Abstract
Mitochondrial genomes are generally thought to be under selection for compactness, due to their small size, consistent gene content, and a lack of introns or intergenic spacers. As more animal mitochondrial genomes are fully sequenced, rearrangements and partial duplications are being identified with increasing frequency, particularly in birds (Class Aves). In this study, we investigate the evolutionary history of mitochondrial control region states within the avian order Psittaciformes (parrots and cockatoos). To this aim, we reconstructed a comprehensive multi-locus phylogeny of parrots, used PCR of three diagnostic fragments to classify the mitochondrial control region state as single or duplicated, and mapped these states onto the phylogeny. We further sequenced 44 selected species to validate these inferences of control region state. Ancestral state reconstruction using a range of weighting schemes identified six independent origins of mitochondrial control region duplications within Psittaciformes. Analysis of sequence data showed that varying levels of mitochondrial gene and tRNA homology and degradation were present within a given clade exhibiting duplications. Levels of divergence between control regions within an individual varied from 0–10.9% with the differences occurring mainly between 51 and 225 nucleotides 3′ of the goose hairpin in domain I. Further investigations into the fates of duplicated mitochondrial genes, the potential costs and benefits of having a second control region, and the complex relationship between evolutionary rates, selection, and time since duplication are needed to fully explain these patterns in the mitochondrial genome.
Keywords: Ancestral state reconstruction, Control region, Control region duplication, Gene duplication, Mitochondrial genomes, Parrots
1. Introduction
Conservation of genome size, consistent gene content, and a lack of introns or intergenic spacers in animal mitochondria are generally interpreted as evidence that mitochondrial genomes are under selection for small size (Brown et al., 1979; Quinn and Wilson, 1993; Rand, 1993). This selection regime suggests that gene duplications in the mitochondria should be very rare or quickly eliminated because smaller genomes can replicate more quickly (Attardi, 1985; Diaz et al., 2002; Gray, 1989; Rand, 2001; Selosse et al., 2001; Sogin, 1997). As more mitochondrial genomes are sequenced, however, duplications of mitochondrial genes have been identified with increasing frequency in diverse species such as birds, lizards, ostracods, fish, arthropods, and snakes (Abbott et al., 2005; Arndt and Smith, 1998; Bensch and Härlid, 2000; Black and Roehrdanz, 1998; Campbell and Barker, 1999; Desjardins and Morais, 1990; Eberhard et al., 2001; Gibb et al., 2007; Kumazawa et al., 1996, 1998; Lee and Kocher, 1995; Lee et al., 2001; Macey et al., 1997; Mindell et al., 1998; Moritz and Brown, 1987; Ogoh and Ohmiya, 2004, 2007; Quinn and Mindell, 1996; Shao and Barker, 2003; Shao et al., 2005). It is now clear that duplications do occur in the mitochondrial genome and are much more common than previously thought. However, understanding the underlying mechanisms, evolutionary dynamics, and fitness consequences of these duplications remains an ongoing challenge for the field of molecular evolution.
Mitochondrial duplications often occur as tandem arrays, with a gene or group of genes repeated one after the other (Campbell and Barker, 1999; Eberhard et al., 2001; Abbott et al., 2005). Several mechanisms have been proposed that would result in this type of structure. Slipped strand mispairing can frequently result in tandem duplications in the presence of repeat units or sequences that form secondary structures (Levinson and Gutman, 1987; Madsen et al., 1993; Mueller and Boore, 2005). During DNA replication, a portion of the DNA strand dissociates between two repeats forming a loop. The polymerase then reassociates at the first repeat and duplicates the looped section (Levinson and Gutman, 1987; Madsen et al., 1993; Boore, 2000). The consistent presence of repeats at either end of the junctions of mitochondrial tandem duplications in parthenogenetic lizards led Fujita et al. (2007) to conclude that slipped strand mispairing was likely the cause of duplications. Over-running the termination signal during DNA replication has also been suggested as a way to form tandem duplications (Boore, 2000; Mueller and Boore, 2005). In this case, the duplications would include the genes flanking the origin of replication, which is often seen in mitochondrial duplications (San Mauro et al., 2006). Initiation of replication at sites of secondary structure other than the origin has also been suggested to result in tandem duplications (Levinson and Gutman, 1987; Madsen et al., 1993; Stanton et al., 1994; Lunt and Hyman, 1997; Macey et al., 1997; Boore, 2000). tRNAs or other sequences that are capable of forming secondary structures often are found at the ends of duplicated regions, suggesting that these structures may cause illicit priming of mitochondrial replication (Stanton et al., 1994; San Mauro et al., 2006). Finally, unequal crossing over could potentially result in tandem duplications when two mitochondrial genomes within a single mitochondrion recombine with one genome donating its copies of a group of genes to the other genome (see Ohno, 1970; Zhang, 2003 for this mechanism in the nucleus).
Consideration of the fates of duplicated nuclear genes suggests four potential fates for duplicated mitochondrial genes: (1) non-functionalization in which one copy becomes a pseudogene and is eventually eliminated from the genome, (2) subfunctionalization, in which copies of a multifunctional gene can each become specialized for one of the different original functions, and will each be stably maintained within the genome because they are under selection to carry out different functions, (3) neofunctionalization, in which a duplicated gene acquires a novel function due to mutations in the regulatory region or within the gene copy, and (4) redundant maintenance, in which multiple copies of a gene are maintained through gene conversion or purifying selection because the extra copies help meet high expression demands (Force et al., 1999; Lynch et al., 2001; Rastogi and Liberles, 2005; Roth et al., 2007; Zhang, 2003). However as mitochondria are believed to be under selection for compactness, it would seem that nonfunctionalization and elimination of extra gene copies would be the most likely fate of mitochondrial duplications (Rand and Harrison, 1986). Depending upon which copy of a gene is eliminated, a new gene arrangement may arise or the original gene order may be restored (Boore, 2000). This hypothesis has come to be known as the tandem duplication/random loss model of mitochondrial genome rearrangement (Boore, 2000; Macey et al., 1997; Mindell et al., 1998; Mortiz, 1991). In this model, loss of duplicated genes is thought to occur rapidly relative to evolutionary time (Mortiz, 1991; Quinn, 1997). Therefore, residual evidence of a previous duplication such as the presence of pseudogenes may be suggestive of a relatively recent event (Mortiz, 1991; Quinn, 1997).
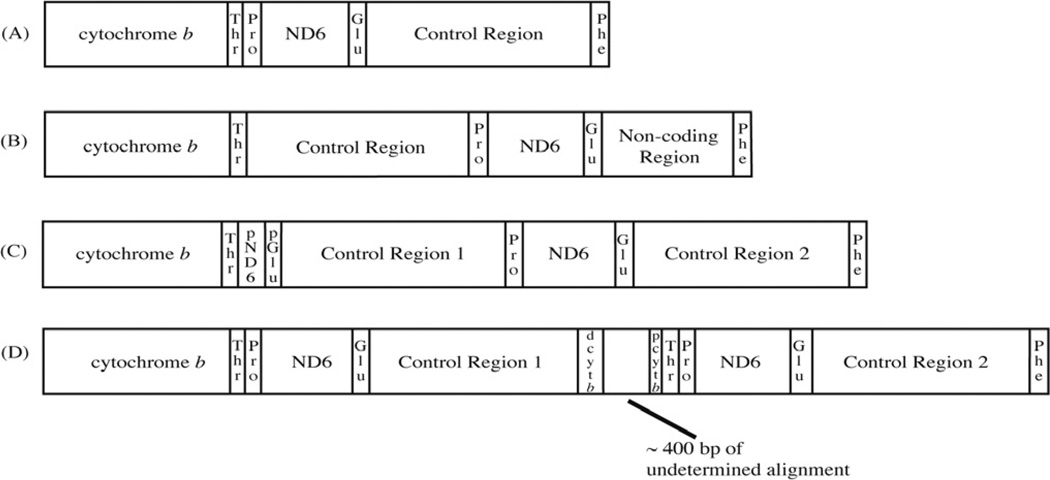
Four different mitochondrial genome arrangements have been identified within birds (Class Aves) that differ from that of the typical vertebrate (Fig. 1). The common avian arrangement, first identified in the chicken by Desjardins and Morais (1990), can be derived from the common vertebrate arrangement (ND6/tRNAGlu/cyt b/tRNAThr/tRNAPro/control region) by one tandem duplication/random loss event (involving cyt b/tRNAThr/tRNAPro/ND6/tRNAGlu/control region). Later, Mindell et al. (1998) described a second arrangement in which a non-coding region of variable length and with some similarity to the control region was found in the typical location of the control region, while the full-length control region was located after tRNAThr. A single tandem duplication/random loss event is necessary to derive this arrangement from the common avian mitochondrial arrangement (Mindell et al., 1998). This second gene order has been found in several diverse orders of birds, such as Piciformes (woodpeckers), Cuculiformes (cuckoos), Falconiformes (falcons), Passeriformes (oscines and suboscines), and Tinamiformes (tinamous) (Bensch and Härlid, 2000; Haddrath and Baker, 2001; Mindell et al., 1998). A third arrangement of mitochondrial genes was found in several species of Amazona parrots (Eberhard et al., 2001). In this case, one degenerate copy of the duplicated ND6 and tRNAGlu was still present making the extent of the duplication more easily defined. Additionally, the second noncoding region showed high similarity with the control region and appeared to be functional. This arrangement has also been found in the osprey (Pandion haliaetus), ivory-billed aracari (Pteroglossus azara) and Philippine hornbills (Aceros waldeni and Penelopides panini) (Gibb et al., 2007; Sammler et al., 2011). The fourth arrangement was identified in Thalassarche albatrosses (Abbott et al., 2005). Here, the genes from cytochrome b to the control region were tandemly duplicated and most appeared to still be functional. However, the second copy of cytochrome b appeared greatly reduced in size with only portions of the 5′and 3′ends (designated as d-cyt b and p-cyt b) being alignable with the full-length copy. The two control regions were also easily alignable and appeared to be functional, but differed in sequence and length of domain III, with control region 1 lacking repeats at the 3′end (Abbott et al., 2005). A similar rearrangement has also been found in the black-faced spoonbill (Platalea minor) (Cho et al., 2009), the ruff (Philomachus pugnax) (Verkuil et al., 2010), three species of booby in the genus Sula (Morris-Pocock et al., 2010) and two species of Philippine hornbills (Sammler et al., 2011).
Fig. 1.
(a) The typical avian mitochondrial gene order identified by Desjardins and Morais (1990). A tandem duplication followed by random loss of the control region and tRNAs can explain the rearrangement from the typical vertebrate gene order. (b) An alternative avian mitochondrial gene order identified by Mindell et al. (1998). A single tandem duplication followed by incomplete loss of the duplicated region may explain this rearrangement from the typical avian mitochondrial gene order. (c) The mitochondrial gene order identified by Eberhard et al. (2001) in Amazona parrots, in which two complete and putatively functional mitochondrial control regions appear to be maintained. (d) The mitochondrial gene order discovered by Abbott et al. (2005) in Thalassarche albatrosses in which cytochrome b to the control region is tandemly duplicated. Only the second copy of cytochrome b shows any degradation as depicted by the labels pcytb and dcytb, which are regions with high similarity to the full-length cytochrome b gene.
Despite the many descriptions of avian mitochondrial gene arrangements that have been published, we still lack a clear understanding of when or how often mitochondrial duplications have occurred in birds. Few orders have been systematically surveyed for gene arrangements or have been paired with a well-sampled phylogeny to allow robust conclusions about the evolutionary history of mitochondrial duplications and genome rearrangements. The order Psittaciformes (parrots and cockatoos, hereafter ‘parrots’), presents an excellent opportunity to identify the frequency with which mitochondrial control region duplications occur within a clade. Eberhard et al. (2001) established that several species of Neotropical parrots contained a duplicated control region, while preliminary data from other parrots suggested that these duplications were not shared by the entire order (T.F. Wright, J.R. Eberhard, unpublished data; E.S. Tavares, C.Y. Miyaki, unpublished data).
The current study seeks to address the following two questions: (1) Does the mitochondrial control region duplication, first identified in Amazona parrots, exist in other parrot genera? (2) If so, was there a single origin or were there multiple independent origins of these duplications? To answer these questions, we surveyed 117 parrot species by PCR for the presence of mitochondrial control region duplications and mapped these results onto a phylogeny reconstructed from mitochondrial and nuclear intron DNA sequences.
2. Materials and methods
2.1. Taxon and character sampling
For the phylogeny and survey of mitochondrial control region duplications, we added 51 new taxa to the dataset of Wright et al. (2008) for a total of 117 parrot species representing 79 of the 82 extant genera (Tables 1S and 2S). We used a stratified sampling method to determine the number of species sampled per genus such that genera with one to four species were represented by a single species or 25–100% coverage, genera with 5–11 species had two representatives (18–40% coverage), genera with 12–16 species were represented by three species (19–25% coverage) and genera with more than 17 species had four representatives (13–24% coverage). The new species included in this study were chosen based upon the accessibility of tissue or blood samples in museum or zoo collections. Samples for three genera (Geopsittacus, Ognorhynchus and Oreopsittacus) were unobtainable. Taxonomic nomenclature follows Forshaw (2006) for Old World species and the 2010 AOU North American and South American checklists for New World species (Chesser et al., 2010; Remsen et al., 2010). Coccyzus americanus (Cuculiformes), Colius colius (Coliiformes), Columbina passerina (Columbiformes), Falco peregrinus (Falconiformes), Otis sunia (Strigiformes), Picus canus (Piciformes), Serinus canarius (Passeriformes) and Tockus flavirostris (Coraciiformes) were included as outgroups as each has been identified as an ally or a sister group of the parrots in previous studies (Ericson et al., 2006; Fain and Houde, 2004; Hackett et al., 2008; Sibley and Ahlquist, 1990; Sorenson et al., 1999).
For the phylogenetic analyses, we sampled two mitochondrial protein-coding loci (cytochrome c oxidase I (COI) and nicatinamide adenosine dehydrogenase subunit 2 (ND2)) and two nuclear introns (tropomyosin intron five (TROP) and transforming growth factor beta 2 intron one (TGFB2)). These genes have proven to be informative in other phylogenetic studies of parrots (Wright et al., 2008; Joseph et al., 2012).
2.2. DNA extraction, PCR and sequencing
We extracted DNA from tissue or blood samples, performed polymerase chain reaction amplification (PCR), and sequenced the PCR products at laboratories in three locations due to legal restrictions on transporting specimens from endangered species. The laboratories were New Mexico State University (NMSU) in Las Cruces, New Mexico; the Instituto Nacional de Toxicologia y Ciencias Forenses (Spain) in Tenerife, Canary Islands, Spain; and the University of São Paulo (Brazil) in São Paulo, Brazil.
At NMSU and in Spain we extracted DNA using the DN easy Blood and Tissue Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol for each tissue type. In Brazil, we extracted DNA from blood samples using a phenol/chloroform protocol (Bruford et al., 1992). At all locations we amplified the four gene regions by PCR using primers, reactions, and cycling conditions as described in Wright et al. (2008). PCR products were checked for correct size and the presence of multiple bands by electrophoresis on a 0.5–2% agarose gel and stained with ethidium bromide.
We cleaned PCR products using a Qia Quick PCR Purification Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions at NMSU and in Spain, while in Brazil we used 1 µL exonuclease I and 0.5 µL of shrimp alkaline phosphatase per 10 µL PCR reaction incubated at 37 °C for 30 min, then 80 °C for 15 min to clean PCR products. We sequenced each PCR product in both directions using the PCR primers and Big Dye v3.1 Terminator Cycle Sequencing chemistry (Applied Biosystems Inc, Foster City, CA). Each sequencing reaction at NMSU and in Spain consisted of 2 µL of Big Dye, 1 µL of 5X sequencing buffer, 3.2 µL of 1uM primer, 2 µL of clean PCR product, and 11.8 µL of water. In Brazil each sequencing reaction consisted of 2 µL of Big Dye, 2–4 µL of clean PCR product, and 1 µL of primer. Sequencing conditions at all locations were 25 cycles of 95 °C for 25 s, 50 °C for 5 s, and 60 °C for 4 min. We cleaned sequencing reactions at NSMU by centrifugation through Sephadex columns. Clean reactions were dried and resuspended in 20 µL of Hi Di Formamide (Applied Biosystems Inc., Foster City, CA) before sequencing on an ABI 3100 Avant automated sequencer. In Brazil sequencing reactions were cleaned by isopropanol/ethanol precipitation, dried, resuspended in 1.8 µL of formamide, heated to 95 °C for 2 min, placed on ice until loaded on an ABI 377 automated sequencer. In Spain, we cleaned sequencing reactions by centrifugation through Centri-Sep columns in a 96 well format (Applied Biosystems Inc., Foster City, CA). The cleaned reaction was dried, resuspended in 20 µL of formamide, and sequenced on an ABI 310 automated sequencer.
2.3. Phylogenetic analysis
Raw sequences were checked for ambiguous base calls in Sequencher 4.7 (Gene Codes, Ann Arbor, MI) and combined into contigs by locus and taxon. Sequences were aligned using Clustal W with default parameters as implemented at http://www.ebi.ac.uk/Tools/clustalw and adjusted by eye. Gaps within introns were coded by the simple indel coding method (Simmons and Ochoterena, 2000) as implemented in Indel Coder 0.5 in the Seq State 1.40 program (Müller, 2005, 2006).
Maximum likelihood methods of phylogenetic reconstruction were implemented in GARLI v0.951 using the default settings (Zwickl, 2006). The General Time Reversible model with a gamma distribution of among site heterogeneity and a proportion of invariant sites was chosen, with parameter values estimated by the software and SPR branch swapping (Zwickl, 2006). To ensure that the tree was not located in a local optimum, 20 independent runs were conducted in GARLI and the tree with the highest likelihood was chosen for subsequent analyses (Zwickl, 2006). Nodal support was evaluated by 100 maximum likelihood bootstraps calculated in GARLI.
Bayesian methods of phylogeny reconstruction were implemented using Mr Bayes 3.1.2 (Ronquist and Huelsenbeck, 2003). A separate evolutionary model was determined for each gene region in Mr Model Test 2.3 under the Akaike Information Criterion (Nylander, 2004). Gaps were coded as restriction sites using the default settings. The analysis consisted of two parallel runs, each with one cold chain and three heated chains with default parameters. The mixed model analysis of the combined dataset was run for 15,000,000 generations with trees sampled every 1000 generations and a burn-in of 25%. Convergence was assumed when the average standard deviation of split frequencies was less than or equal to 0.01 and when the effective sample size (ESS) value for parameter values was greater than 200 when viewed in Tracer v1.4 (Rambaut and Drummond, 2007).
2.4. Mitochondrial control region survey
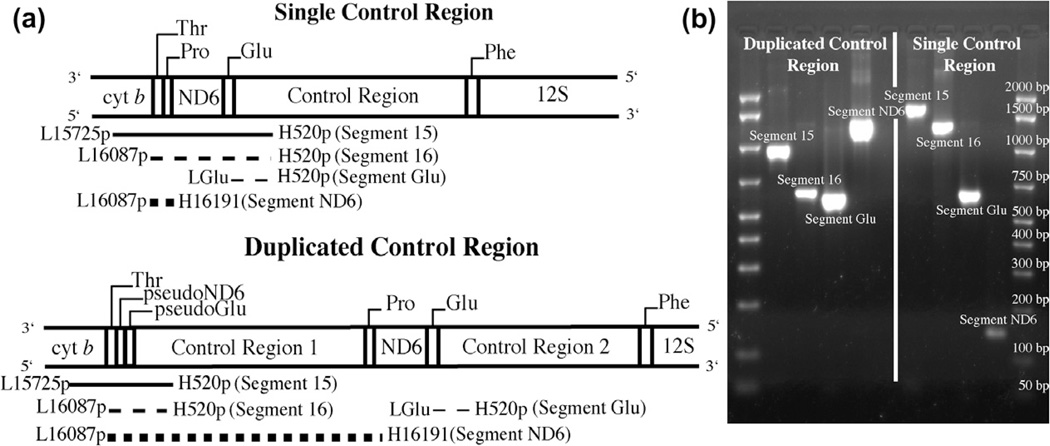
Of the 117 species included in the phylogeny 112 were surveyed at NMSU for the presence of a mitochondrial control region duplication using PCR to amplify diagnostic fragments from the regions predicted to differ in length depending on the presence or absence of a duplicated control region. We could not survey five taxa using the PCR approach (Strigops habroptilus, Cyanoramphus auriceps, Cyanopsitta spixii, Enicognathus leptorhynchus and Triclaria malachitacea) because tissue samples were not available at NMSU. We used the primer pairs L15725p – AAACCAGARTGATAYTTYC TMTTYGCAT (modified from Sorenson et al., 1999) and H520p – TGKSCCTGACCKAGGAACCAG (modified from Sorenson et al., 1999), L16087p – TGGYCTTGTAARCCAAARRAYGAAG (modified from Sorenson et al., 1999) and H520p and L16087p with H16191 – TCTCGDGGGGCDATTCGGGC (Sorenson et al., 1999) to amplify diagnostic Segments 15, 16 and ND6 respectively. An additional primer pair, LGlu – GCCCTGAAAARCCATCGTTG (Eberhard et al., 2001) in conjunction with H520p was used to amplify Segment Glu to verify the presence of a second control region that is usually flanked by the intact tRNAGlu. The expected location of each primer and the genes contained within each segment are depicted in Fig. 2. PCR reaction conditions were the same as for the phylogeny except for the cycling conditions which were 94 °C for 4 min followed by 29 cycles of 94 °C for 25 s, 60 °C for 30 s on the initial cycle with a decrease of 0.4 °C each cycle, and 72 °C for 2 min. This was followed by 6 cycles of 94 °C for 25 s, 45 °C for 30, 72 °C for 2 min, with a final extension of 72 °C for 10 min. PCR products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide. Gels were imaged on a Bio Rad Gel Doc XR and band sizes for each segment were estimated using the Bio Rad Amplisize 50–2000 bp Molecular Ruler and the Band Analysis protocol in the Quantity One software (Bio Rad Life Sciences, Hercules, CA).
Fig. 2.
Location of primer pairs and relative fragment lengths used to classify control region duplication state. (a) Schematic of fragment sizes based upon the presence of a single or duplicated mitochondrial control region. (b) A representative agarose gel of fragment sizes for a duplicated and single control region.
Expected sizes of the diagnostic segments used to score each species as a single or duplicated control region were as follows: Single Control Region, Segment 15 = 1579–1896 base pair (bp), Segment 16 = 1225–1542 bp, Segment ND6 = 132 bp; Duplicated Control Region, Segment 15 = 800–1250 bp, Segment 16 = 600–1000 bp, Segment ND6 ≥ 1200 bp. These segment sizes were derived from preliminary studies of parrot control region duplications and then further refined by analysis of the GenBank parrot mitochondrial sequences (Table 3S and T.F. Wright, J.R. Eberhard, E.E. Schirtzinger, unpublished data). Band sizes were expected to show some variation due to the variation in size of domains I and III of the control region (Baker and Marshall, 1997) as determined by alignment of primer H520p to parrot control region sequences available on GenBank. Another source of variation is the presence and size of intergenic spacers. This variation was evaluated by counting the base pairs between the 3′end of one annotated gene and the 5′end of the next annotated gene in the region of cytochrome b to tRNAPhe from three parrot mitochondrial genomes on GenBank (Strigops habroptilus, Agapornis roseicollis and Melopsittacus undulatus). These ranges were added to the sizes of the genes included in each segment to get the total estimated range of variation. A species was scored as having a single or duplicated control region based on the correspondence of its measured segment sizes to the expected sizes for each segment (Fig. 1S). Taxa that did not amplify at least two diagnostic segments were classified as unscorable.
2.5. Sequencing of selected taxa
Because the expected band sizes encompass a large range, diagnostic Segment 16 and Segment Glu, from selected species were sequenced at NMSU or Brazil to confirm the status of the mitochondrial control region as classified by our PCR survey. At least one representative of each clade that contained an inferred duplicated control region was sequenced. In addition, species that were ambiguous in their classification were also sequenced. Finally, the GenBank mitochondrial sequences for Agapornis roseicollis, Strigops habroptilus (single control regions) and Melopsittacus undulatus, Amazona farinosa, Amazona ochrocephala and Psittacus erithacus (duplicated control regions) were also used as confirmation of the PCR survey results.
At NMSU, PCR products were amplified, cleaned as described above, and sent to the University of Chicago Cancer Sequencing Facility for sequencing on an ABI 3730 automated sequencer using Big Dye chemistry. In Brazil, amplifications for sequencing were performed in 10 µL reactions with 1X buffer (GE Healthcare or Biotools), 2 µM of dNTP, 1 µM of each primer, 0.5 U of Taq polymerase, and 20–50 nanograms (ng) of template DNA, or in 25 µL reactions with 1X buffer (Biotools), 2 µM of dNTP, 1 µM of each primer, 1 U of Taq polymerase, and 25–50 ng of DNA. PCR conditions were: initial denaturation 96 °C for 5 min, 30 cycles of 95 °C for 60 s, 50–54 °C for 25 s, and 65 °C for 40–80 s, with a final extension of 65 °C for 5 min. The size and quality of PCR products were verified, purified as described above or bands were excised from agarose gels and the product was isolated by centrifugation through filter tips (Axigen). Sequencing reactions were prepared, cleaned, and run as described above.
Raw sequences were proofread as previously described and combined into a consensus sequence by taxon and segment using Sequencher 4.7 (Gene Codes, Ann Arbor, MI). The location of tRNAThr, tRNAPro, ND6, and tRNAGlu were identified by comparison with the homologous genes from the mitochondrial genome of Melopsittacus undulatus (NC_009134), while the control region was identified by the presence of the goose hairpin (C7TAC7) near the 5′ end. The identity of pseudogenes was based on similarity with known sequences from the Melopsittacus undulatus mitochondrial genome or comparison with the pseudogenes defined by Eberhard et al. (2001). Functionality of tRNAs was assessed by simulation in tRNA scan-SE (Lowe and Eddy, 1997).
For each species sequenced, the gene order from tRNAThr through domain I of the control region was identified by similarity with previously described avian gene orders (Abbott et al., 2005; Desjardins and Morais, 1990; Eberhard et al., 2001; Mindell et al., 1998). For those species with a duplication, we measured the length of the non-coding region (calculated as the number of nucleotides from the end of tRNAThr to the goose hairpin), and the number of nucleotide differences between the two control regions (calculated as the number of differences when the two control region fragments were aligned divided by the total length of the aligned control region segment). Because the 5′end of the control region does not have a definitive starting motif and the 3′ end of the tRNAGlu could not be identified for all species, the goose hairpin was used as a proxy for the beginning of the control region.
2.6. Ancestral state reconstruction
Using the classifications from the PCR survey, sequences from selected taxa and GenBank sequences, the presence of a single (0) or duplicated (1) control region was coded into a matrix and mapped onto the phylogeny. Three species (Aprosmictus erythropterus, Eos reticulata, Pionites melanocephala) were coded as unscorable because they either failed to amplify the diagnostic fragments by PCR or they produced ambiguous results and no confirmatory sequence was available. Ancestral state reconstructions were undertaken in Mesquite 2.01 (Maddison and Maddison, 2007) under parsimony (Fitch) and the maximum likelihood criterion. Likelihood reconstructions were undertaken using the symmetric MK1 model, which is a generalization of the Jukes–Cantor model with equal probability of changing states and the AsymmMK model, which uses different rates of change between states (Maddison and Maddison, 2007). To test the robustness of the likelihood analysis, separate reconstructions were undertaken using the AsymmMK model and rates translating to (a) gains five times as likely as losses and (b) gains 1/5 as likely as losses.
3. Results
3.1. Phylogenetic analysis
The phylogenetic dataset consisted of 117 parrot species and eight non-parrot outgroups. The mitochondrial sequences, COI and ND2, showed no insertions/deletions (indels). The COI sequences were 570 base pairs (bp) in length while the ND2 sequences were 1041 bp. The intron sequences, TROP and TGFB2, were more variable in length due to the presence of 48 and 78 indels respectively. The TROP sequences were 498–533 bp with a total of 554 aligned bp. The TGFB2 sequences were 611–630 bp with a total of 817 aligned bp. The maximum likelihood dataset consisted of 2982 concatenated bp. The Bayesian analysis was performed on the 2982 characters included in the concatenated dataset (partitioned by gene region) and the 126 coded indels for a total of 3108 characters. Table 4S includes the genetic details for each gene region and the concatenated dataset. All new sequences have been deposited in GenBank (see Table 1S).
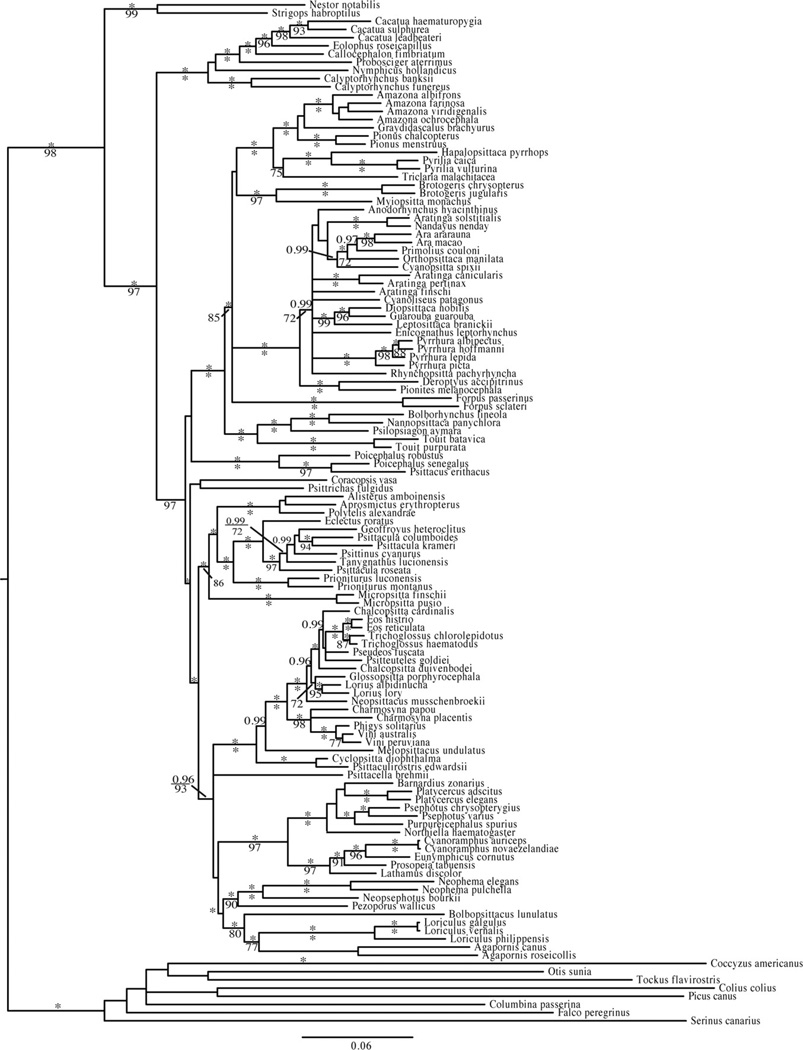
The most likely tree (−ln L = −63846.569) from 20 independent maximum likelihood (ML) analyses in GARLI of the concatenated dataset (Fig. 2S) and the consensus tree from the Bayesian analysis (Fig. 3) were broadly congruent, differing only in the placement of two genera, Micropsitta and Graydidascalus. The Bayesian analysis places Micropsitta with high support (posterior probability = 1) as sister to the clade consisting of Alisterus, Aprosmictus, Polytelis, Eclectus, Geoffroyus, Psittacula, Psittinus, Tanygnathus and Prioniturus while in the ML analysis Micropsitta is sister to the clade of Alisterus, Aprosmictus and Polytelis with poor support (ML boot-strap 6 50). The Bayesian analysis places Graydidascalus as sister to Amazona while in the ML analysis Graydidascalus is sister to the clade consisting of Amazona and Pionus. However, in both analyses the position of Graydidascalus is poorly supported. Nucleotide substitution models, priors for the Bayesian analysis, and final estimates of parameters for each analysis are listed in Table 5S.
Fig. 3.
The partitioned Bayesian analysis of 117 parrot species reconstructed using two mitochondrial protein-coding genes (COI and ND2), two nuclear introns (TROP and TGFB2) and coded gaps. Posterior probabilities >0.95 are given above the branches and maximum likelihood bootstraps >70 are given below the branches. Asterisks indicate a posterior probability of 1.0 or a maximum likelihood bootstrap value of 100.
3.2. Mitochondrial control region survey
One hundred and twelve parrot species were surveyed for the status of its mitochondrial control region by PCR of three diagnostic segments that show variation in size when a duplicated control region is present. Table 1 reports the control region status of each species surveyed and the length of each amplicon. Segment 15 was amplifiable for 96 species, while Segment 16 and Segment ND6 were amplifiable for 108 and 110 species respectively (see Table 1). Aprosmictus erythropterus, Guarouba guaroouba, Nandayus nenday and Rhynchopsitta pachyrhyncha could not be scored by PCR at NMSU due to a lack of amplification. Five species (Eos reticulate, raydidascalus brachyurus, Hapalopsittaca pyrrhops, Pionites melanocephala and Psittrichas fulgidus) all amplified ND6 bands indicative of a single control region while the sizes of Segment 15 and Segment 16 suggested that a duplicated control region was present. These species are listed as ambiguous in Table 1.
Table 1.
Measured PCR fragment lengths and control region status for each parrot species in the phylogeny.
| Measured fragment size (nucleotides) |
|||||
|---|---|---|---|---|---|
| Scored by PCR | Control region status | 15 | 16 | Glu | ND6 |
| Agapornis canus | 1 | 1781 | 1394 | 708 | 141 |
| Agapornis roseicollis | 1 | 1912 | 1399 | 689 | 144 |
| Alisterus amboinensis | 1 | 1652 | 1253 | 488 | 141 |
| Amazona albifrons | 2 | 1042 | 756 | 609 | 501 |
| Amazona farinosa | 2 | 1134 | 769 | 618 | 137, 1943 |
| Amazona ochrocephala | 2 | 1109 | 779 | 637 | 1883 |
| Amazona viridigenalis | 2 | 1182 | 781 | 626 | 153, 508, 2146 |
| Anodorhynchus hyacinthinus | 1 | 1672 | 1316 | 645 | 148 |
| Aprosmictus erythropterus | UN | * | * | * | 144 |
| Ara ararauna | 1 | 1742 | 1344 | 627 | 147 |
| Ara macao | 1 | 1738 | 1302 | 685 | 152 |
| Aratinga canicularis | 1 | 1833 | 1357 | 648 | 152 |
| Aratinga finschi | 1 | 1658 | 1341 | 630 | 144 |
| Aratinga pertinax | 1 | 2121 | 1697 | 640 | 145 |
| Aratinga solstitialis | 1 | * | 1368 | 627 | 145 |
| Barnardius zonarius | 1 | 1963 | 1576 | 847 | 149 |
| Bolbopsittacus lunulatus | 1 | * | 1681 | 901 | 153 |
| Bolborhynchus lineola | 1 | 1580 | 1255 | 543 | 149 |
| Brotogeris chrysopterus | 1 | 1809 | 1376 | 667 | 149 |
| Brotogeris jugularis | 1 | 1717 | 1343 | 677 | 145 |
| Cacatua haematuropygia | 1 | 1627 | 1236 | 640 | 148 |
| Cacatua leadbeateri | 1 | 1722 | 1245 | 629 | 149 |
| Cacatua sulphurea | 1 | * | 1227 | 604 | 144 |
| Callocephalon fimbriatum | 1 | 1505 | 1198 | 606 | 148 |
| Calyptorhynchus banksii | 1 | 1543 | 1215 | 549 | 148 |
| Calyptorhynchus funereus | 1 | 1765 | 1320 | 553 | 150 |
| Chalcopsitta cardinalis | 2 | 990 | 656 | 602 | 1346 |
| Chalcopsitta duivenbodei | 2 | 943 | 602 | 680 | 144, 1387 |
| Charmosyna papou | 2 | * | 1049 | 625 | 141, 315, 1164 |
| Charmosyna placentis | 2 | 998 | 628 | 625 | 1383 |
| Coracopsis vasa | 1 | 1572 | 1237 | 562 | 143 |
| Cyanoliseus patagonus | 1 | 1805 | 1368 | 671 | 147 |
| Cyanoramphus novaezelandiae | 1 | 2047 | 1626 | 875 | 144 |
| Cyclopsitta diophthalma | 2 | * | 673 | 636 | 475 |
| Deroptyus accipitrinus | 2 | 1261 | 952 | 642 | 147, 2244 |
| Diopsittaca nobilis | 1 | * | 1294 | 631 | 155 |
| Eclectus roratus | 1 | 1658 | 1253 | 561 | 153 |
| Eolophus roseicapillus | 1 | 1621 | 1187 | * | 152 |
| Eos histrio | 2 | 1010 | 622 | 573 | 1465 |
| Eos reticulata | 2? | * | 627 | 575 | 145 |
| Eunymphicus cornutus | 1 | 1928 | 1583 | 861 | 150 |
| Forpus passerinus | 2 | 1200 | 810 | 670 | 138, 1813 |
| Forpus sclateri | 2 | 1234 | 650 | 672 | 2980 |
| Geoffroyus heteroclitus | 1 | * | 1498 | 781 | 152 |
| Glossopsitta porphyrocephala | 2 | 1088 | 720 | 687 | 1440 |
| Graydidascalus brachyurus | 2? | 1410 | 889 | 700 | 154 |
| Guarouba guarouba | UN | * | * | * | * |
| Hapalopsittaca pyrrhops | 2? | 1321 | 786 | 663 | 143 |
| Lathamus discolor | 1 | 1962 | 1577 | 840 | 147 |
| Leptosittaca branickii | 1 | 1726 | 1296 | 620 | 146 |
| Loriculus galgulus | 1 | 1571 | 1269 | 631 | 146 |
| Loriculus philippensis | 1 | 1598 | 1279 | 640 | 147 |
| Loriculus vernalis | 1 | 1636 | 1295 | 628 | 143 |
| Lorius albidinuchas | 2 | 998 | 640 | 618 | 1350 |
| Lorius lory | 2 | 984 | 645 | 580 | 1472 |
| Melopsittacus undulatus | 2 | 968 | 610 | 585 | 1612 |
| Micropsitta finschii | 1 | 1555 | 1180 | 502 | 149 |
| Micropsitta pusio | 1 | 1505 | 1142 | 479 | 142 |
| Myiopsitta monachus | 1 | 1639 | 1259 | 596 | 152 |
| Nandayus nenday | UN | * | * | * | * |
| Nannopsittaca panychlora | 1 | 1620 | 1254 | 556 | 151 |
| Neophema elegans | 1 | * | 1379 | 693 | 164 |
| Neophema pulchella | 1 | 1673 | 1288 | 640 | 160 |
| Neopsephotus bourkii | 1 | 1657 | 1313 | 628 | 154 |
| Neopsittacus musschenbroekii | 2 | 1004 | 640 | 584 | 1423 |
| Nestor notabilis | 1 | 1615 | 1264 | 596 | 156 |
| Northiella haematogaster | 1 | 1914 | 1322 | 592 | 147 |
| Nymphicus hollandicus | 1 | 1633 | 1200 | 591 | 146 |
| Orthopsittaca manilata | 1 | 1727 | 1363 | 640 | 152 |
| Pezoporus wallicus | 1 | 1900 | 1500 | 800 | 155 |
| Phigys solitarius | 2 | 1028 | 646 | 550 | 155, 1524 |
| Pionites melanocephala | 2? | * | 971 | 626 | 154 |
| Pyrilia caica | 2? | 1136 | 782 | 638 | 158 |
| Pyrilia vulturina | 2 | 982 | 615 | 600 | 166, 2223 |
| Pionus chalcopterus | 2 | 1150 | 781 | 614 | 157, 525, 2128 |
| Pionus menstruus | 2 | 1167 | 793 | 640 | 490 |
| Platycercus adscitus | 1 | 1919 | 1521 | 809 | 149 |
| Platycercus elegans | 1 | 1829 | 1449 | 771 | 148 |
| Poicephalus robustus | 2 | 1058 | 666 | 560 | 153, 1521, 1804 |
| Poicephalus senegalus | 2 | 978 | 639 | 531 | 1549 |
| Polytelis alexandrae | 1 | 1265 | 635 | 145 | |
| Primolius couloni | 1 | 1685 | 1313 | 630 | 147 |
| Prioniturus luconensis | 2 | 1231 | 761 | 500 | 145, 2328 |
| Prioniturus montanus | 2 | 1130 | 795 | 492 | 148 |
| Probosciger aterrimus | 1 | 1601 | 1244 | 594 | 144 |
| Prosopeia tabuensis | 1 | 1878 | 1465 | 758 | 148 |
| Psephotus chrysopterygius | 1 | 1872 | 1494 | 858 | 138 |
| Psephotus varius | 1 | 1924 | 1517 | 837 | 148 |
| Pseudeos fuscata | 2 | 988 | 619 | 578 | 1370 |
| Psilopsiagon aymara | 1 | 1555 | 1210 | 546 | 151 |
| Psittacella brehmii | 1 | 1640 | 1476 | 604 | 150 |
| Psittacula columboides | 1 | * | 1211 | 550 | 150 |
| Psittacula krameri | 1 | * | 1343 | 582 | 136 |
| Psittacula roseata | 1 | 1503 | 1198 | 492 | 144 |
| Psittaculirostris edwardsii | 2 | 817 | 620 | 586 | 391, 680 |
| Psittacus erithacus | 2 | 984 | 617 | 545 | 144, 1589 |
| Psitteuteles goldiei | 2 | 1341 | 638 | 599 | 1455 |
| Psittinus cyanurus | 1 | 1554 | 1192 | 486 | 132 |
| Psittrichas fulgidus | 2? | 1244 | 907 | 535 | 135 |
| Purpureicephalus spurius | 1 | 1893 | 1563 | 849 | 142 |
| Pyrrhura albipectus | 1 | 1712 | 1331 | 626 | 145 |
| Pyrrhura hoffmanni | 1 | 1563 | 1297 | 600 | 142 |
| Pyrrhura lepida | 1 | 1665 | 1306 | 640 | 136 |
| Pyrrhura picta | 1 | 1724 | 1310 | 622 | 154 |
| Rhynchopsitta pachyrhyncha | UN | * | * | 638 | 137 |
| Tanygnathus lucionensis | 1 | 1585 | 1262 | 550 | 133 |
| Touit batavica | 1 | 1700 | 1200 | 500 | 137 |
| Touit purpurata | 1 | 1700 | 1100 | 500 | 148 |
| Trichoglossus chlorolepidotus | 2 | 842 | 664 | * | 143, 175, 772, 1501 |
| Trichoglossus haematodus | 2 | 1009 | 643 | 586 | 1491 |
| Vini australis | 2 | 1000 | 650 | 600 | 142. 1348 |
| Vini peruviana | 2 | 1000 | 650 | 600 | 1336 |
| N=112 | N=96 | N=108 | N=107 | N=110 | |
|
Scored from Sequences (BR) |
|||||
| Cyanopsitta spixii | 1 | ||||
| Enicognathus leptorhynchus | 1 | ||||
| Guaruba guarouba | 1 | ||||
| Nandayus nenday | 1 | ||||
| Rhynchopsitta pachyrhyncha | 1 | ||||
| Triclaria malachitacea | 2 | ||||
| N=6 | |||||
|
Scored from GB Sequences or Literature |
|||||
| Cyanoramphus auriceps | 1 | Boon 2000 | |||
| Strigops habroptilus | 1 | AY309456 | |||
| N=2 | |||||
fragment did not amplify.
UN = unscorable by PCR.
ambiguous by PCR.
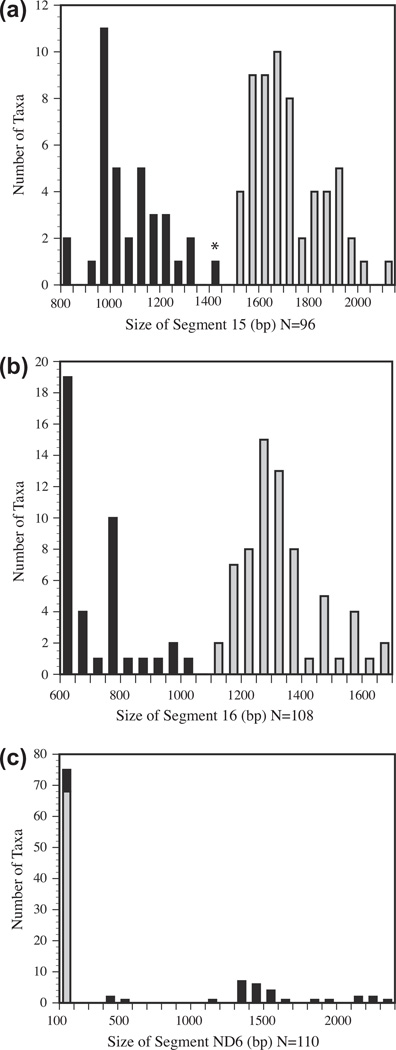
The band sizes for each segment are plotted in Fig. 4. Segment 15 showed a bimodal distribution, with most species clearly falling into either the single or duplicated control region size categories. However, Gradydidascalus brachyurus fell between the expected ranges. Segments Glu and 16 from this species were subsequently sequenced. In four taxa, Segment 15 amplicons were larger than the expected size of 1896 bp for a single control region. Lathamus discolor and Barnardius zonarius were approximately 75 bp larger than expected, while Aratinga pertinax and Cyanoramphus novaezelandiae exceeded the single control region size range by 225 bp and 150 bp. These sizes may be stochastic effects of slightly different gel conditions that affect the distance run by the size marker used to measure the bands. The other bands for these species were also larger than expected but within the range of single control region sizes.
Fig. 4.
Histograms of the number of species per 50 base pair bins for each amplified segment. Black bars indicate duplicate control regions. Gray bars indicate a single control region. An asterisk indicates species that fall outside of the expected fragment size ranges based on preliminary surveys of control region lengths. (a) lengths of Segment 15, (b) lengths of Segment 16, and (c) lengths of Segment ND6.
The size distribution for Segment 16 amplicons was also bimodal with all taxa falling within the two expected range sizes. Segment ND6 showed two different patterns: a tight cluster of species from 130 to 160 bp and a scatter of species with segment sizes from 1150 bp to over 2200 bp. Pionus menstruus and Amazona albifrons had values of 450 bp and 550 bp respectively. Based upon the other band sizes, Amazona albifrons and Pionus menstruus were classified as having a duplicated control region. The presence of a duplicated control region in Amazona albifrons was later confirmed by sequencing.
Of the 112 species surveyed by PCR, 68 were classified as having a single control region and 35 species were classified as having a duplicated control region. Four species, Aprosmictus erythropterus, Guarouba, Nandayus nenday and Rhynchopsitta pachyrhyncha were unscorable due to a lack of PCR amplification and five species (Eos reticulata, Graydidascalus brachyurus, Hapalopsittaca pyrrhops, Pionites melanocephala and Psittrichas fulgidus) produced ambiguous results.
3.3. Sequencing of selected taxa
In order to validate the control region classifications from the PCR survey we sequenced 44 parrot species in the phylogeny (Table 2). The common avian gene order with a single control region was confirmed in 19 species (Desjardins and Morais, 1990), while the gene order with a duplicated control region previously described by Eberhard et al. (2001) for Amazona and Pionus species was confirmed for 25 species. Of the five species that produced ambiguous results in the PCR survey, two species (Graydidascalus brachyurus, Pseudoes fuscata) were shown to have a duplicated control region by sequencing. The aberrant size of the ND6 fragment was due to a lack of retained homology in the non-coding region. Therefore, only the functional ND6 was amplified. Sequencing of Cyanopsitta spixii, Enicognathus leptorhynchus, Guarouba, Nandayus nenday, Rhynchopsitta pachyrhyncha and Triclaria malachitacea in Brazil found that only Triclaria malachitacea had a duplicated control region. Three species were listed as unscorable (Aprosmictus erythropterus, no PCR amplification) or ambiguous (Eos reticulata and Pionites melanocephala, no sequence available to confirm status).
Table 2.
Confirmation of PCR Classified Control Region Status by Sequencing of Selected Species or by GenBank Sequences.
| Scored by PCR | Control region status |
Confirmed by Sequence source |
GenBank # |
|---|---|---|---|
| Agapornis rosiecollis | 1 | GB | EU410486 |
| Amazona albifrons | 2 | NMSU | JQ341164, JQ360543 |
| Amazona farinosa | 2 | GB | AF228821 |
| Amazona ochrocephala | 2 | NMSU/GB | AF338819, AF338820, JQ341165, JQ360544 |
| Anodorhynchus hyacinthinus | 1 | BR | EF104124 |
| Ara ararauna | 1 | BR | EF104127 |
| Aratinga solstitialis | 1 | BR | EF104138 |
| Bolborhynchus lineola | 1 | BR | EF104137 |
| Calyptorhynchus banksii | 1 | NMSU | JQ360567 |
| Chalcopsitta cardinalis | 2 | NMSU | JQ341170, JQ360549 |
| Chalcopsitta duivenbodei | 2 | NMSU | JQ360545 |
| Charmosyna papou | 2 | NMSU | JQ341166 |
| Charmosyna placentis | 2 | NMSU | JQ341167, JQ360546 |
| Coracopsis vasa | 1 | NMSU | JQ341168, JQ360570 |
| Cyanoliseus patagonus | 1 | BR | EF104136 |
| Cyanopsitta spixii | 1 | BR | EF104128 |
| Cyclopsitta diophthalma | 2 | NMSU | JQ241169, JQ36-547 |
| Deroptyus accipitrinus | 2 | BR/NMSU | AF365437, JQ360548 |
| Diopsittaca nobilis | 1 | BR | EF104121 |
| Enicognathus leptorhynchus | 1 | BR | EF104139 |
| Eos histrio | 2 | NMSU | JQ341171, JQ360550 |
| Forpus sclateri | 2 | NMSU | JQ341172, JQ360551 |
| Glossopsitta porphyrocephala | 2 | NMSU | JQ341173, JQ360552 |
| Graydidascalus brachyurus | 2 | BR | EF104148 |
| Guaruba guarouba | 1 | BR | EF104123 |
| Lorius albidinucha | 2 | NMSU | JQ341174, JQ360553 |
| Melopsittacus undulatus | 2 | GB | NC_009134 |
| Micropsitta pusio | 1 | NMSU | JQ360568 |
| Myiopsitta monachus | 1 | BR | EF104118 |
| Nandayus nenday | 1 | BR | EF104131, EF104149 |
| Neopsittacus musschenbroekii | 2 | NMSU | JQ341175, JQ360554 |
| Orthopsittaca manilata | 1 | BR | EF104119 |
| Phigys solitarius | 2 | NMSU | JQ341176, JQ360555 |
| Pyrilia caica | 2 | NMSU | JQ341178, JQ360556 |
| Pionus chalcopterus | 2 | NMSU/GB | AF338817, AF338818, JQ360557 |
| Poicephalus robustus | 2 | NMSU | JQ360558 |
| Prioniturus montanus | 2 | NMSU | JQ341180, JQ360566 |
| Pseudeos fuscata | 2 | NMSU | JQ341183, JQ360561 |
| Psittacula roseata | 1 | NMSU | JQ360569 |
| Psittaculirostris edwardsii | 2 | NMSU | JQ341181, JQ360559 |
| Psittacus erithacus | 2 | NMSU/GB | DQ335468, JQ341182, JQ360560 |
| Psitteuteles goldiei | 1 | NMSU | JQ341184, JQ360562 |
| Pyrrhura picta | 1 | BR | EF104130, EF104150 |
| Rhynchopsitta pachyrhyncha | 1 | BR | EF104135 |
| Trichoglossus haematodus | 2 | NMSU | JQ341186, JQ360564 |
| Triclaria malachitacea | 2 | BR | EF104143, EF104146 |
| Vini australis | 2 | NMSU | JQ341187, JQ360565 |
| Related Species Sequenced | |||
| Aratinga aurea | 1 | BR | EF104132, EF104151 |
| Aratinga leucophthalmus | 1 | BR | EF104133 |
| Brotogeris chirri | 1 | BR | EF104117 |
| Primolius auricollis | 1 | BR | EF104126 |
| Pionites leucogaster | 2 | BR | JQ749718, JQ749719 |
| Pyrilia barrabandi | 2 | BR/NMSU | JQ341177, EF104141, EF104144 |
| Forpus xanttopterygius | 2 | BR | EF104140, EF104147 |
| Nannopsittaca dachillae | 1 | BR | EF104134 |
| Psittaculirostris salvadorii | 2 | NMSU | JQ341185, JQ360563 |
In the set of species with duplicated control regions, non-coding regions of various sizes were found between tRNAThr and the first control region. These non-coding regions were examined with tRNA-Scan (Lowe and Eddy, 1997) to determine if copies of tRNAPro or tRNAGlu retained enough homology to be identifiable. In most cases no homology could be determined due to the extent of degeneration. The control regions were identified by the presence of conserved sequences: the goose hairpin at the 5′ end of domain I and the D-Box in domain II (Eberhard et al., 2001). The F-Box sequence of Gallus gallus was not identified in all species. An additional eight species (Aratinga aurea, Aratinga leucophthalmus, Brotogeris chirri, Forpus xanthopterygius, Nannopsittaca dachillae, Pyrilia barrabandi, Pionites leucogaster, and Primolius auricollis) were sequenced in Brazil for a different study (E. Tavares, C. Miyaki unpublished data), and served as additional confirmation of the distribution of control region duplications. Although these species were not included in the phylogeny, all are known from a broader phylogenetic survey to cluster with their congeners included in this study (E.E. Schirtzinger, unpublished data). GenBank sequences or published literature was used to confirm gene order for Cyanoramphus auriceps, Strigops habroptilus, Amazona ochrocephala, Amazona farinosa, Pionus chalcopterus, Agapornis roseicollis, Melopsittacus undulatus, and Psittacus erithacus (see Table 2 and Boon, 2000). The control region status of Chalcopsitta duivenbodei and Charmosyna papou could not be confirmed due to poor sequencing reactions.
Within the Neotropical parrots, a variation of the Amazona gene order was observed in Deroptyus accipitrinus and Pionites leucogaster. In contrast to all of the other species with control region duplications that we sequenced, Deroptyus accipitrinus and Pionites leucogaster retain a potentially functional copy of tRNAPro before the non-coding region 5′ to the first control region.
3.4. Ancestral state reconstruction
To determine if mitochondrial control region duplications in parrots originated multiple times, the character states of single (0) or duplicated control region (1) were mapped onto the Bayesian tree using parsimony and maximum likelihood methods (Fig. 5). These states were assigned using our PCR classifications (68 single control region, 35 duplicated control region, three unscorable/ambiguous), sequences from selected taxa (six single control region and three duplicated control region), GenBank sequences (one single control region), or publications (one single control region). Both methods of reconstruction identified the ancestral control region state in parrots as a single control region, and indicated that control region duplications have originated at least six times (Clades A–F in Fig. 5). No reversions from a duplicated control region to a single control region state were reconstructed by either method. Likelihood reconstructions using the symmetric rate (MK1) model and the asymmetric rate (AsymmMK) model did not affect the number of reconstructed independent origins or result in considerable differences in likelihoods of states at interior nodes. Similarly, changing the transition rate between states in the AsymmMK model did not affect our conclusions (See Fig. 3S and Table 6S for proportional likelihood values of each reconstructed state for interior nodes for each model analyzed).
Fig. 5.
Ancestral state reconstruction of the parrot mitochondrial control region duplications on the Bayesian tree under the maximum likelihood criterion using the MK1 model. White circles indicate species classified as having a single mitochondrial control region by the PCR fragment length analysis and/or sequencing. Black circles indicate species classified as having a duplicated mitochondrial control region. Gray circles indicate an unscorable state at the terminals and ambiguous ancestral states at interior nodes. The circles at nodes toward the interior of the tree are representative of the likelihood of each state at that node. * = GenBank sequences, ^ = species sequenced at BR, ° = related species sequenced at BR, + = species sequenced at NMSU and α = species that have published gene orders. Letters to the right indicate clades defined by a single origin event.
3.5. Comparison of control regions
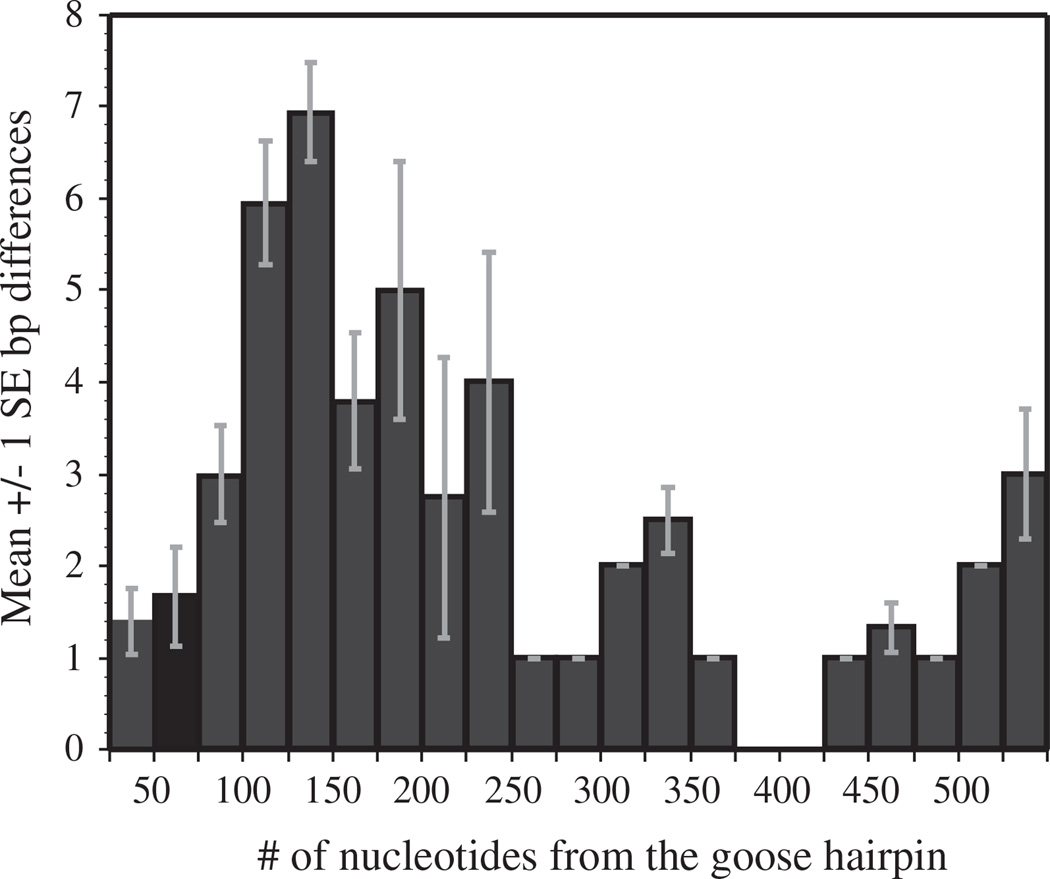
To determine if either of the two control regions had degenerated in species with a duplicated control region, the two control region fragments were aligned and nucleotide differences were calculated. These alignments found that the two control region sequences were typically highly similar, with sequence divergences ranging from 0–10.9% between the two copies within an individual. Most nucleotide differences were found between 51 and 225 nucleotides from the goose hairpin (Fig. 6). In domain I the only conserved sequence of known function is the termination-associated sequence (Baker and Marshall, 1997; Quinn, 1997; Sbisa et al., 1997). Graydidascalus brachyurus was not included in these calculations because neither of its control regions contained a goose hairpin.
Fig. 6.
Means (±1 standard error) for nucleotide differences in 25 bp non-overlapping windows of aligned duplicated control regions. The figure shows that the greatest number of nucleotide differences occurs 51–225 bp from the goose hairpin in domain I of the control region.
4. Discussion
We reconstructed the phylogenetic relationships of 117 parrot species and classified their mitochondrial control region state from PCR fragment length analysis, DNA sequences or GenBank accessions to investigate the origins and distribution of mitochondrial control region duplications within the order Psittaciformes. A total of 76 parrot species were determined to have a single control region, while 38 parrot species were determined to have a duplicated control region. One species was unscorable and two species produced ambiguous results for which no sequence confirmation was available. Mapping the control region states onto the resulting phylogeny identified at least six independent origins of the duplicated control region state. Below we discuss the implications of these results for parrot evolutionary relationships and present two alternative hypotheses for the evolution of mtDNA duplications in parrots.
4.1. Parrot phylogeny
The phylogeny reconstructed in this study is the most taxonomically comprehensive phylogeny to date of the Psittaciformes. The Bayesian tree and the maximum likelihood tree are generally well resolved and largely congruent in topology. Disagreement between the two analyses occurs solely on the location of the genus Micropsitta and Graydidascalus brachyurus (Fig. 2S and Fig. 3). The Bayesian phylogeny is broadly consistent with other published studies of parrot genus level relationships, in which a clade composed of the New Zealand endemics, Strigops and Nestor, was the sister group to all other parrots, and the Cacatuoidea (cockatoos) was the second oldest extant clade (Tavares et al., 2006; Wright et al., 2008; Schweizer et al., 2010; Joseph et al., 2012). Other well-supported parrot clades consistently recovered across various studies include the Neotropical parrots (Arinae), the African Psittacinae, the Australasian Psittaculidae and the Platycercinae from Australia, New Zealand, Oceania and Africa (de Kloet and de Kloet, 2005; Juniper and Parr, 1998; Tavares et al., 2006; Wright et al., 2008; Schweizer et al., 2010; Joseph et al., 2012). In agreement with previous studies by Tavares et al. (2006) and Wright et al. (2008), three Neotropical clades were recovered here: the parrotlets, including Bolborhynchus, Nannopsittaca, Touit, and Psilopsiagon; amazons and allies, including Amazona, Pionus, Pyrilia, Triclaria and Graydidascalus; and macaws and allies, including Ara, Cyanopsitta, Aratinga, Orthopsittaca, Pyrrhura, Pionites and Anodorhynchus.
4.2. Mitochondrial gene order in parrots
Among the 114 parrot species for which data was available there is evidence for two of the four described avian mitochondrial gene orders. The typical avian mitochondrial gene order originally described by Desjardins and Morais (1990) was inferred for 76 species by PCR, sequencing and examination of GenBank sequences. In contrast, duplicate control regions and the gene order described in Amazona parrots by Eberhard et al. (2001) was found in 38 of the species surveyed and/or sequenced. In this genome arrangement, a non-coding region, that in some species has apparent similarity to ND6 and tRNAGlu, is located between tRNAThr and the first control region. Two species, Deroptyus accipitrinus and Pionites leucogaster, which was not in the phylogeny, were shown by sequencing to have a variant of the Amazona gene order, with a functional tRNAPro located between tRNAThr and the first control region.
4.3. Evolutionary patterns of mtDNA control region duplications
Three striking patterns are apparent in our reconstructions of the evolution of control region duplications and sequencing of duplicate control regions in parrots. First, the mapping of control region duplication states onto the phylogeny of parrots identifies at least six independent origins of the control region duplications. Second, there were no reversions to a single control region state in any of these six clades. Third, although there was considerable interspecific variation, levels of sequence similarity between duplicated control regions within an individual were typically high (89–100% similarity in the species examined), at least for the first 400–500 nucleotides of domain I that was examined. This similarity would appear to be unusual due to the fact that this segment of the control region is often very different between species with many small insertions, deletions and mutations that are thought to occur as a result of the D-loop being single-stranded and accessible to mutagenic agents, such as reactive oxygen species (Shokolenko et al., 2007).
One hypothesis to explain these patterns is that a duplication of the control region and neighboring sequences occurred in the ancestors of these six clades and was retained in all descendent species of each clade. This hypothesis begs the question of what maintains the generally high degree of sequence similarity between the duplicated control regions within each taxon. The within-individual divergences that we observed between control region copies of 0–10.9% (Table 3) fall within the range of divergences observed in other taxa with duplicated control regions such as Amazona parrots, albatrosses, killifish, snakes, ticks, and ostracods (Abbott et al., 2005; Campbell and Barker, 1999; Eberhard et al., 2001; Kumazawa et al., 1996, 1998; Lee et al., 2001; Ogoh and Ohmiya, 2007; Tatarenkov and Avise, 2007). A high degree of sequence similarity is often interpreted as evidence for the maintenance of function in both duplicated control regions. Portions of the control region are under selection for the ability to bind with nuclear-encoded replication factors, and for functional control of replication and transcription (Doda et al., 1981; Gensler et al., 2001; He et al., 2007; Lee and Clayton, 1998; Schultz et al., 1998; Shadel and Clayton, 1997); such functionality may provide stabilizing selection on duplicated control regions. Alternatively, several studies of organisms with a duplicated control region have explained the high degree of similarity between the two control regions as evidence of gene conversion (Eberhard et al., 2001; Kumazawa et al., 1996, 1998; Ogoh and Ohmiya, 2007, Tatarenkov and Avise, 2007; Verkuil et al., 2010), but the molecular mechanisms responsible remain unclear. In either case, the maintenance of duplicated control regions concurrent with the elimination of duplicated mitochondrial genes and tRNAs suggests an advantage to having a second control region that overrides selection for compactness. Potential advantages include faster replication (Kumazawa et al., 1996) or protection against age-related deterioration of mitochondrial function (T.F. Wright and J.R. Eberhard, unpublished data).
Table 3.
Gene order, non-coding region length and percent control region differences for parrot species with a duplicated control region.
| Taxa | Source | Cladea | Gene order | Non-coding region lengthb | %CR differencesc |
|---|---|---|---|---|---|
| Amazona albifrons | NMSU | A | Amazona | 141 | 0.6 |
| Amazona farinosa | AF338821 | A | Amazona | 158 | 0 |
| Amazona ochrocephala | NMSU | A | Amazona | 127 | 2.1 |
| Pyrilia caica | NMSU | A | Amazona | 155 | 0 |
| Pyrilia barrabandi | BR/NMSU | A | Amazona | 160 | 6 |
| Pionus chalcopterus | NMSU/AF338817-18 | A | Amazona | 180 | 0.2 |
| Triclaria malachitacea | BR | A | Amazona | 112 | 2.5 |
| Deroptyus accipitrinus | BR | B | Variant A | 319 | 4.7 |
| Pionites leucogaster | BR | B | Variant A | 325 | 10.9 |
| Forpus xanthopterygius | BR | C | Amazona | 129 | 0.2 |
| Forpus sclateri | NMSU | C | Amazona | 117 | 5 |
| Poicephalus robustus | NMSU | D | Amazona | 129 | 7.7 |
| Psittacus erithacus | NMSU | D | Amazona | 88 | 10.8 |
| Prioniturus montanus | NMSU | E | Amazona | 360 | 1.9 |
| Chalcopsitta cardinalis | NMSU | F | Amazona | 84 | 7.9 |
| Charmosyna placentis | NMSU | F | Amazona | 45 | 2.7 |
| Eos histrio | NMSU | F | Amazona | 84 | 2.3 |
| Glossopsitta porphyrocephala | NMSU | F | Amazona | 68 | 3.3 |
| Lorius albidinucha | NMSU | F | Amazona | 73 | 3.3 |
| Melopsittacus undulatus | NC009134 | F | Amazona | 58 | 4.6 |
| Neopsittacus musschenbroekii | NMSU | F | Amazona | 88 | 3.3 |
| Phigys solitarius | NMSU | F | Amazona | 32 | 2.7 |
| Psitteuteles goldiei | NMSU | F | Amazona | 81 | 2.6 |
| Pseudeos fuscata | NMSU | F | Amazona | 71 | 3.5 |
| Trichoglossus haematodus | NMSU | F | Amazona | 86 | 3.3 |
| Vini australis | NMSU | F | Amazona | 58 | 3.5 |
| Cyclopsitta diophthalma | NMSU | F | Amazona | 62 | 5.1 |
| Psittaculirostris edwardsii | NMSU | F | Amazona | 60 | 5.5 |
Clades labeled in Fig. 5.
Non-coding region length was calculated as the number of nucleotides from the 3′ end of tRNATHR to the 5′ end of the goose hairpin.
Percent control region differences was calculated as the number of nucleotides that differed between the 2 aligned control regions divided by the total number of overlapping nucleotides multiplied by 100. Differences were only counted if they occurred 3′ of the goose hairpin.
An alternative hypothesis for the observed patterns is that a propensity for duplications to occur such as through replication slippage due to secondary structure in the control region was present in the common ancestor of each of the six clades, leading to repeated duplications of the region from cytochrome b to the control region with lineage specific degradation and deletion events passing to each of the descendent taxa (Verkuil et al., 2010; Zhuang and Cheng, 2010). Zhuang and Cheng (2010) found a similar pattern within Notothenioid fish and suggested that within each clade with control region duplications if these mutations are neutral, each descendant species of a specific ancestor should have approximately the same amount of divergence between its two control regions. Table 3 shows that while species within the Australasian clade (labeled F in Fig. 5) exhibit similar levels of divergence between their duplicated control regions, the other clades show considerable variation among member species in the degree of divergence exhibited between their two control regions. Further investigation into patterns of sequence divergence over the entire length of the duplicated control region, coupled with functional studies of mitochondrial replication in species with and without control region duplications, should help distinguish between these alternative hypotheses for the evolution of duplicate control regions.
5. Conclusions
The presence of multiple mitochondrial gene orders within Psittaciformes supports the idea that the avian mitochondrial genome is a dynamic molecule. This study has shown that mitochondrial control region duplications have occurred many times in parrots, with ancestral state reconstructions suggesting six independent origins of the duplicated control region state and no reversions to a single control region state. Further investigations into the fates of duplicated mitochondrial genes, the potential costs and advantages of having a second control region, and the complex relationship between evolutionary rates, selection and time since duplication are needed to fully explain these patterns in the mitochondrial genome.
Supplementary Material
Acknowledgments
We thank the following institutions and people for the generous loan of tissue specimens: American Museum of Natural History (P. Sweet), Academy of Natural Sciences of Philadelphia (L. Joseph), the African Safari Zoo, the Australian National Wildlife Collection (L. Joseph), the Field Museum of Natural History (D. Willard), Fundação Crax, University of Kansas Museum of Natural History (M. Robbins), Loro Parque Fundación, Louisiana State University Museum of Natural Science (R. Brumfield), Museu Paraense Emílio Goeldi, Parque Ecológico do Tietê, the San Diego Zoological Park (N. Lamberski), Universidade Estadual Paulista (UNESP), the US National Museum of Natural History, the New Mexico State University Vertebrate Museum (P. Houde), Zoológico de Sorocaba, Zoológico de Americana, Zoológico Cyro-Gevaerd and several breeders. We particularly thank David Waugh, Sara Cappelli and Julia Scharpegge of the Loro Parque Fundación and Tania Matsumoto for generous assistance in obtaining data from critical specimens. We thank Rogério Lourenço for providing some of the sequences, Sergio L. Pereira for suggestions, and Elizabeth Moseman for many useful comments on the manuscript. Research support was provided by NIH grant S06 GM008136 (TFW), Sigma Xi Grant-in-Aid-of-Research (EES), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (EST), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (EST) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (EST). CYM has a CNPq research productivity fellowship. This study was approved by the Institutional Animal Care and Use Committee of New Mexico State University.
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ympev.2012.04.009.
References
- Abbott CL, Double MC, Trueman JWH, Robinson A, Cockburn A. An unusual source of apparent mitochondrial heteroplasmy: duplicate mitochondrial control regions in Thalassarche albatrosses. Mol. Ecol. 2005;14:3605–3613. doi: 10.1111/j.1365-294X.2005.02672.x. [DOI] [PubMed] [Google Scholar]
- Arndt A, Smith MJ. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol. Biol. Evol. 1998;15:1009–1016. doi: 10.1093/oxfordjournals.molbev.a025999. [DOI] [PubMed] [Google Scholar]
- Attardi G. Animal mitochondrial DNA: an extreme example of genetic economy. Int. Rev. Cyt. 1985;93:93–141. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- Baker AJ, Marshall HD. Mitochondrial control region sequences as tools for understanding evolution. In: Mindell DP, editor. Avian Molecular Evolution and Systematics. San Diego: Academic Press; 1997. pp. 51–83. [Google Scholar]
- Bensch S, Härlid A. Mitochondrial genomic rearrangements in songbirds. Mol. Biol. Evol. 2000;17:107–113. doi: 10.1093/oxfordjournals.molbev.a026223. [DOI] [PubMed] [Google Scholar]
- Black WC, Roehrdanz RL. Mitochondrial gene order is not conserved in arthropods: prostriate and metastriate tick mitochondrial genomes. Mol. Biol. Evol. 1998;15:1772–1785. doi: 10.1093/oxfordjournals.molbev.a025903. [DOI] [PubMed] [Google Scholar]
- Boon WM. Ph.D. dissertation. Wellington, Australia: Victoria University of Wellington; 2000. Molecular systematics and conservation of the Cyanoramphus parakeet complex and the evolution of parrots. [Google Scholar]
- Boore JL. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. In: Sankoff D, Nadeau JH, editors. Comparative Genomes: Empirical and Analytical Approaches to Gene Order Dynamics, Map Alignment and the Evolution of Gene Families. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 133–148. [Google Scholar]
- Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruford M, Hanotte O, Brookfield JY, Burke T. Single-locus and multilocus DNA fingerprinting. In: Hoelzel AR, editor. Molecular Genetic Analysis of Populations: A Practical Approach. Oxford, UK: IRL Press; 1992. pp. 225–269. [Google Scholar]
- Campbell NJH, Barker SC. The novel mitochondrial gene arrangement of the cattle tick, Boophilus microplus : fivefold tandem repetition of a coding region. Mol. Biol. Evol. 1999;16:732–740. doi: 10.1093/oxfordjournals.molbev.a026158. [DOI] [PubMed] [Google Scholar]
- Chesser RT, Banks RC, Barker FK, Cicero C, Dunn JL, Kratter AW, Lovette IJ, Rasmussen PC, Remsen JV, Jr, Rising JD, Stotz DF, Winker K. Fifty-first supplement to the American Ornithologists’ Union check-list of North American Birds. The Auk. 2010;127:726–744. [Google Scholar]
- Cho H-J, Eda M, Nishida S, Yasukochi Y, Chong J-R, Koike H. Tandem duplication of mitochondrial DNA in the black-faced spoonbill, Platalea minor Genes Genet. Syst. 2009;84:297–305. doi: 10.1266/ggs.84.297. [DOI] [PubMed] [Google Scholar]
- de Kloet RS, de Kloet SR. The evolution of the spindlin gene in birds: sequence analysis of an intron of the spindlin W and Z gene reveals four major divisions of the Psittaciformes. Mol. Phylogenet. Evol. 2005;36:706–721. doi: 10.1016/j.ympev.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Desjardins P, Morais R. Sequence and gene organization of the chicken mitochondrial genome: a novel gene order in higher vertebrates. J. Mol. Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Diaz F, Bayona-Bafaluy MP, Rana M, Mora M, Hao H, Moraes CT. Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 2002;30:4626–4633. doi: 10.1093/nar/gkf602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doda JN, Wright CT, Clayton DA. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl. Acad. Sci. USA. 1981;78:6116–6120. doi: 10.1073/pnas.78.10.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard JR, Wright TF, Bermingham E. Duplication and concerted evolution of the mitochondrial control region in the parrot genus Amazona. Mol. Biol. Evol. 2001;18:1330–1342. doi: 10.1093/oxfordjournals.molbev.a003917. [DOI] [PubMed] [Google Scholar]
- Ericson PGP, Anderson CL, Britton T, Elzanowski A, Johansson US, Källersjö M, Ohlson JI, Parsons TJ, Zuccon D, Mayr G. Diversification of Neoaves: integration of molecular sequence data and fossils. Biol. Lett. 2006;2:543–547. doi: 10.1098/rsbl.2006.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fain M, Houde P. Parallel radiations in the primary clades of birds. Evolution. 2004;58:2558–2573. doi: 10.1111/j.0014-3820.2004.tb00884.x. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y-L, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshaw JM. Parrots of the World: An Identification Guide. Princeton, NY: Princeton University Press; 2006. [Google Scholar]
- Fujita MK, Boore JL, Moritz C. Multiple origins and rapid evolution of duplicated mitochondrial genes in parthenogenetic geckos (Heteronotia binoei ; Squamata Gekkonidae) Mol. Biol. Evol. 2007;24:2775–2786. doi: 10.1093/molbev/msm212. [DOI] [PubMed] [Google Scholar]
- Gensler S, Weber K, Schmitt WE, Perez-Martos A, Enriquez JA, Montoya J, Wiesner RJ. Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res. 2001;29:3657–3663. doi: 10.1093/nar/29.17.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol. Biol. Evol. 2007;24:269–280. doi: 10.1093/molbev/msl158. [DOI] [PubMed] [Google Scholar]
- Gray MW. Origin and evolution of mitochondrial DNA. Annu. Rev. Cell Biol. 1989;5:25–50. doi: 10.1146/annurev.cb.05.110189.000325. [DOI] [PubMed] [Google Scholar]
- Hackett S, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han K-L, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Haddrath O, Baker AJ. Complete mitochondrial DNA genome sequences of extinct birds: ratite phylogenetics and the vicariance biogeography hypothesis. Proc. R. Soc. Lond. B. 2001;268:939–945. doi: 10.1098/rspb.2001.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Mao C-C, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, Reichelt S, Spelbrink JN, Walker, Holt IJ. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Toon A, Schirtzinger EE, Wright TF, Schodde R. A revised nomenclature and classification for family-group taxa of parrots (Psittaciformes) Zootaxa. 2012;3205:26–40. [Google Scholar]
- Juniper T, Parr M. Parrots: A Guide to Parrots of the World. New Haven, CT: Yale University Press; 1998. [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T. Gene rearrangements in snake mitochondrial genomes: highly concerted evolution of control-region-like sequences duplicated and inserted into a tRNA gene cluster. Mol. Biol. Evol. 1996;13:1242–1254. doi: 10.1093/oxfordjournals.molbev.a025690. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T. The complete nucleotide sequence of a snake (Dinodon semicarinatus) mitochondrial genome with two identical control regions. Genetics. 1998;150:313–329. doi: 10.1093/genetics/150.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DU, Clayton DA. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem. 1998;273:30614–30621. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- Lee JS, Miya M, Lee YS, Kim CG, Park EH, Aoki Y, Nishida M. The complete DNA sequence of the mitochondrial genome of the self-fertilizing fish Rivulus marmoratus (Cyprinodontiformes, Rivulidae) and the first description of duplication of a control region in fish. Gene. 2001;280:1–7. doi: 10.1016/s0378-1119(01)00765-x. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Kocher TD. Complete sequence of a sea lamprey (Petromyzon marinus) mitochondrial genome: early establishment of the vertebrate genome organization. Genetics. 1995;139:873–887. doi: 10.1093/genetics/139.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt DH, Hyman BC. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- Lynch M, O’Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macey JR, Larson A, Ananjeva NB, Fang Z, Papenfuss TJ. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 1997;14:91–104. doi: 10.1093/oxfordjournals.molbev.a025706. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.01. 2007 < http://www.mesquiteproject.org>. [Google Scholar]
- Madsen CS, Ghivizzani SC, Hauswirth WW. Protein-binding to a single termination-associated sequence in the mitochondrial DNA D-loop region. Mol. Cell. Biol. 1993;13:2162–2171. doi: 10.1128/mcb.13.4.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell DP, Sorenson MD, Dimcheff DE. Multiple independent origins of mitochondrial gene order in birds. Proc. Natl. Acad. Sci. USA. 1998;95:10693–10697. doi: 10.1073/pnas.95.18.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Pocock JA, Taylor SA, Birt TP, Friesen VL. Concerted evolution of duplicated mitochondrial control regions in three related seabird species. BMC Evol. Biol. 2010;10:14. doi: 10.1186/1471-2148-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortiz C. Evolutionary dynamics of mitochondrial DNA duplications in parthenogenetic geckos, Heteronotia binoei. Genetics. 1991;129:221–230. doi: 10.1093/genetics/129.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Brown WM. Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc. Natl. Acad. Sci. USA. 1987;84:7183–7187. doi: 10.1073/pnas.84.20.7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RL, Boore JL. Molecular mechanisms of extensive mitochondrial gene rearrangment in plethodontid salamanders. Mol. Biol. Evol. 2005;22:2104–2112. doi: 10.1093/molbev/msi204. [DOI] [PubMed] [Google Scholar]
- Müller K. Seqstate: primer design and sequence statistics for phylogenetic DNA datasets. Appl. Bioinf. 2005;4:65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]
- Müller K. Incorporating information from length-mutational events into phylogenetic analysis. Mol. Phylogenet. Evol. 2006;38:667–676. doi: 10.1016/j.ympev.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Ogoh K, Ohmiya Y. Complete mitochondrial DNA sequence of the sea-firefly, Vargula hilgendorfii (Crustacea, Ostracoda) with duplicate control regions. Gene. 2004;327:131–139. doi: 10.1016/j.gene.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Ogoh K, Ohmiya Y. Concerted evolution of duplicated control regions within an ostracod mitochondrial genome. Mol. Biol. Evol. 2007;24:74–78. doi: 10.1093/molbev/msl132. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York, NY: Springer-Verlag; 1970. [Google Scholar]
- Quinn TW. Molecular evolution of the mitochondrial genome. In: Mindell DP, editor. Avian Molecular Evolution and Systematics. San Diego, CA: Academic Press; 1997. pp. 4–29. [Google Scholar]
- Quinn TW, Wilson AC. Sequence evolution in and around the mitochondrial control region in birds. J. Mol. Evol. 1993;37:417–425. doi: 10.1007/BF00178871. [DOI] [PubMed] [Google Scholar]
- Quinn TW, Mindell DP. Mitochondrial gene order adjacent to the control region in crocodile, turtle and tuatara. Mol. Phylogenet. Evol. 1996;5:344–351. doi: 10.1006/mpev.1996.0029. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4. 2007 < http://www.beast.bio.ed.ac.uk/Tracer>. [Google Scholar]
- Rand DM. Endotherms, ectotherms and mitochondrial genome-size variation. J. Mol. Evol. 1993;37:281–295. doi: 10.1007/BF00175505. [DOI] [PubMed] [Google Scholar]
- Rand DM. The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Syst. 2001;32:415–448. [Google Scholar]
- Rand DM, Harrison RG. Mitochondrial DNA transmission genetics in crickets. Genetics. 1986;114:955–970. doi: 10.1093/genetics/114.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 2005;5:28. doi: 10.1186/1471-2148-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remsen JV, JV, Cadena CD, Jaramillo A, Nores M, Pacheco JF, Robbins MB, Schulenberg TS, Stiles FG, Stotz DF, Zimmer KJ. A classification of the bird species of South America Version 7. American Ornithologists’ Union. 2010 < http://www.museum.lsu.edu/~Remsen/SACCBaseline.html>. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roth C, Rastogi S, Arvestad L, Dittmar K, Light S, Ekman D, Liberles DA. Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. J. Exp. Zool. (Mol. Dev. Evol.) 2007;308B:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- Sammler S, Bleidorn C, Tiedemann R. Full mitochondrial genomes of two endemic Philippine hornbill species (Aves: Bucerotidae) provide evidence for pervasive mitochondrial DNA recombination. BMC Genomics. 2011;12:35. doi: 10.1186/1471-2164-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Mauro D, Gower DJ, Zardoya R, Wilkinson M. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol. Biol. Evol. 2006;23:227–234. doi: 10.1093/molbev/msj025. [DOI] [PubMed] [Google Scholar]
- Sbisa E, Tanzariello F, Reyes A, Pesole G, Saccone C. Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and the functional and evolutionary implications. Gene. 1997;205:125–140. doi: 10.1016/s0378-1119(97)00404-6. [DOI] [PubMed] [Google Scholar]
- Schultz RA, Swoap SJ, McDaniel LD, Zhang B, Koon EC, Garry DJ, Li K, Williams RS. Differential expression of mitochondrial DNA replication factors in mammalian tissues. J. Biol. Chem. 1998;273:3447–3451. doi: 10.1074/jbc.273.6.3447. [DOI] [PubMed] [Google Scholar]
- Schweizer M, Seehausen O, Güntert M, Hertwig ST. The evolutionary diversification of parrots supports a taxon pulse model with multiple transoceanic dispersal events and local radiations. Mol. Phylogenet. Evol. 2010;54:984–994. doi: 10.1016/j.ympev.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Selosse M-A, Albert B, Godelle B. Reducing the genome size of organelles favors gene transfer to the nucleus. Trends Ecol. Evol. 2001;16:135–141. doi: 10.1016/s0169-5347(00)02084-x. [DOI] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Ann. Rev. Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- Shao R, Barker SC. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. [DOI] [PubMed] [Google Scholar]
- Shao R, Barker SC, Mitani H, Aoki Y, Fukunaga M. Evolution of duplicate control regions in the mitochondrial genomes of Metazoa: a case study with Australasian Ixodes ticks. Mol. Biol. Evol. 2005;22:620–629. doi: 10.1093/molbev/msi047. [DOI] [PubMed] [Google Scholar]
- Shokolenko IN, Ledoux SP, Wison GL. Mitochondrial DNA damage and repair. In: Schaffer SW, Suleiman M-S, editors. Mitochondria: The Dynamic Organelle. New York, NY: Springer; 2007. pp. 323–348. [Google Scholar]
- Sibley C, Ahlquist J. Phylogeny and Classification of Birds. New Haven, CT: Yale University Press; 1990. [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 2000;49:369–381. [PubMed] [Google Scholar]
- Sogin ML. History assignment: when was the mitochondrion founded? Curr. Opin. Genetics Dev. 1997;7:792–799. doi: 10.1016/s0959-437x(97)80042-1. [DOI] [PubMed] [Google Scholar]
- Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol. Phylogenet. Evol. 1999;12:105–114. doi: 10.1006/mpev.1998.0602. [DOI] [PubMed] [Google Scholar]
- Stanton DJ, Daehler LL, Moritz CC, Brown WM. Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics. 1994;137:233–241. doi: 10.1093/genetics/137.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarenkov A, Avise JC. Rapid concerted evolution in animal mitochondrial DNA. Proc. R. Soc. B. 2007;264:1795–1798. doi: 10.1098/rspb.2007.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares ES, Baker AJ, Pereira SL, Miyaki CY. Phylogenetic relationships and historical biogeography of Neotropical parrots (Psittaciformes: Psittacidae: Arini) inferred from mitochondrial and nuclear DNA sequences. Syst. Biol. 2006;55:454–470. doi: 10.1080/10635150600697390. [DOI] [PubMed] [Google Scholar]
- Verkuil YI, Piersma T, Baker AJ. A novel mitochondrial gene order in shorebirds (Scolopacidae, Charadriiformes) Mol. Phylogent. Evol. 2010;57:411–416. doi: 10.1016/j.ympev.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Wright TF, Schirtzinger EE, Matsumoto T, Eberhard JR, Graves GR, Sanchez JJ, Capelli S, Müller H, Scharpegge J, Chambers GK, Fleischer RC. A multilocus molecular phylogeny of the parrots (Psittaciformes): support for a Gondwanan origin during the Cretaceous. Mol. Biol. Evol. 2008;25:2141–2156. doi: 10.1093/molbev/msn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: AN update. Trends Ecol. Evol. 2003;18:292–298. [Google Scholar]
- Zhuang X, Cheng C-H. ND6 gene “lost” and found: evolution of mitochondrial gene rearrangement in Antarctic Notothenoids. Mol. Biol. Evol. 2010;27:1391–1403. doi: 10.1093/molbev/msq026. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. Ph.D. dissertation. Austin, Texas: The University of Texas at Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.