analysis of the AIEOP-BFM and UK NCRI-CCLG study groups. Br J Haematol. 2012;158(6):772–777. [DOI] [PubMed] [Google Scholar]
- 6.Cario G, Zimmermann M, Romey R, et al. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010; 115(26):5393–5397. [DOI] [PubMed] [Google Scholar]
- 7.Chen IM, Harvey RC, Mullighan CG, et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2012;119(15):3512–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ensor HM, Schwab C, Russell LJ, et al. Demographic, clinical, and outcome features of children with acute lymphoblastic leukemia and CRLF2 deregulation: results from the MRC ALL97 clinical trial. Blood. 2011;117(7):2129–2136. [DOI] [PubMed] [Google Scholar]
- 9.Palmi C, Vendramini E, Silvestri D, et al. Poor prognosis for P2RY8-CRLF2 fusion but not for CRLF2 over-expression in children with intermediate risk B-cell precursor acute lymphoblastic leukemia. Leukemia. 2012;26(10):2245–2253. [DOI] [PubMed] [Google Scholar]
- 10.van der Veer A, Waanders E, Pieters R, et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood. 2013;122(15):2622–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isaksen DE, Baumann H, Trobridge PA, Farr AG, Levin SD, Ziegler SF. Requirement for stat5 in thymic stromal lymphopoietin-mediated signal transduction. J Immunol. 1999;163(11):5971–5977. [PubMed] [Google Scholar]
- 12.Levin SD, Koelling RM, Friend SL, et al. Thymic stromal lymphopoietin: a cytokine that promotes the development of IgM+ B cells in vitro and signals via a novel mechanism. J Immunol. 1999;162(2):677–683. [PubMed] [Google Scholar]
- 13.Rochman Y, Kashyap M, Robinson GW, et al. Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7-induced signaling. Proc Natl Acad Sci USA. 2010;107(45):19455–19460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasian SK, Doral MY, Borowitz MJ, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120(4):833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]


