Abstract
Murine γ-herpesvirus 68 (MHV-68) infection of Mus musculus-derived strains of mice is an established model of γ-herpesvirus infection. We have previously developed an alternative system using a natural host, the wood mouse (Apodemus sylvaticus), and shown that the MHV-68 M3 chemokine-binding protein contributes significantly to MHV-68 pathogenesis. Here we demonstrate in A. sylvaticus using high-density micro-arrays that M3 influences the expression of genes involved in the host response including Scgb1a1 and Bpifa1 that encode potential innate defense proteins secreted into the respiratory tract. Further analysis of MHV-68-infected animals showed that the levels of both protein and RNA for SCGB1A1 and BPIFA1 were decreased at day 7 post infection (p.i.) but increased at day 14 p.i. as compared with M3-deficient and mock-infected animals. The modulation of expression was most pronounced in bronchioles but was also present in the bronchi and trachea. Double staining using RNA in situ hybridization and immunohistology demonstrated that much of the BPIFA1 expression occurs in club cells along with SCGB1A1 and that BPIFA1 is stored within granules in these cells. The increase in SCGB1A1 and BPIFA1 expression at day 14 p.i. was associated with the differentiation of club cells into mucus-secreting cells. Our data highlight the role of club cells and the potential of SCGB1A1 and BPIFA1 as innate defense mediators during respiratory virus infection.
Murine γ-herpesvirus 68 (MHV-68, Murid herpesvirus 4 (MuHV-4)) infection of laboratory mice is a tractable and widely used small-animal model of γ-herpesvirus infection,1, 2, 3, 4, 5, 6, 7 specifically for the human pathogens Epstein–Barr virus and Kaposi's sarcoma-associated herpesvirus. Laboratory mice (M. musculus) are not a natural host for MHV-68.8, 9, 10, 11 So, to define host–pathogen interactions in an authentic system, we have developed infection of wood mice (A. sylvaticus), which are a natural host of MHV-68, as an alternative model.12 The host response to MHV-68 in wood mice differs from that seen in laboratory mice. Intra-nasal infection of wood mice with MHV-68 leads to productive replication in lung epithelial cells and in macrophages within granulomatous infiltrates that peaks at day 7 post infection (p.i.) and is resolved by day 21 p.i.12 This is followed by a latent infection of B cells in the spleen and in the lung within perivascular and peribronchiolar lymphocyte accumulations that leads to the formation of iBALT. The expansion of latently infected B cells peaks at day 14 p.i. but MHV-68 then establishes a latent infection in the spleen and lung that persists long term.12 The M3 gene of MHV-68 encodes a viral chemokine-binding protein, known to bind a range of chemokines in vitro; however, its in vivo function could not be discerned when using laboratory mice that are a non-natural host.13, 14 Our previous experiments in wood mice using an MHV-68 mutant deficient in M3 showed that although M3 is not essential for infection, there is a marked alteration in the cellular response to the M3 mutant. Specifically, in wood mice infected with MHV-68 lacking M3, there is an alteration in the chemokine and cytokine environment, loss of the B-cell-dominated infiltrate in lungs at day 7 p.i., absence of iBALT formation at day 14 p.i., and a significantly reduced latent infection,15 further highlighting the significance of our wood mouse system.
The airway epithelium secretes multiple proteins that function in innate defense. Two highly expressed proteins that are thought to have this role are secretoglobin, family 1A, member 1 (SCGB1A1; also called uteroglobin, club (Clara) cell secretory protein or CC10) and BPI fold-containing family A1 (BPIFA1; also called SPLUNC1).16, 17, 18, 19, 20 SCGB1A1 is produced by non-ciliated epithelial, ie, club cells (Clara cells) in the airways.16 The precise role of SCGB1A1 has not been clearly defined and is likely to be multifactorial. However, Scgb1a1−/− mice show both increased viral loads and increased pulmonary inflammation following respiratory syncytial virus infection21 and increased inflammatory, cytokine, and chemokine responses following infection with an E1- and E3-deleted adenoviral vector.22
BPIFA1 is found in the respiratory epithelium and submucosal glands of the upper airways and in the salivary glands in mice and humans.17, 18, 19, 23, 24, 25 The interspecies diversity and rapid evolution of BPIF genes26, 27 suggest a role for the proteins in host defense. BPIFA1 has a surfactant-like function28 and is involved in the regulation of the amiloride-sensitive epithelial sodium channel, ENaC.29 Infection with Mycoplasma spp. induces Bpifa1 expression in murine airways30 and, importantly, BPIFA1 enhances IL-8 production and bacterial clearance.30 Recent data also suggest that the protein is important in the defense against Klebsiella pneumoniae infection31 and acts through modulation of macrophage function.32
As part of a study to identify transcriptional signatures associated with the MHV-68 M3 protein during infection, we identified a modulation of Scgb1a1 and Bpifa1 expression. Here we describe the temporal and spatial changes in the expression of these two genes and their products during MHV-68 infection of wood mice.
METHODS
Cell Culture and Virus
Stocks of MHV-68, clone g2.4,33 and previously published, genetically engineered mutant MHV-68 viruses vM3.stop and vM3.MR13 were grown and titrated by infection of baby hamster kidney cells (BHK-21), as previously described.34 vM3.stop contains a stop codon inserted into the M3 locus so as to disrupt the production of M3 protein. vM3.MR is a marker-rescue control virus derived from vM3.stop that expresses M3. BHK-21 cells were maintained in Glasgow's Modified Minimal Essential Medium with 10% newborn calf serum and 10% tryptose-phosphate broth, 2 mM L-glutamine, 70 μg/ml penicillin, and 10 μg/ml streptomycin.
Wood Mice
Animal work was reviewed by the local University of Liverpool ethics committee and performed under UK Home Office Project Licence 40/2483 and personal licence 60/6501. MHV-68-negative, laboratory bred wood mice (Apodemus sylvaticus) were obtained from the established colony at the University of Liverpool, housed, and maintained as previously described.12, 15
Virus Infection
Animals were randomly assigned into multiple cohorts, anesthetized lightly with halothane, and separate cohorts inoculated intra-nasally with either 4 × 105 PFU of MHV-68 or recombinants vM3.stop (M3 deficient) or vM3.MR (marker-rescue control; kind gifts of Samuel Speck, Herbert W. Virgin IV and Victor van Berkel13) in 40 μl sterile PBS, or were mock infected with PBS. They were killed on day 7 or 14 p.i. by cervical dislocation. Lungs and trachea were removed immediately and samples subjected to RNA purification or fixed for histology and transmission electron microscopy (TEM). A sample size of n=3 was used and was determined using power calculations and previous experience of experimental infection with these viruses. Separate cohorts of mice were used for microarray analysis, RNA (quantitative reverse transcriptase-PCR (qRT-PCR)) analysis, histopathology (RNA in situ, immunohistology (IH)), and electron microscopy.
Microarray Analysis
Microarray work was performed by the University of Liverpool Centre for Genomic Research (http://www.liv.ac.uk/genomic-research/). Cohorts of three wood mice were infected with MHV-68 vM3.MR or vM3.stop35 and euthanized on day 14 p.i. Total cellular RNA was purified from the lungs using an RNeasy mini kit (Qiagen). We used one lung sample from each mouse per chip, ie, three biological replicates for each infection group. RNA quality was assessed using a 2100 bioanalyzer (Agilent Technologies) and 5 μg used for array hybridization. RNA was reverse transcribed, amplified in vitro, biotinylated, cRNA fragmented, hybridized to Mouse Genome 430A V2 arrays, and scanned following Affymetrix protocols (http://www.affymetrix.com). Image processing and normalizations were performed using the Affymetrix MAS 5.0 software before input into GeneSpring (Agilent Technologies). The derived signal value was globally normalized and targeted to all probe sets equal to 100 before comparative analysis. Data were normalized per chip and to the 50th percentile. Signal intensities represent the abundance of RNA transcripts. Genes (probe sets) showing greater than twofold change in value and with a P<0.001 with respect to ‘Present' or ‘Absent' flags were chosen as changed genes. Genes whose expression was regulated more than twofold are described in Table 1.
Table 1. Genes whose expression was modulated in the lungs of wood mice infected with vM3.MR compared with those infected with vM3.stop.
| Symbol | Description | Tissue/cell specificity | Biological processa | Ratiob |
|---|---|---|---|---|
| Upregulated in vM3.MR-infected wood mice | ||||
| Immune/inflammatory response | ||||
| Igha | Immunoglobulin heavy constant α | Lymph node, intestine | Mucosal immune response | 2.6 |
| Ighg1 | Immunoglobulin heavy constant gamma 1 | Lymph node | Humoral immune response mediated by circulating Ig | 4.5 |
| Ighg2b | Immunoglobulin heavy constant gamma 2B | Lymph node | Humoral immune response mediated by circulating Ig | 2.4 |
| Ighm | Immunoglobulin heavy constant mu | Lymph node | Humoral immune response mediated by circulating Ig | 2.8 |
| Ighm | Immunoglobulin heavy constant mu | Lymph node | Humoral immune response mediated by circulating Ig | 3 |
| Ighv1–72 | Immunoglobulin heavy variable 1–72 | Lymph node, small intestine | Humoral immune response mediated by circulating Ig | 2.7 |
| Igk-V8 | Immunoglobulin κ chain | B cells, lymph node | Humoral immune response mediated by circulating Ig | 5.7 |
| Igkv8–30 | Immunoglobulin κ chain variable 8–30 | B cells, lymph node | Humoral immune response mediated by circulating Ig | 3.4 |
| Igkc | Immunoglobulin κ constant | B cells, lymph node | B-cell differentiation | 2.4 |
| Oas2 | 2'-5' Oligoadenylate synthetase 2 | Macrophage | Immune response/Rnase L induction | 2 |
| C1qb | Complement component 1, q subcomponent, β polypeptide | Macrophage, microglia, spleen, lymph nodes | Complement activation, classical pathway | 2.1 |
| Itgb6 | Integrin β 6 | Kidney, lung, skeletal muscle | Cell adhesion/signaling/inflammation | 3.1 |
| Cell signaling | ||||
| Dusp1 | Dual specificity phosphatase 1 | Macrophage, lung, dendritic cell | Inactivation of MAPK activity | 2 |
| Rem1 | Rad- and gem-related GTP-binding protein 1 | Myoblasts | Signal transduction | 2 |
| Map3k4 | Mitogen-activated protein kinase kinase kinase 4 | Ubiquitous | Intracellular protein kinase cascade | 4.8 |
| Tprkb | Tp53rk-binding protein | liver, kidney, eye | protein catabolic process | 2.3 |
| Ywhaq | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Embryonal stem cells, neuronal tissue | Signal transduction | 2 |
| Regulation of expression | ||||
| Crx | Cone-rod homeobox-containing gene | Retina | Regulation of transcription | 3.5 |
| Myb | Myeloblastosis oncogene | Myeloid progenitor, T cell | G1/S transition of mitotic cell cycle | 2.2 |
| Bcas2 | Breast carcinoma amplified sequence 2 | Embryonal stem cells | mRNA processing | 2.3 |
| Paip1 | Polyadenylate-binding protein-interacting protein 1 | stem cell, pro-B cell | Positive regulation of translation | 7.4 |
| Cell adhesion/morphogenesis | ||||
| Vcl | Vinculin | Lung, mast cells | Focal adhesion plaque formation | 3 |
| Cdh15 | Cadherin 15 | Muscle | Cell adhesion | 2.6 |
| Npnt | Nephronectin | Lung, kidney | Cell–cell adhesion mediated by integrin | 2.8 |
| Parva | Parvin, α | Osteoblasts, stem cells, lung | Cell adhesion | 2.9 |
| Lung/respiratory tract | ||||
| Bpifa1 | BPI fold-containing family A, member 1 | Lung, heart | 17.6 | |
| Bpifb1 | BPI fold-containing family B, member 1 | Lung, stomach | 7.3 | |
| Scgb1a1 | Secretoglobin, family 1A, member 1 | Lung | 10.3 | |
| Scgb3a2 | Secretoglobin, family 3A, member 2 | Lung | 3.2 | |
| Detoxification | ||||
| Cyp2f2 | Cytochrome P450, family 2, subfamily f, polypeptide 2 | Lung, lacrimal gland, salivary gland, liver | Response to toxinoxidation-reduction process | 2.2 |
| Cyp2a4 | Similar to cytochrome P450, family 2, subfamily a, polypeptide 4 | Lacrimal gland | Oxidation-reduction process | 12.8 |
| Myeloid cell activation | ||||
| Snca | Synuclein, α | Bone marrow, neuronal tissue | Myeloid leukocyte activation | 2.1 |
| Trim10 | Tripartite motif-containing 10 | Bone marrow | Myeloid cell haemopoiesis | 4.5 |
| Ion transport | ||||
| Slc4a1 | Solute carrier family 4 (anion exchanger), member 1 | Bone marrow | Anion transport | 2.2 |
| Lrrc26 | Leucine-rich repeat containing 26 | Salivary gland, lacrimal gland | Ion transport | 2 |
| Miscellaneous | ||||
| Ap2a2 | Adaptor protein complex AP-2, α 2 subunit | CD4 T cell, granulocytes | Endocytosis | 3 |
| Bc1 | Brain cytoplasmic RNA 1 | Translational repressor activity | 2.7 | |
| Mus81 | MUS81 endonuclease homolog (yeast) | Adipose tissue, muscle | DNA repair | 2.1 |
| Mccc2 | Methylcrotonoyl-Coenzyme A carboxylase 2 (β) | Adipose tissue, liver, kidney | Coenzyme A metabolic process | 2 |
| Spa17 | Sperm autoantigenic protein 17 | Testis | Ciliary or flagellar motility | 2.1 |
| Myl4 | Myosin, light polypeptide 4 | Heart, lung | Muscle contraction | 2.5 |
| Agr2 | Anterior gradient homolog 2 | Intestine, lacrymal gland | ER secreory pathway | 4.2 |
| Agr3 | Anterior gradient homolog 3 | Lung, testis | ER secreory pathway | 10 |
| Downregulated in vM3.MR-infected wood mice | ||||
| Regulation of expression | ||||
| Rnpc3 | RNA-binding region (RNP1, RRM) containing 3 | Ubiquitous | Regulation of alternative mRNA splicing, via spliceosome | 2.5 |
| Malat1 | Metastasis-associated lung adenocarcinoma transcript 1 (non-coding RNA) | Adrenal gland | Regulation of alternative mRNA splicing, via spliceosome | 2.2 |
| Malat1 | Metastasis-associated lung adenocarcinoma transcript 1 (non-coding RNA) | Regulation of alternative mRNA splicing, via spliceosome | 3.0 | |
| Nsa2 | NSA2 ribosome biogenesis homolog | B cells, mast cells and macrophages | rRNA processing | 4.3 |
| Ddx6 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | B cells, T cells | Cytoplasmic mRNA processing body assembly | 2.0 |
| C1d | C1D nuclear receptor co-repressor | Myeloid, dendritic cells, osteoblasts | Regulation of transcription | 2.9 |
| Mtf1 | Metal response element-binding transcription factor 1 | Lacrimal gland, testis | Regulation of transcription | 2.7 |
| Ankrd1 | Ankyrin repeat domain 1 (cardiac muscle) | Heart | Regulation of transcription | 2.2 |
| Fubp1 | Far upstream element (FUSE)-binding protein 1 | Embryonal stem cells, T cells, B cells | Regulation of transcription | 6.0 |
| Nfic | Nuclear factor I/C | Muscle, adipose tissue | Regulation of transcription | 3.3 |
| Yap1 | Yes-associated protein 1 | Embryonal Stem cells, placenta | Regulation of transcription | 2.0 |
| Hist1h1c | Histone cluster 1, H1c | Pancreas, intestine, osteoblast | Chromatin organization | 4.0 |
| Srp9 | Signal recognition particle 9 | Lacrimal gland, prostate | Negative regulation of translational elongation | 3.0 |
| Ion transport | ||||
| Slc39a8 | Solute carrier family 39 (metal ion transporter), member 8 | Lung, uterus, cornea | Zinc ion transport | 2.0 |
| Nipal2 | NIPA-like domain containing 2 | Salivary, lacrimal glands | Magnesium ion transport | 5.9 |
| Mfsd7c | Major facilitator superfamily domain containing 7C | Placenta | Haeme transport | 2.0 |
| Sln | Sarcolipin | Macrophage, thymocyte | Regulation of calcium ion transport | 7.3 |
| Atp6v0b | ATPase, H+ transporting, lysosomal V0 subunit B | Macrophage, microglia | Proton transport | 2.0 |
| Cell signaling | ||||
| Rnd3 | Rho family GTPase 3 | Dendritic cell, fibroblast, osteoblast | Small GTPase-mediated signal transduction | 3.0 |
| Calm1 | Calmodulin 1 | Ubiquitous | Response to calcium ion | 7.0 |
| Leprot | Leptin receptor overlapping transcript | Mast cell, macrophage, osteoblast | Negative regulation of JAK-STAT cascade | 2.0 |
| Igbp1 | Immunoglobulin (CD79A)-binding protein 1 | Granulocytes | Negative regulation of stress-activated MAPK cascade | 2.4 |
| Camk1d | Calcium/calmodulin-dependent protein kinase ID | Dendritic cells, neuronal tissue | Protein modification process | 2.0 |
| Immune/inflammatory response | ||||
| Igk-V1 | Immunoglobulin κ chain variable 1 (V1) | Intestine, spleen, B cell | 2.4 | |
| Bcap29 | B-cell receptor-associated protein 29 | B cells, macrophage, testis | Apoptosis/intracellular transport | 3.0 |
| Sod2 | Superoxide dismutase 2, mitochondrial | Macrophage, microglia | Response to reactive oxygen species | 5.0 |
| Retnla | Resistin like α | Adipose tissue, lung | Hormone activity/Inflammatory response | 2.1 |
| Tbk1 | TANK-binding kinase 1 | Macrophage | Positive regulation of interferon-α/β | 2.0 |
| Cell adhesion/morphogenesis | ||||
| Itgav | Integrin α V | Macrophage, osteoblast | Cell adhesion/phagocytosis | 4.8 |
| Sirpa | Signal-regulatory protein α | Macrophage | Phagocytosis | 2.5 |
| Sytl1 | Synaptotagmin-like 1; similar to synaptotagmin-like 1 | Salivary, lacrimal glands | Exocytosis | 4.7 |
| Cyfip1 | Cytoplasmic FMR1-interacting protein 1 | Macrophage, microglia | Cell morphogenesis | 6.8 |
| Cell cycle | ||||
| Cdc123 | Cell division cycle 123 | Ubiquitous | G1 phase of mitotic cell cycle | 6.6 |
| Commd5 | COMM domain containing 5 | Macrophage | Cell cycle arrest | 2.2 |
| Miscellaneous | ||||
| Zdhhc12 | zinc finger, DHHC domain containing 12 | macrophage | 3.0 | |
| Ssfa2 | Sperm-specific antigen 2 | epidermis, mast cells, macrophage | 2.7 | |
| Ndufa2 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 2 | Ubiquitous | Electron transport chain | 2.0 |
| Tpm3 | Tropomyosin 3, γ | NK cells, mast cells | Nervous system development | 2.4 |
| Pgk1 | Phosphoglycerate kinase 1 | Macrophage, skeletal muscle | Glucose catabolic process | 3.8 |
| Bgn | Biglycan | Osteoblast, fibroblast, lung | Extracellular matrix | 11.7 |
| 2700089E24Rik | RIKEN cDNA 2700089E24 gene | Salivary gland, mammary gland | 4.3 | |
| Ckap4 | Cytoskeleton-associated protein 4 | Osteoblast, fibroblast | 2.7 | |
| Ttn | Titin | Heart, mammary gland, skeletal muscle | Sarcomere organization | 3.9 |
| Mb | Myoglobin | Ubiquitous | Oxygen transport | 2.7 |
| Acaa1a | Acetyl-Coenzyme A acyltransferase 1A | Liver, kidney | Fatty acid metabolic process | 4.2 |
| Hsd17b7 | Hydroxysteroid (17-β) dehydrogenase 7 | Ovary, liver, macrophage | Steroid biosynthetic process | 4.8 |
Gene symbol, description, tissue/cell specificity, and Biological Process were annotated using DAVID (http://david.abcc.ncifcrf.gov/)56, 57 and BioGPS (http://biogps.org/).58
Ratio of the normalized signal intensities obtained after Affymetrix genechip analysis for animals infected with vM3.MR vs vM3.stop. Only genes (probe sets) showing greater than twofold change in value and with a P<0.001 with respect to Present or Absent flags are represented.Bolded values are transcripts that were significantly upregulated and the focus of this manuscript.
Cloning and Sequencing of Wood Mouse cDNAs
Total RNA was purified from lung tissue of uninfected A. sylvaticus using the RNeasy Mini Kit (Qiagen) and DNA contamination removed by treating RNA with amplification grade DNase I (Life Technologies) according to the manufacturers' recommendations. Reverse transcription was performed at 50 °C for 30 min with 2 μg RNA in a 20-μl reaction volume containing 200 U Superscript III reverse transcriptase (Invitrogen), 500 ng oligo(dT)15 primer (Roche), 0.5 mM dNTP mix (Promega), 5 mM DTT, 40 U RNase inhibitor (RNaseOUT; Life Technologies) in First-Strand buffer (50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2; Life Technologies). Afterwards, 2 μl was used as template for RT-PCR in 20 μl reaction volumes. The oligodeoxynucleotide primers used for PCR are provided in Table 2. The cycling parameters were initially 95 °C for 10 min, and then for each cycle: 94 °C for 10 s, 60 °C for 20 s, and 72 °C for 60 s. The products were inserted into pCR2.1 using a TOPO-TA cloning kit (Life Technologies) according to the manufacturer's instructions. DNA from these cultures was extracted by alkaline lysis and sequenced commercially (Eurofins MWG, Germany). Six clones were sequenced for each gene and the sequence obtained compared with existing sequences using BLASTN.36 The sequences have been submitted to Genbank and have accession numbers HM008619 (Scgb1a1) and HM008620 (Bpifa1).
Table 2. Primers used for amplification of A. sylvaticus Scgb1a1 and Bpifa1 cDNA for cloning.
| Primer | Gene | Sequence 5′→3′ | Product length (bp) |
|---|---|---|---|
| SCGB1A1-f | Scgb1a1 | CCTCTGGCCTCTACCATGAA | 351 |
| SCGB1A1-r | Scgb1a1 | GACAGGGGCCTTTAGCAGTA | |
| BPIFA1-f | Bpifa1 | ACTCAGACACCAAGAGAGAT | 1011 |
| BPIFA1-r | Bpifa1 | CGTGAGGAGAAGGAAGACAT |
Quantitative Reverse Transcriptase-PCR
qRT-PCR was performed as previously described12 using total RNA purified from the lungs (see above). Each sample was amplified in triplicate and the means from three animals were used and expressed relative to the copy number of the house-keeping gene 60S ribosomal protein L8 (Rpl8) cDNA. The oligodeoxynucleotide primers used for PCR are provided in Table 3.
Table 3. Oligodeoxynucleotide primers used in quantitative RT-PCR.
| Primer | Gene | Sequence 5′→3′ | Product length (bp) |
|---|---|---|---|
| RPL8int-f | Rpl8 | ACAGAGCCGTTGTTGGTGTTGT | 108 |
| RPL8int-r | Rpl8 | CAGTTCCTCTTTGCCTTGTACT | |
| bSCGB1A1-f | Scgb1a1 | GATCGCCATCACAATCACTGTGG | 156 |
| bSCGB1A1-r | Scgb1a1 | GTCTGAGCCAGGGTTGAAAGG | |
| bBPIFA1-f | Bpifa1 | TGGCAGCCTGAAAATCAGCTTGC | 161 |
| bBPIFA1-r | Bpifa1 | TGCACCAGGGTGACATCCAAAC |
Histology, IH, and In Situ Hybridization
Lung and trachea were fixed in 4% buffered paraformaldehyde for 24–48 h and routinely paraffin wax embedded. Consecutive sections (3–5 μm) were either stained with hematoxylin and eosin, used for IH or RNA in situ hybridization (RNA-ISH) and double stains.
IH was performed using the peroxidase anti-peroxidase method as previously described.37, 38 Primary antibodies used were rabbit anti-mSCGB1A1 (a kind gift of Barry Stripp)39 and rabbit anti-mBPIFA1 that was generated previously to an epitope localized in the N-terminal portion of the protein that is unique to the rodent lineage.25 The specificity of these targets had been determined previously. Papanicolaou's hematoxylin or the alcian blue/periodic acid–Schiff (AB-PAS) reaction for the demonstration of mucins were used as counterstains for the IH.
Detection of RNA by RNA-ISH followed a previously described protocol using digoxigenin-labeled sense and antisense probes, which were generated by in vitro transcription (digoxigenin RNA Labelling Kit (SP6/T7), Roche Applied Science, Mannheim, Germany).12, 40, 41 RNA-ISH probes were generated using IMAGE clones of mouse Scgb1a1 (MGC:41130, IMAGE:1434396) and Bpifa1 (MGC:62586, IMAGE:6314015) in pBluescript SK that were obtained via the Mammalian Gene Collection.42 Control sense strand probes were consistently found to show no reaction when hybridized to sections of wood mouse lung.
In addition, a combination of two techniques was used in which RNA-ISH was followed by IH.
Quantitative Histopathological Assessment
The percentage area and density of DAB staining within airway epithelium was quantified using whole slide images scanned and analyzed using the Chromavision automated cellular imaging system (ACIS II) and ACIS Product version 2.4.8.0 (Clarient, Inc.) and Matrox® Imaging Library. Regions of interest comprising the airway epithelium and excluding other cells were outlined and defined as trachea, bronchus, or bronchiole. The ACIS system analyses thresholds of hue, luminosity, and saturation, which were set relative to the chromogen (DAB) utilized for BPIFA1 and SCGB1A1 localization. These data provided an average density of staining for each region of interest. The percentage area stained was defined as the brown area (positive immunostaining) divided by the total brown area plus blue area (hematoxylin; negative staining) × 100. The data from each anatomical area are presented as the mean±standard error of the mean (s.e.m.) for each time point of infection. Statistical significance was determined by Student's t-test and values of P<0.05 considered significant, calculated using Minitab v.15 (Minitab Inc.).
Transmission Electron Microscopy
Sections of lung and trachea were fixed in 4% paraformaldehyde with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, and were routinely embedded in epoxy resin. Semi-thin sections (0.5 μm) were prepared to select areas of interest. Ultrathin (60 nm) sections were cut, mounted on copper grids, stained with Reynold's lead citrate, and viewed with an H600 TEM (Hitachi).
Statistical Analysis
Data were analyzed by one-way ANOVA with Bonferroni post-tests using the minitab v16 statistical package; P-values were set at 95% confidence interval.
RESULTS
Transcriptional Signatures Associated with MHV-68 M3 Expression During Infection
Our earlier study revealed that M3 is critical for modulating the host response wood mice. Thus, the efficient establishment of virus latency and formation of iBALT in the lung are dependent on M3 expression.9, 15 To gain further insight into the mechanism of M3 action, we identified genes that were differentially expressed in the presence and absence of M3. RNA extracted from the lungs of wood mice at day 14 p.i. with recombinant MHV-68 vM3.stop (M3-deficient) or vM3.MR (marker-rescue expressing M3) was analyzed using high-density microarrays. This revealed a limited number of genes (n=89) whose expression was at least twofold changed (Table 1). Functional annotation and grouping of these genes was performed. Of note were the obvious upregulation of genes encoding immunoglobulins and a downregulation of a high proportion of macrophage-specific genes in wood mice infected with vM3.MR as compared with vM3.stop infection. Of particular interest, several genes expressed in the airway epithelium encoding secreted glycoproteins were also upregulated in vM3.MR-infected wood mice. These included two members of the secretoglobin family (SCGB1A1 (club cell secretory protein or uteroglobin) and SCGB3A2 (uteroglobin-related protein 2)) and two members of the BPI fold-containing protein family (BPIFA1 (short PLUNC1, SPLUNC1) and BPIFB1 (long PLUNC1, LPLUNC1)). Scgb1a1 and Bpifa1 were highly differentially expressed (10.3- and 17.6-fold, respectively; Table 1 bold print). As it had been suggested that both these proteins were involved in airway defense,19, 21 we decided to analyze these proteins further during MHV-68 infection.
To validate the microarray results we performed qRT-PCR. However, to ensure accurate quantification, cDNAs corresponding to Scgb1a1 and Bpifa1 were generated from RNA extracted from wood mouse lung tissue using degenerate primers and cloned into pCR2.1. The DNA sequence of these clones revealed that the wood mouse Scgb1a1-coding sequence shares 93% identity at the nucleotide level with M. musculus Scgb1a1 and that the wood mouse Bpifa1-coding sequence shares 94% identity at the nucleotide level with M. musculus Bpifa1. An alignment of the predicted amino-acid sequences of SCGB1A1 and BPIFA1 from wood mouse and other mammalian species was made using ClustalW2.043 (Supplementary Figures E1 and E2). This revealed a high level of conservation of these proteins between wood mouse and house mouse. Of particular note is the conservation of an extended N-terminal glycine/proline-rich region between wood mouse and mouse BPIFA1 (Supplementary Figure E2) that has previously been shown to be rodent specific.
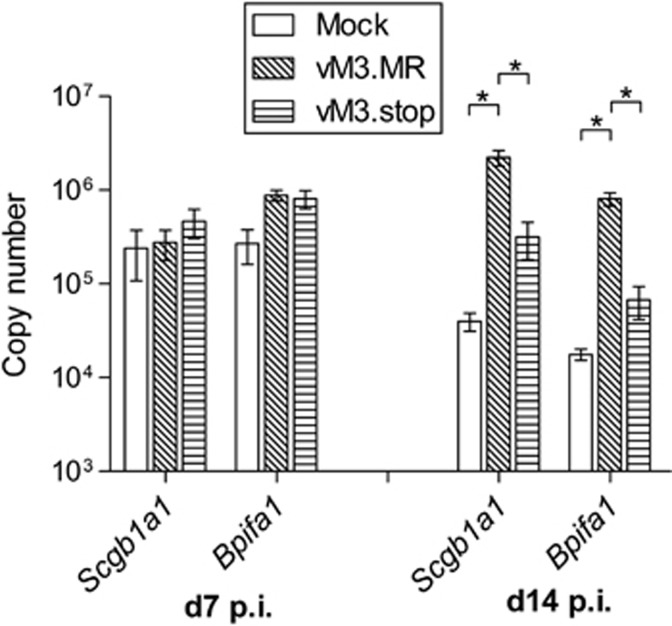
To quantify Scgb1a1 and Bpifa1 mRNA, cohorts of wood mice were either mock-infected, infected with vM3.MR or with vM3.stop for either 7 or 14 days. These time points were chosen as we have shown previously that they correspond to the peak of productive virus replication and peak of latency expansion (and iBALT) in the wood mice.12 RNA was then extracted from the lung tissues and analyzed by qRT-PCR using wood mouse-specific primers (Table 3). At day 7 p.i., there were no significant differences in the levels of Scgb1a1 and Bpifa1 between any of the groups of wood mice (Figure 1). However, at day 14 p.i., the levels of both Scgb1a1 and Bpifa1 transcripts were significantly elevated (P<0.05) in the lungs of wood mice infected with vM3.MR as compared with both mock- and vM3.stop-infected animals. Both transcripts were also elevated in animals infected with vM3.stop as compared with mock-infected wood mice at this time point, although these changes were not statistically significant. This confirms our microarray data whereby expression of Scgb1a1 and Bpifa1 was elevated at day 14 p.i. in vM3.MR- as compared with vM3.stop-infected animals, but also show that there is a greater change when vM3.MR-infected are compared with mock-infected wood mice, suggesting that M3 was partially responsible for elevation of the expression of these host genes. Our previously published results15 have shown that vM3.stop is attenuated in wood mice, the viral load at day 14 p.i. being ca 1 log lower in M3.stop as compared with vM3.MR-infected animals. In addition, the chemokine and cytokine levels were shown in our previous study to vary between the two groups so it is equally possible that other factors are responsible for differences in SCGB1A1 and BPIFA1 expression. However, the modulation of SCGB1A1 and BPIFA1 expression does suggest a role for these factors during the viral infection. As these represented novel findings, and little is known about the function of these proteins in the context of viral infection, we decided to concentrate on how they behave during MHV-68 infection.
Figure 1.
MHV-68 modulates the transcription of Scgb1a1 and Bpifa1 in the lungs of wood mice. RNA was extracted from the lungs of infected wood mice at days 7 and 14 p.i. as indicated and analyzed by qRT-PCR using primers specific for wood mouse Bpifa1 and Scgb1a1. The copy number of mRNA was normalized to the copy number of cellular Rpl8. Bars represent mean±s.e.m. (n=3). Statistically significant differences (one-way ANOVA with Bonferroni post-tests) between groups are represented by square brackets above. *P<0.05.
SCGB1A1 Expression is Modulated after MHV-68 Infection
To determine any anatomical- and/or temporal-based variation in SCGB1A1 after infection, trachea and lungs from wood mice that were either mock-infected or infected with MHV-68 were analyzed at day 7 and 14 p.i. (Figure 2).
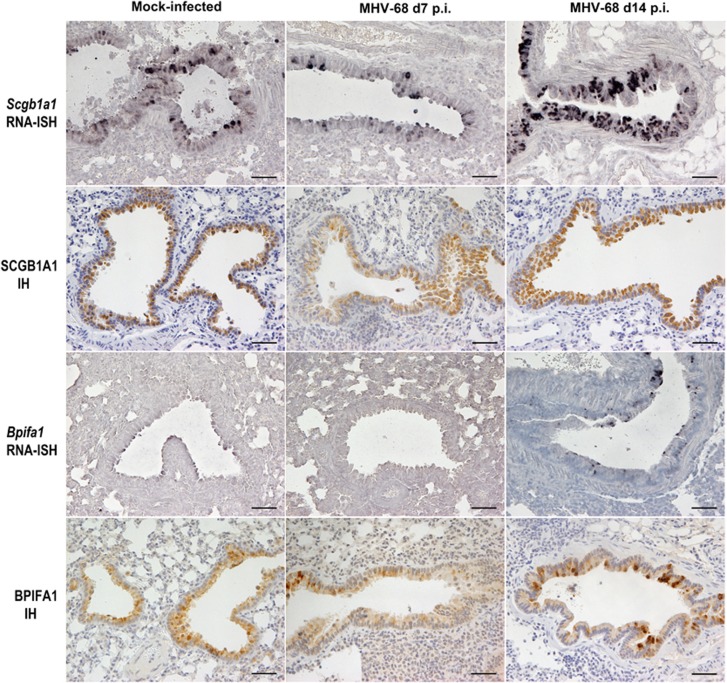
Figure 2.
MHV-68 modulates the expression of SCGB1A1 and BPIFA1 RNA and antigen in bronchiolar epithelial cells. Lungs were harvested from wood mice that were either mock- or MHV-68-infected at either day 7 or day 14 p.i. as indicated. RNA specific for Scgb1a1 and Bpifa1 was detected by RNA-ISH using specific riboprobes, visualized with BCIP/NBT (dark blue–black), and counterstained with hematoxylin. Antigen was detected by immunohistology (IH) analysis using antibodies specific for SCGB1A1 and BPIFA1, visualized with DAB, and counterstained with hematoxylin. Scale bar represents 50 μm.
Histopathological analysis of hematoxylin and eosin-stained sections revealed a pattern of changes after infection with MHV-68 consistent with our previous observations, represented predominantly by mutifocal mononuclear perivascular and peribronchiolar as well as multifocal granulomatous infiltration on day 7 p.i., and perivascular and peribronchiolar lymphocyte-dominated infiltration with iBALT formation at day 14 p.i. (not shown).
Expression of Scgb1a1 was detected by RNA-ISH within mock-infected wood mice in rare epithelial cells in the trachea, but was seen more frequently in the bronchial epithelium. Cells in the larger proximal bronchioles stained most frequently for Scgb1a1, with numerous cells showing Scgb1a1 transcripts. Smaller (terminal) bronchioles showed less frequent staining for Scgb1a1 and the alveoli were negative. After infection with MHV-68, at both days 7 and 14 p.i. the trachea and bronchi showed a distribution of Scgb1a1 expression similar to that observed in control wood mice (not shown). However, in the bronchioles, there was a downregulation of Scgb1a1 expression at day 7 p.i. and an upregulation at day 14 p.i. (Figure 2, top row).
The presence of SCGB1A1 protein was determined by IH staining, which showed that the anatomical distribution of SCGB1A1 was similar to that of Scgb1a1 RNA and to that previously described for M. musculus44 in both mock- and MHV-68-infected mice at both time points p.i. (Figure 2, second row).
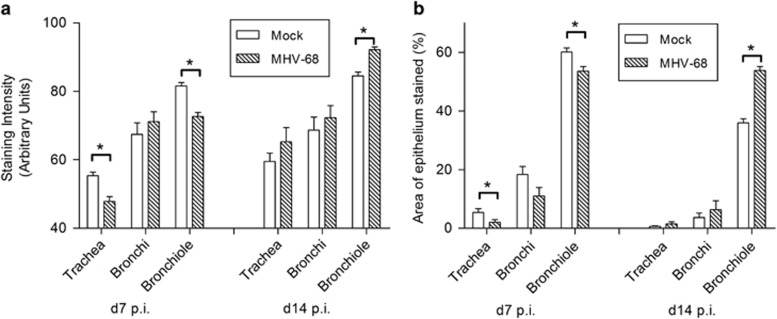
To assess further the amount of epithelial SCGB1A1 protein present across the respiratory tract, we performed quantitative analysis of the IH. At day 7 p.i., the intensity of staining for SCGB1A1 was significantly lower in the trachea (P<0.005) and bronchioles (P<0.05) in MHV-68-infected mice as compared with mock-infected control mice (Figure 3a). The percentage area of epithelium that stained positively for SCGB1A1 was similarly significantly decreased (Figure 3b). SCGB1A1 protein changes in the bronchi were not significant. At day 14 p.i., there were no significant alterations in the intensity of staining or the percentage of tissue stained in either the trachea or bronchi (Figure 3a and b), but within the bronchioles (closest to the site of viral replication) there were significant increases (P<0.005) in both parameters in MHV-68-infected wood mice compared with mock-infected wood mice (Figure 3).
Figure 3.
SCGB1A1 protein levels are modulated after MHV-68 infection. Lungs were harvested from wood mice that were either mock- or MHV-68-infected at either day 7 or day 14 p.i. as indicated. SCGB1A1 was detected by IH analysis using antibodies specific for SCGB1A1, visualized with DAB, and counterstained with hematoxylin. The intensity of staining (a) and percentage area of epithelium stained (b) in the trachea, bronchi, and bronchioles were assessed by image analysis. Data are for three mice per group presented as mean±s.e.m. (n=3). Significant differences from mock-infected mouse values are shown by an asterisk.
BPIFA1 Expression is Modulated after MHV-68 Infection
Anatomical and temporal changes in Bpifa1 RNA and BPIFA1 protein expression following MHV-68 infection were similarly determined in the same wood mice used for SCGB1A1 analysis. The BPIFA1 antibody used was generated to an epitope localized in the N-terminal portion of the protein that is unique to the rodent lineage (Supplementary Figure E2). This epitope shares 13/16 amino acids with the mouse sequence used to generate the antibody.
In mock-infected wood mice, Bpifa1 RNA was most notable in the trachea and bronchi, within the respiratory epithelium (non-ciliated cells; Figure 2, third row) and the submucosal glands. Within the bronchioles there were scattered Bpifa1-positive cells. We did not detect Bpifa1 expression in the alveoli. Following infection with MHV-68, there were similar, high levels of Bpifa1 transcription in the trachea and bronchi, and the proximal bronchioles showed an increase in transcription at day 14 p.i. (Figure 2, third row). The smaller terminal bronchioles were frequently negative, showing no changes from control animals.
IH analysis demonstrated that BPIFA1 protein was distributed in a similar pattern to the RNA, ie, within the non-ciliated epithelium of the trachea and bronchi and submucosal glands and to a lesser extent in the bronchiolar epithelium in both mock- and MHV-68-infected animals (Figure 2, bottom row).
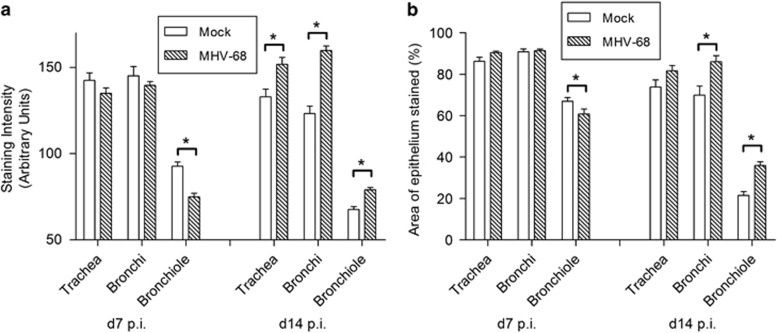
Quantitative analysis (Figure 4) showed that although there was no significant difference in intensity of staining or percentage of tissue stained in the trachea and bronchi at day 7 p.i., there was a significant decrease in the intensity of staining (P<0.005) and in the percentage area stained (P<0.05) in the bronchioles at this time point. In contrast, at day 14 p.i., this pattern was reversed, as increases in BPIFA1 staining intensity was seen at all levels of the respiratory tract in MHV-68-infected compared with mock-infected animals (trachea P<0.05, bronchi and bronchioles P<0.05) and a significant increase in the area stained was seen in the bronchi and bronchioles. This increase did not correlate well with the staining for Bpifa1 transcripts by RNA-ISH.
Figure 4.
BPIFA1 protein levels are modulated after MHV-68 infection. Lungs were harvested from wood mice that were either mock- or MHV-68-infected at either day 7 or day 14 p.i. as indicated. BPIFA1 was detected by IH analysis using antibodies specific for BPIFA1, visualized with DAB, and counterstained with hematoxylin. The intensity of staining (a) and percentage area of epithelium stained (b) in the trachea, bronchi, and bronchioles were assessed by image analysis. Data are for three mice per group presented as mean±s.e.m. (n=3). Significant differences from mock-infected mouse values are shown by an asterisk.
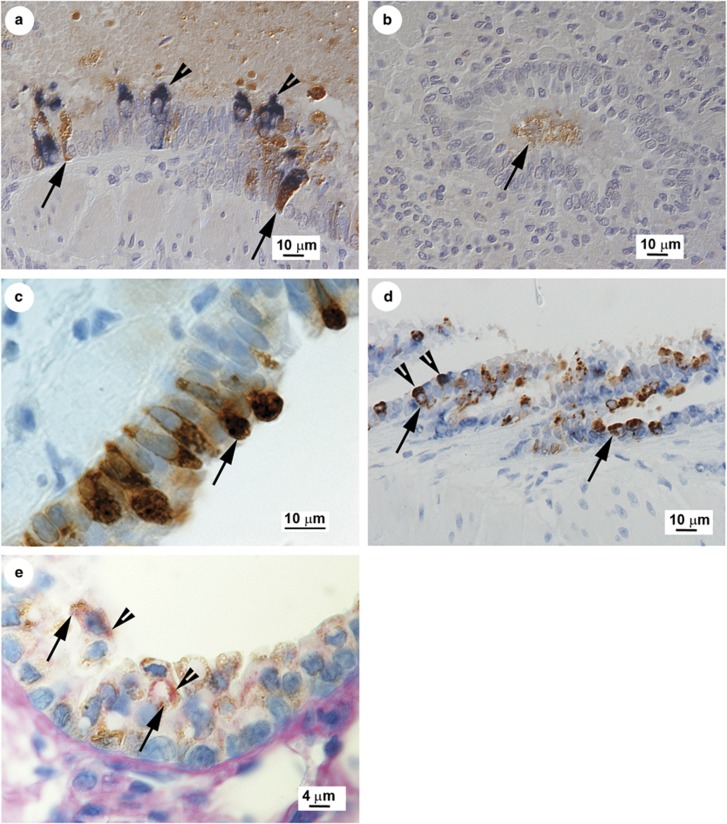
To investigate further the apparent discrepancy between the detection of Bpifa1 RNA and BPIFA1 protein in bronchioles, double staining (IH and RNA-ISH) was performed on the same slide. The results showed co-localization of RNA and protein in trachea, bronchi, and proximal bronchioles (Figure 5a), but the presence of BPIFA1 protein without evidence of Bpifa1 RNA in the terminal bronchioles (Figure 5b). In addition, BPIFA1 protein was localized predominantly intracellularly in larger airways (Figure 5a) within what appear to be storage granules (Figure 5c). In contrast, the strong staining observed in the terminal bronchioles was present not only within the airway epithelium but also in the airway lumen or apical surface of the cells (Figure 5b).
Figure 5.
SCGB1A1 and BPIFA1 RNA and protein are co-expressed with mucus in club cells. (a, b) Combined IH analysis for BPIFA1 protein (visualized with DAB, arrow) and RNA-ISH with Bpifa1-specific probes (visualized with NBT/BCIP, arrowhead) demonstrates the presence of BPIFA1 protein in the distal terminal bronchioles (b) without evidence of Bpifa1 transcription at this site, which occurred in the proximal bronchioles (a) and upper airways in MHV-68-infected wood mice at day 14 p.i. Hematoxylin counterstain. (c) IH analysis for BPIFA1 antigen (DAB, arrow) demonstrates granular staining in club cells in the cytoplasm consistent with intra-vesicular storage. (d) IH analysis for SCGB1A1 protein (DAB, arrows) was used to identify club cells and subsequent RNA-ISH with Bpifa1-specific probes demonstrated that club cells also transcribe Bpifa1 (arrowheads). Hematoxylin counterstain. (e) Club cells identified by staining for SCGB1A1 protein (DAB, arrows) also contained AB-PAS-positive vesicles within the cytoplasm (arrowheads), consistent with mucous vesicles, suggesting that club cells also have the potential to produce mucus.
Club Cells can be Multi-Functional
Club cells by definition secrete SCGB1A1.45, 46 To investigate if the same cells that produced SCGB1A1 also secreted BPIFA1, we used a combination of RNA-ISH and IH to determine co-localization. An RNA probe for Bpifa1 was used followed by detection of SCGB1A1 by IH. This identified club cells in the bronchioles that expressed both Bpifa1 transcripts and SCGB1A1 antigen (Figure 5d). To determine if club cells also produced mucins, IH using the SCGB1A1 antibody was performed as before and the AB-PAS reaction to highlight carbohydrates (neutral mucins: magenta, acidic mucins: blue) was applied as counterstain. The results (Figure 5e) identified club cells positive for SCGB1A1 that also contained AB-PAS-positive granules in the cytoplasm, consistent with mucus vesicles. Thus, we show for the first time that club cells producing SCGB1A1 not only can produce mucus after virus infection but also produce increased amounts of BPIFA1.
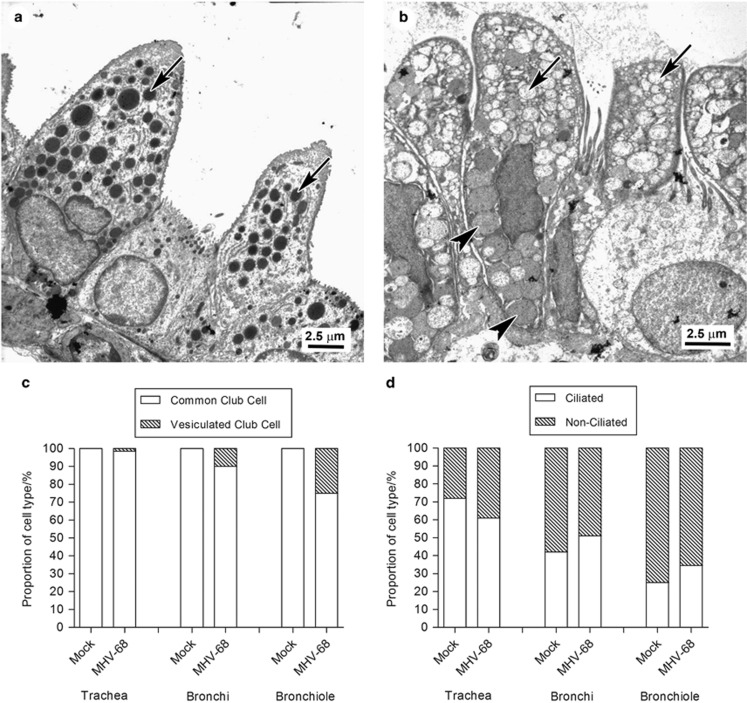
Club Cells in the Bronchiole become Vesiculated after Infection
As MHV-68 infection lead to an induction of mucus production by club cells, we wished to determine if this was associated with a corresponding alteration in their morphological phenotype. TEM was performed on lung tissue of mock- or MHV-68-infected wood mice at day 14 p.i. The proportion of ciliated vs non-ciliated epithelial cells was assessed as was the proportion of club cells with either a common or more vesiculated morphology indicative of mucus secretion, as previously defined.44 In mock-infected animals, the overwhelming majority of club cells demonstrated the typical or common morphology, including cytoplasm of moderate electron density, organized arrays of smooth endoplasmic reticulum, and electron-dense secretory vesicles in the apical part of the cell (Figure 6a and c) at all levels of the respiratory tract. In wood mice infected with MHV-68, in addition to the common type of club cell, vesiculated club cells were observed. These were infrequent in the trachea but increased in number in the distal airways (Figure 6b and c) making up ca 10% of bronchial and ca 25% of bronchiolar club cells. They exhibited marked vesiculation of the apical part or all of the cytoplasm and the vesicles contained moderately electron-dense material characteristic of mucin. There was no significant difference in the proportion of ciliated and non-ciliated epithelial cells revealed no significant difference after infection (Figure 6d).44
Figure 6.
Transmission electron microscopic analysis of the morphology of club cells before and after infection. Transmission electron microscopy was used to quantify the morphological type and frequency of epithelial cells in the respiratory tract of wood mice. (a) Bronchiole. In mock-infected wood mice, the common type of club cell morphology vastly predominated with numerous electron-dense secretory vesicles in the apical part of the cell (arrow). (b) Bronchiole. A proportion of club cells, particularly in the bronchioles of MHV-68-infected wood mice at day 14 p.i. exhibited a distinct morphology with marked vesiculation of the apical part or all of the cytoplasm (arrows) and vesicles containing moderately electron-dense material characteristic of mucin (arrowheads). (c) The proportion of types of club cells in the airways was determined by enumeration of cell types present in electron micrograph images. (d) The proportion of ciliated and non-ciliated cells in the airways was determined by enumeration of cell types present in electron micrograph images.
DISCUSSION
We have developed MHV-68 infection of wood mice as an authentic system to study γ-herpesvirus infection. MHV-68 encodes a chemokine-binding protein termed M3, and in the current study, we used our wood mouse system along with a virus deficient in M3 expression (vM3.stop) to identify transcriptional signatures associated with the M3 protein during infection of the lung. Among other changes, this identified a significant upregulation of Scgb1a1 and Bpifa1 expression that was not only associated with expression of M3 but also with MHV-68 infection in general. We further examined the differential temporal and spatial expression of these two genes and their products in airways during MHV-68 infection of wood mice. This showed that BPIFA1 can be expressed and stored in granules within club cells along with SCGB1A1 and that there is a decrease in expression of both proteins in the bronchioles at day 7 p.i. consistent with release of stored protein. At day 14 p.i., the expression of both proteins increased. This is associated with the production of mucus in club cells.
Global analysis indicated differential expression of 89 genes between vM3.stop and vM3.MR-infected wood mice at day 14 p.i. (Table 1). Of note is the upregulation of a number of genes encoding immunoglobulins in mice infected with vM3.MR, which in likely a consequence of iBALT formation in these animals and its absence in vM3.stop-infected wood mice.15 It is also interesting that the expression of a high proportion of macrophage-specific genes was downregulated in vM3.MR-infected mice as compared with vM3.stop infection. This suggests disruption of macrophage function by M3 in vivo and fits well with the in vitro evidence for M3 binding to chemokines that affect macrophage function13, 14 and the in vivo evidence that M3 results in lower levels of macrophage-specific chemokines.15
The most highly differentially expressed genes at day 14 p.i. were Scgb1a1 and Bpifa1. Differential expression was confirmed by qRT-PCR and in addition it was found that expression of both genes was lower in mock-infected animals than after either vM3.MR or vM3.stop virus infection (Figure 1). However, at day 7 p.i., there were no significant differences in the expression of either gene between any of the groups. A reduction in the level of expression of both genes in the mock control between day 7 and day 14 is likely due to alterations in expression caused by anesthesia and administration of PBS carrier and underlines the importance of using this control at both time points. The potential function of the M3 chemokine-binding protein in modulation of Scgb1a1 and Bpifa1 expression is interesting, but likely to be complex, and made more so by the M3-deficient virus being less pathogenic than vM3.MR, and hence, harboring a much higher viral load in the lung.15 Modulation of Scgb1a1 and Bpifa1 expression after MHV-68 infection clearly suggests a role for these factors in the host response. However, future studies will concentrate on if or how M3 might have a role in modulating expression of these interesting proteins.
The expression of Scgb1a1 RNA was most frequently observed in the bronchiolar epithelium in mock-infected wood mice (Figure 2), but was also present throughout the respiratory epithelium, similar to that seen in M. musculus.44 At day 7 p.i. with MHV-68, there was a decrease in Scgb1a1 RNA in the bronchioles and a significant decrease in both intensity of staining and percentage area stained for SCGB1A1 protein in the trachea and bronchioles (Figures 2 and 3). In contrast, there was no difference in expression of Scgb1a1 mRNA detected by qRT-PCR between mock- and MHV-68-infected wood mice in the lungs at this time point (Figure 1). This discrepancy is probably due to the decreased RNA expression within bronchioles (as detected by RNA-ISH) being rendered undetectable by qRT-PCR analysis in a background of RNA extracted from whole lung tissue, including trachea and bronchi, where there was no difference in Scgb1a1 RNA expression. This highlights the value of RNA-ISH in our analysis. However, at day 14 p.i., there was an increase in Scgb1a1 expression in infected animals within the bronchiolar epithelium (Figures 1 and 2). In line with this, there were significant increases in SCGB1A1 protein in the bronchioles in MHV-68-infected wood mice (Figures 2 and 3).
The precise function of SCGB1A1 is not known, although it has been shown to inhibit phospholipase A2 and the production of IFN-γ, IL-1, and TNF-α.16 In addition, Scgb1a1−/− mice infected with respiratory syncytial virus display an increased inflammatory response and also, increased viral titer when compared with wild-type animals.21 Mice deficient in SCGB1A1 that are infected with adenovirus also show a more intense inflammatory response.22 Thus, our observations of modulation of SCGB1A1 expression after infection with MHV-68 fit with a role in host defense and/or immunomodulation.
In laboratory mice, BPIFA1 is found predominantly in the upper respiratory tract and palate47 as a product of the airway epithelium23, 47, 48 and the submucosal glands.48, 49 In humans, it is found predominantly in the non-ciliated cells of the upper respiratory tract and submucosal glands.50 We observed a similar pattern of expression in wood mice (Figure 2). After infection, we detected Bpifa1 RNA in non-ciliated epithelial cells in the trachea, bronchi, and proximal bronchioles but not terminal bronchioles (Figure 5a and b). When we analyzed BPIFA1 protein by IH, however, we found its presence in the entire respiratory tract, although the staining was less intense in the terminal bronchioles than in the trachea and bronchi (Figure 2), where the staining was mostly within granules in club cells (Figure 5c). Interestingly, IH analysis also identified the protein cell-free within the lumen of some terminal bronchioles (Figure 5b). Quantitative analysis showed that BPIFA1 levels were decreased in bronchioles at day 7 p.i. but increased throughout the entire respiratory tract in response to infection at day 14 p.i. (Figure 4). Our results show for the first time that BPIFA1 is produced and stored in club cells. The presence of BPIFA1 protein in the terminal bronchioles in the apparent absence of RNA is enigmatic. There is no precedence for secreted proteins to travel from higher up in the respiratory tract to terminal bronchioles, so BPIFA1 protein in this region could represent limited local production and rapid secretion from amounts of RNA that we did not detect by RNA-ISH. A recent report has described a decrease in BPIFA1 in the bronchio-alveolar lavage fluid after challenge of mice with bacteria and influenza A virus up to 7 days p.i. and suggested that BPIFA1 may act as a sensor of exposure to pathogens.51 The site of production of BPIFA1 was not analyzed in this study, nor were later time points, but the results concur with our observation of its reduced expression in bronchioles at day 7 p.i. during acute infection. In turn, the increased levels of BPIFA1 protein that we observed throughout the entire respiratory tract at day 14 p.i. may indicate a distinct role in the resolution of infection.
The cellular locations of SCGB1A1 and BPIFA1 overlap, but are not identical. We have shown here that cells positive for SCGB1A1 protein in bronchioles also transcribe Bpifa1 (Figure 5d). Thus, we show for the first time that club cells are able to produce BPIFA1 and also appear to retain the protein within granules (Figure 5c). We also found the morphology of a proportion of club cells changed in response to infection and that the ‘vesiculated type' of club cell44 became much more frequent in the bronchioles of infected wood mice (Figure 6). The club cells also appeared to produce mucin following infection, as indicated by the AB-PAS reaction (Figure 5e). Club cells have been shown to produce mucus in response to antigen challenge,52 although other reports dispute that they have the capacity to store mucin and rather undergo metaplasia, becoming goblet-like cells.53 Boers et al.,54 in a survey of club cells in humans, found that SCGB1A1 expression and a positive PAS reaction within the same cell occurred in between 25 and 45% of goblet cells, depending on the location within the respiratory tract. This was defined as an ‘intermediate' cell type with characteristics of both goblet cells and club cells.54 Our results suggest that a proportion of club cells may undergo differentiation toward a goblet cell morphology, and may be in part the mechanism of mucous metaplasia observed in response to viral infection.55 The increase in BPIFA1 expression seen here at day 14 p.i. may also be associated with the phenotypic alteration of club cells into mucin-secreting cells.
Like SCGB1A1, the precise function of BPIFA1 is unknown. It has a surfactant-like effect28 and is involved in the regulation of the amiloride-sensitive epithelial sodium channel, ENaC.29 Infection with Mycoplasma spp. induces Bpifa1 expression in murine airways30 and, importantly, BPIFA1 enhances IL-8 production and bacterial clearance.30 Recent data also suggest that the protein may be implicated in the mediation of host responses through modulation of macrophage function.32 Thus, the modulation of BPIFA1 expression after MHV-68 infection is consistent with a function in host defense and/or immune modulation including, possibly, sensing of infection.51
Our data show that the quantitative modulation of two proteins, SCGB1A1 and BPIFA1, is associated with MHV-68 infection. Although there are subtle differences, the temporal and spatial distribution of these two molecules throughout the respiratory tract is generally coordinated suggesting some commonality of regulation. Both proteins are stored in granules in club cells and their decrease in expression at day 7 p.i. is consistent with release of stored protein during the phase of acute MHV-68 virion production and epithelial cell damage. An increase in both proteins along with mucus and a change in morphology of a proportion of club cells likewise suggests a role for both proteins during the resolution of virion production and the establishment of latency. This also highlights a potentially important role of the club cell during virus infection in terms of secretion of factors involved in host defense.
Acknowledgments
We thank Bahram Ebrahimi and the Liverpool Microarray Facility for help with performing the microarray experiments. We thank Samuel H Speck, Herbert W Virgin IV and Victor van Berkel for the generous gift of vM3.stop and vM3.MR, and Barry Stripp for the generous gift of anti-SCGB1A1. We thank the technical staff in the Histology Laboratories and the Electron Microscopy Unit, Veterinary Laboratory Services, School of Veterinary Science, University of Liverpool, for excellent technical assistance. This work was supported by a Royal Society (London) University Research Fellowship (to JPS), by Biotechnology and Biological Sciences Research Council (UK) grants BB/K009664/1 (to JPS, AK, and GHL) and BB/K009737/1 (CDB and LB), US Public Health Service grant CA090208, the Penn State Hershey Cancer Institute, and a Biotechnology and Biological Sciences Research Council studentship (to GHL).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.laboratoryinvestigation.org)
Supplementary Material
References
- Nash AA, Dutia BM, Stewart JP, et al. Natural history of murine gamma-herpesvirus infection. Philos Trans R Soc Lond B Biol Sci. 2001;356:569–579. doi: 10.1098/rstb.2000.0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck SH, Virgin HW. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr Opin Microbiol. 1999;2:403–409. doi: 10.1016/s1369-5274(99)80071-x. [DOI] [PubMed] [Google Scholar]
- Flano E, Kim IJ, Woodland DL, et al. gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J Exp Med. 2002;196:1363–1372. doi: 10.1084/jem.20020890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty PC, Christensen JP, Belz GT, et al. Dissecting the host response to a gamma-herpesvirus. Philos Trans R Soc Lond B Biol Sci. 2001;356:581–593. doi: 10.1098/rstb.2000.0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simas JP, Efstathiou S. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 1998;6:276–282. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- Barton E, Mandal P, Speck SH. Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu Rev Immunol. 2011;29:351–397. doi: 10.1146/annurev-immunol-072710-081639. [DOI] [PubMed] [Google Scholar]
- Wu TT, Blackman MA, Sun R. Prospects of a novel vaccination strategy for human gamma-herpesviruses. Immunol Res. 2010;48:122–146. doi: 10.1007/s12026-010-8172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers B, Kuchler J, Yasmum N, et al. Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus. J Virol. 2007;81:8091–8100. doi: 10.1128/JVI.00255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DJ, Kipar A, Milligan SG, et al. Characterization of a novel wood mouse virus related to murid herpesvirus 4. J Gen Virol. 2010;91:867–879. doi: 10.1099/vir.0.017327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DJ, Kipar A, Leeming G, et al. Experimental infection of laboratory-bred bank voles (Myodes glareolus) with murid herpesvirus 4. Arch Virol. 2012;157:2207–2212. doi: 10.1007/s00705-012-1397-5. [DOI] [PubMed] [Google Scholar]
- Blasdell K, McCracken C, Morris A, et al. The wood mouse is a natural host for Murid herpesvirus 4. J Gen Virol. 2003;84:111–113. doi: 10.1099/vir.0.18731-0. [DOI] [PubMed] [Google Scholar]
- Hughes DJ, Kipar A, Sample JT, et al. Pathogenesis of a model gammaherpesvirus in a natural host. J Virol. 2010;84:3949–3961. doi: 10.1128/JVI.02085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berkel V, Barrett J, Tiffany HL, et al. Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J Virol. 2000;74:6741–6747. doi: 10.1128/jvi.74.15.6741-6747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry CM, Simas JP, Smith VP, et al. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J Exp Med. 2000;191:573–578. doi: 10.1084/jem.191.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DJ, Kipar A, Leeming GH, et al. Chemokine binding protein M3 of murine gammaherpesvirus 68 modulates the host response to infection in a natural host. PLoS Pathog. 2011;7:e1001321. doi: 10.1371/journal.ppat.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Katyal SL. Clara cell proteins. Ann N Y Acad Sci. 2000;923:43–58. doi: 10.1111/j.1749-6632.2000.tb05518.x. [DOI] [PubMed] [Google Scholar]
- LeClair EE. Four reasons to consider a novel class of innate immune molecules in the oral epithelium. J Dent Res. 2003;82:944–950. doi: 10.1177/154405910308201202. [DOI] [PubMed] [Google Scholar]
- Bingle CD, Craven CJ. PLUNC: a novel family of candidate host defense proteins expressed in the upper airways and nasopharynx. Hum Mol Genet. 2002;11:937–943. doi: 10.1093/hmg/11.8.937. [DOI] [PubMed] [Google Scholar]
- Bingle CD, Gorr SU. Host defense in oral and airway epithelia: chromosome 20 contributes a new protein family. Int J Biochem Cell Biol. 2004;36:2144–2152. doi: 10.1016/j.biocel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Broeckaert F, Clippe A, Knoops B, et al. Clara cell secretory protein (CC16): features as a peripheral lung biomarker. Ann N Y Acad Sci. 2000;923:68–77. doi: 10.1111/j.1749-6632.2000.tb05520.x. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Rosenberger CL, Bao YX, et al. Clara cell secretory protein modulates lung inflammatory and immune responses to respiratory syncytial virus infection. J Immunol. 2003;171:1051–1060. doi: 10.4049/jimmunol.171.2.1051. [DOI] [PubMed] [Google Scholar]
- Harrod KS, Mounday AD, Stripp BR, et al. Clara cell secretory protein decreases lung inflammation after acute virus infection. Am J Physiol. 1998;275:L924–L930. doi: 10.1152/ajplung.1998.275.5.L924. [DOI] [PubMed] [Google Scholar]
- LeClair EE, Nguyen L, Bingle L, et al. Genomic organization of the mouse plunc gene and expression in the developing airways and thymus. Biochem Biophys Res Commun. 2001;284:792–797. doi: 10.1006/bbrc.2001.5024. [DOI] [PubMed] [Google Scholar]
- LeClair EE, Nomellini V, Bahena M, et al. Cloning and expression of a mouse member of the PLUNC protein family exclusively expressed in tongue epithelium. Genomics. 2004;83:658–666. doi: 10.1016/j.ygeno.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Musa M, Wilson K, Sun L, et al. Differential localisation of BPIFA1 (SPLUNC1) and BPIFB1 (LPLUNC1) in the nasal and oral cavities of mice. Cell Tissue Res. 2012;350:455–464. doi: 10.1007/s00441-012-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle CD, LeClair EE, Havard S, et al. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Sci. 2004;13:422–430. doi: 10.1110/ps.03332704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle CD, Bingle L, Craven CJ. Distant cousins: genomic and sequence diversity within the BPI fold-containing (BPIF)/PLUNC protein family. Biochem Soc Trans. 2011;39:961–965. doi: 10.1042/BST0390961. [DOI] [PubMed] [Google Scholar]
- Gakhar L, Bartlett JA, Penterman J, et al. PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One. 2010;5:e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Caballero A, Rasmussen JE, Gaillard E, et al. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc Natl Acad Sci USA. 2009;106:11412–11417. doi: 10.1073/pnas.0903609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HW, Thaikoottathil J, Rino JG, et al. Function and regulation of SPLUNC1 protein in Mycoplasma infection and allergic inflammation. J Immunol. 2007;179:3995–4002. doi: 10.4049/jimmunol.179.6.3995. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bartlett JA, Di ME, et al. SPLUNC1/BPIFA1 contributes to pulmonary host defense against Klebsiella pneumoniae respiratory infection. Am J Pathol. 2013;182:1519–1531. doi: 10.1016/j.ajpath.2013.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaikoottathil JV, Martin RJ, Di PY, et al. SPLUNC1 deficiency enhances airway eosinophilic inflammation in mice. Am J Respir Cell Mol Biol. 2012;47:253–260. doi: 10.1165/rcmb.2012-0064OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou S, Ho YM, Minson AC. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol. 1990;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- Sunil-Chandra NP, Efstathiou S, Arno J, et al. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J Gen Virol. 1992;73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- van Berkel V, Levine B, Kapadia SB, et al. Critical role for a high-affinity chemokine-binding protein in gamma-herpesvirus-induced lethal meningitis. J Clin Invest. 2002;109:905–914. doi: 10.1172/JCI14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JP, Kipar A, Cox H, et al. Induction of tachykinin production in airway epithelia in response to viral infection. PLoS One. 2008;3:e1673. doi: 10.1371/journal.pone.0001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipar A, Kohler K, Leukert W, et al. A comparison of lymphatic tissues from cats with spontaneous feline infectious peritonitis (FIP), cats with FIP virus infection but no FIP, and cats with no infection. J Comp Pathol. 2001;125:182–191. doi: 10.1053/jcpa.2001.0501. [DOI] [PubMed] [Google Scholar]
- Chen H, Matsumoto K, Brockway BL, et al. Airway epithelial progenitors are region specific and show differential responses to bleomycin-induced lung injury. Stem Cells. 2012;30:1948–1960. doi: 10.1002/stem.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LA, Stewart JP, Nash AA, et al. Murine gammaherpesvirus-68 infection of and persistence in the central nervous system. J Gen Virol. 2000;81:2635–2643. doi: 10.1099/0022-1317-81-11-2635. [DOI] [PubMed] [Google Scholar]
- Dutia BM, Stewart JP, Clayton RA, et al. Kinetic and phenotypic changes in murine lymphocytes infected with murine gammaherpesvirus-68 in vitro. J Gen Virol. 1999;80:2729–2736. doi: 10.1099/0022-1317-80-10-2729. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Pack RJ, Al-Ugaily LH, Morris G. The cells of the tracheobronchial epithelium of the mouse: a quantitative light and electron microscope study. J Anat. 1981;132:71–84. [PMC free article] [PubMed] [Google Scholar]
- Bedetti CD, Singh J, Singh G, et al. Ultrastructural localization of rat Clara cell 10 KD secretory protein by the immunogold technique using polyclonal and monoclonal antibodies. J Histochem Cytochem. 1987;35:789–794. doi: 10.1177/35.7.2438324. [DOI] [PubMed] [Google Scholar]
- Singh G, Singh J, Katyal SL, et al. Identification, cellular localization, isolation, and characterization of human Clara cell-specific 10 KD protein. J Histochem Cytochem. 1988;36:73–80. doi: 10.1177/36.1.3275712. [DOI] [PubMed] [Google Scholar]
- Weston WM, LeClair EE, Trzyna W, et al. Differential display identification of plunc, a novel gene expressed in embryonic palate, nasal epithelium, and adult lung. J Biol Chem. 1999;274:13698–13703. doi: 10.1074/jbc.274.19.13698. [DOI] [PubMed] [Google Scholar]
- Leclair EE. Four BPI (bactericidal/permeability-increasing protein)-like genes expressed in the mouse nasal, oral, airway and digestive epithelia. Biochem Soc Trans. 2003;31:801–805. doi: 10.1042/bst0310801. [DOI] [PubMed] [Google Scholar]
- Di YP, Harper R, Zhao Y, et al. Molecular cloning and characterization of spurt, a human novel gene that is retinoic acid-inducible and encodes a secretory protein specific in upper respiratory tracts. J Biol Chem. 2003;278:1165–1173. doi: 10.1074/jbc.M210523200. [DOI] [PubMed] [Google Scholar]
- Bingle L, Cross SS, High AS, et al. SPLUNC1 (PLUNC) is expressed in glandular tissues of the respiratory tract and in lung tumours with a glandular phenotype. J Pathol. 2005;205:491–497. doi: 10.1002/path.1726. [DOI] [PubMed] [Google Scholar]
- Britto CJ, Liu Q, Curran DR, et al. Short palate, lung, and nasal epithelial clone-1 is a tightly regulated airway sensor in innate and adaptive immunity. Am J Respir Cell Mol Biol. 2013;48:717–724. doi: 10.1165/rcmb.2012-0072OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CM, Williams OW, Tuvim MJ, et al. Mucin is produced by clara cells in the proximal airways of antigen-challenged mice. Am J Respir Cell Mol Biol. 2004;31:382–394. doi: 10.1165/rcmb.2004-0060OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CW, Dickey BF. Regulated airway goblet cell mucin secretion. Annu Rev Physiol. 2008;70:487–512. doi: 10.1146/annurev.physiol.70.113006.100638. [DOI] [PubMed] [Google Scholar]
- Boers JE, Ambergen AW, Thunnissen FB. Number and proliferation of clara cells in normal human airway epithelium. Am J Respir Crit Care Med. 1999;159:1585–1591. doi: 10.1164/ajrccm.159.5.9806044. [DOI] [PubMed] [Google Scholar]
- Walter MJ, Morton JD, Kajiwara N, et al. Viral induction of a chronic asthma phenotype and genetic segregation from the acute response. J Clin Invest. 2002;110:165–175. doi: 10.1172/JCI14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.