Abstract
To address how the configuration of conjugated ubiquitins determines the recognition of substrates by the proteasome, we analyzed the degradation kinetics of substrates with chemically defined ubiquitin configurations. Contrary to the view that a tetraubiquitin chain is the minimal signal for efficient degradation, we find that distributing the ubiquitins as diubiquitin chains provides a more efficient signal. To understand how the proteasome actually discriminates among ubiquitin configurations, we developed single-molecule assays that distinguished intermediate steps of degradation kinetically. The level of ubiquitin on a substrate drives proteasome-substrate interaction, whereas the chain structure of ubiquitin affects translocation into the axial channel on the proteasome. Together these two features largely determine the susceptibility of substrates for proteasomal degradation.
Introduction
The propensity of a protein for degradation is largely encoded in its state of ubiquitylation(1, 2). The ubiquitylation process results in highly diverse configurations of ubiquitin chains on target proteins(3). The 26S proteasome recognizes ubiquitylated substrates and degrades them into short peptides(4). Tests of defined configurations of ubiquitins on substrates for their influence on recognition and degradation showed that a substrate protein must generally be conjugated with a chain of at least four ubiquitins to interact tightly enough with the proteasome for degradation(5). However, no compelling evidence supports the existence of a tetraubiquitin chain receptor on the proteasome. Alternative explanations for the tetraubiquitin-chain selection rule mostly rely on the geometric distance between a pair of proteasomal subunits to gauge the length of a ubiquitin chain(6, 7). The requirement for a tetraubiquitin chain for degradation also differs from substrate to substrate, without a predictable relationship to a substrate’s function or structure(8–11).
Exit from mitosis and passage into the G1 phase of the cell cycle requires ubiquitylation mediated by the Anaphase-Promoting Complex (APC) (12). Substrates, such as cyclinB, securin, and geminin, typically contain multiple lysine residues, to which ubiquitin moieties are conjugated, providing a very large number of possible combinations of ubiquitin chain configurations(3, 8, 13, 14). Mass spectrometry studies indicate that ubiquitin chains on cyclinB molecules generated in reconstituted reactions contain on average only 2 ubiquitins(3). Moreover, multi-monoubiquitylation (referring to ubiquitylation on multiple lysine residues without chain formation) on cyclinB leads to efficient degradation. Multiple ubiquitylation sites are commonly found on substrates of other E3 enzymes(15, 16); at least 56% of ubiquitylated human proteins contain more than one ubiquitylation site, though the relevance of these ubiquitylations to protein degradation has not been fully demonstrated(17).
Ubiquitins conjugated to substrate promotes interaction with the proteasome; however, binding by itself is not sufficient to initiate degradation. Several known proteasome-interacting proteins (Usp14, Rad23, etc) are stable(18), and autoubiquitylated Cdc34 is not degraded despite its strong interaction with the proteasome (19). To facilitate initiation of substrate translocation, it has been proposed that substrates must have an unstructured terminal region of at least 30AA(20); multiple steps occur in the proteasomal degradation process, as suggested by cryoEM structures(6, 7, 21). Before or coincident with peptide translocation, conjugated ubiquitins are removed by deubiquitylating enzymes(DUB) Rpn11, Usp14, and Uch37 on the proteasome(22). Whereas Usp14 and Uch37 may have an editing role to tune the rate of proteasomal degradation, they are dispensable for the degradation process. The DUB Rpn11, by contrast, is required for efficient proteasomal activity. Rpn11 is located close to the substrate entry port, where it removes ubiquitin chains en bloc from the translocating peptide(6, 23). It is unclear how the proteasome might use these features to establish selectivity in substrate recognition.
To understand the requirements for efficient protein degradation, we examined the kinetics of degradation of substrates with defined ubiquitin configurations. We found that, for APC substrates with multiple ubiquitylated lysine residues, tetraubiquitin chains were not required for efficient degradation. Rather, given the same number of total ubiquitins on a substrate molecule, diubiquitin chains were more efficient than tetraubiquitin chains in promoting degradation. To understand the molecular basis through which some ubiquitin configurations promote more efficient degradation than others, we investigated the intermediate steps in the degradation pathway using single-molecule(SM) methods. For substrates containing multiple lysines, the strength of interaction with the proteasome was determined largely by the total number of ubiquitins and was less sensitive to the ubiquitin configuration. However, substrate binding alone was not sufficient for rapid degradation for most substrates. Rather, degradation depended strongly on the initiation rate of passage into the substrate translocation channel; and this transition was promoted by the presence of ubiquitin chain structures on substrates.
Results
Degradation of defined multiple ubiquitylated substrates
Most, if not all, APC substrates contain multiple ubiquitylatable lysine residues. To reduce the complexity of these mixed configurations, we sought to control the number, length and linkage of conjugated ubiquitin chains on wt-substrates. Purified APC was used as the ubiquitin ligase to conjugate preformed di- or tetra-ubiquitin chains of K48 linkage, which support most proteasome-mediated degradation in cells(24) (Fig. 1A). The chains were methylated on lysines before conjugation to prevent secondary elongation. We then separated the reaction products having different numbers of conjugated ubiquitins by electrophoresis. This approach limits the heterogeneity of ubiquitin configurations on the substrate to combinations of sites accepting a known number of defined ubiquitin chains. We measured the degradation rate of the ubiquitylated substrate for each electrophoretically resolved species by exposure to purified human 26S proteasomes that were free of reversibly-associating ubiquitin receptors such as Rad23 (Fig. 1A). Success of this strategy required the absence of interconversion of different ubiquitylated species generated by partial deubiquitylation by the proteasome; this was ensured by removing the DUB Usp14 on the proteasome by salt wash (25). Usp14-free proteasomes are known to efficiently degrade substrates, such as cyclinB and Sic1, without generating partially deubiquitylated products (Fig. S1)(8, 25, 26). Another DUB on the proteasome, Uch37, does not appreciably deubiquitylate the substrates used here(25). Controls using a general DUB inhibitor (not active against Rpn11) further confirmed that DUB-driven interconversion of ubiquitylated species was unlikely (Fig. S2).
Figure 1. Quantitative degradation assay.

A. The assay strategy. Preformed, methylated ubiquitin chains were conjugated to PKA-labeled securin using purified APC and E2 UbcH10. After reaction, the product was subject to degradation by purified 26S human proteasome. The decay constant for each ubiquitylated species, separated on a gel, was measured from a time series. Signal from unmodified substrates (Ub0) was used as a control for loading and nonspecific dephosphorylation which is the main reason for the decrease of Ub0 signals(Methods). B–D. 160nM geminin, securin, cyclinB-NT(Xenopus) were ubiquitylated using indicated constructs of ubiquitin. Their rates of degradation by 3nM purified human 26S proteasome were measured, and shown as a function of total ubiquitins per substrate molecule. Errorbars represent the standard deviation of three experimental replicates. ‘*’ the rate for this species is 1.4. The lack of data for certain species is either due to these species not having been tested or to their signals being too weak to quantify. E–F. Human cyclinB-NT mutants carrying lysines only at indicated positions were ubiquitylated with either methylated ubiquitin (M-Ub) or wt-Ub, and tested in a quantitative degradation assay. Original autoradiography for retrieving the rate information is compiled in Fig. S8. In E, the inset shows the location of D-box on cyclinB-NT and relevant ubiquitylatable lysine residues identified by mass spectrometry.
We analyzed the rates of degradation of ubiquitylated securin, geminin, and cyclinB-NT (N-terminal fragment from cyclinB), each with a known number of ubiquitin chains of defined length. We incubated these substrates with the 26S proteasome at a non-limiting concentration (25)(Fig. S3). The measured degradation rates were similar to rates observed in cells(26, 27). The degradation rates were substrate-dependent, even for substrates with the same number of conjugated ubiquitins. For example, highly ubiquitylated securin was degraded fastest, followed by cyclinB and geminin (Fig. 1B–D). Both methylated K48-diubiquitin chains and wt-ubiquitins promoted efficient proteasomal degradation. Conjugation with K48 diubiquitin chains supported a higher rate of degradation than K48-tetraubiquitin chains, when the same number of total ubiquitins were conjugated to a substrate molecule(Fig. 1B–D). The slower degradation associated with tetraubiquitin chains was not due to methylation(Fig. S4), nor to potential competition from free tetraubiquitin chains at the experimental concentration, as multi-diubiquitylated securin was degraded at the same rate even after free tetraubiquitin chains were added to the degradation reaction (Fig. S4). Faster degradation was observed with diubiquitin chains even when the two chains were closely apposed (Fig. S5); this also held for K11-linked chains (Fig. S6). By contrast to cyclinB, multi-monoubiquitylated securin and geminin, generated with either methylated ubiquitin or lysine-free ubiquitin (Ub0K) precluding chain formation, were degraded very slowly, compared to constructs with the same ubiquitin stoichiometry but containing chains (Fig. 1B,C; S7).
The distribution of ubiquitylated lysines on the substrate may set the degradation rate. Because our method to control ubiquitin configurations does not constrain which lysine residues receive the ubiquitin chains, we explicitly tested the contribution of the sites of ubiquitylation to degradation rate. Multi-monoubiquitylated cyclinB-NT was degraded as efficiently as wt-Ub conjugated cyclinB-NT(8)(Fig. 1D), perhaps because of the specific configurations of lysine targets on cyclinB. We made cyclinB-NT mutants that contain only 3 or 4 lysine residues at their original locations, with other lysines mutated to arginines, and studied rates of degradation of the multi-monoubiquitylated products of those mutants. There was a clear pattern: given the same number of total conjugated ubiquitins, the degradation rate strongly depended on the position of the most N-terminal, ubiquitylated lysine residue; the closer to N-terminus, the higher the rate of degradation (Fig. 1E). This pattern was not explained by some mutants being inherently nondegradable, because all mutants were degraded similarly when conjugated to wt-Ub at high ubiquitin multiplicity (Fig. 1F, Ub=7~8). Therefore, moving lysine residues away from the N-terminus appeared to progressively convert cyclinB into a substrate whose degradation was more similar to that of securin and geminin. Single-molecule studies presented below provide mechanistic insights into these properties.
Single-molecule studies of the kinetics of proteasomal degradation
Proteasome-substrate interactions
To understand the molecular steps that distinguish different ubiquitin configurations, we monitored the interactions of single ubiquitylated substrate molecules with the proteasome. Purified 26S proteasomes from human 293 cells were immobilized onto slides using with an antibody to the core 20S proteasome (Fig. 2A); the surface was passivated with polyethylene-glycol (PEG) and albumin to reduce nonspecific binding. To correlate the SM behavior with the extent of ubiquitin conjugation, each ubiquitin molecule was chemically labeled with a dylight550 fluorophore at the N-terminus. Fluorescent labeling of ubiquitin had no measurable effect on the kinetics of ubiquitylation and degradation in bulk reactions (Fig. S9). After ubiquitylation, the total fluorescence intensity of a substrate molecule was measured by TIRF microscopy, from which the number of conjugated ubiquitins was calculated. Accuracy and linearity of this method were assessed and confirmed by photobleaching experiments, a process that randomly inactivates single fluorophores(28). Background fluctuation was less than 0.2 ubiquitin level; more than 90% of substrate binding events can be identified with less than 30% uncertainty in measuring the number of conjugated ubiquitins (Fig. S10). This uncertainty was principally due to residual uneven illumination.
Figure 2. Proteasome-substrate interaction kinetics by the SM method.
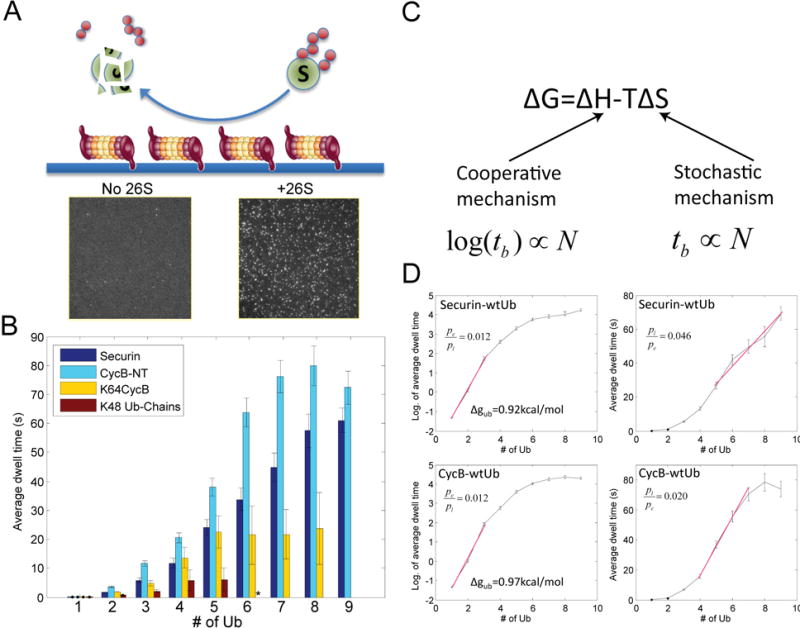
A. Schematics showing the experimental design, where purified 26S proteasome was immobilized on passivated coverslip using anti-20S antibody. Ubiquitin was fluorescently labeled and conjugated to substrates in solution. Sample pictures capture ubiquitin signals on the surface with either 26S proteasome (+26S) or antibody only (no 26S). B. The average dwell time on the proteasome for different substrates (or free ub-chain) with X-number of conjugated ubiquitin, measured by the SM method. The distributions of individual dwell times are shown in Fig. S15. Errorbars represent standard deviation of the mean. ‘*’: data for Ub-chain from 6~9 is not shown due to insufficient events. C. Cooperative and stochastic mechanisms affect binding enthalpy and entropy respectively. By each mechanism, the expected relationship between dwell time tb and the number of ubiquitins N is shown below. D. Dwell time on the proteasome(right), or its logarithm(left), for securin and cyclinB vs. the number of conjugated ubiquitins. The red line shows a linear fitting. The ratio of p-values by fitting the red segment (in linear scale) with either a linear model (pl) or an exponential model (pe) is shown. Δgub is the binding free energy per ubiquitin on the proteasome, calculated from the slope of the binding curve.
Ubiquitylated substrates transiently interacted with the 26S proteasome. Only background levels of binding were observed, if either the 20S proteasome was substituted for the 26S proteasome or if the substrate was omitted (Fig. S11). Interaction with the proteasome requires a hydrophobic patch spanning Leu8-Ile44-Val70 on ubiquitin and is compromised by substitutions for Ile44(29). In the SM assay, this mutation abrogated the binding of fluorescent ubiquitin to the proteasome (Fig. S11–12). These results indicate that the interaction occurred predominately at ubiquitin receptors on the 26S proteasome; the kinetics depended on the amount of ubiquitylation of the substrate, and was insensitive to excitation laser intensities, excluding low-level fluorophore photobleaching as a complicating factor (Fig. S13–14). To minimize systematic errors, each experiment and its controls were performed on the same slide with the same batch of proteasome and substrates.
To further validate the SM assay, we compared the affinity of ubiquitylated substrates with the proteasome using SM methods to published results measured in bulk assays. We determined the average dwell time of ubiquitylated cyclinB, securin, single-lysine K64cyclinB, or free ubiquitin chains on the proteasome, as a function of the total number of conjugated ubiquitins(Fig. 2B). At the same total number of ubiquitins, substrate-anchored ubiquitin chains interacted more strongly with the proteasome than did free chains, consistent with published results(5).
Higher ubiquitin stoichiometry consistently led to longer dwell times but the quantitative relationship between dwell time and the amount of ubiquitin unexpectedly differed from substrate to substrate. Thus, although the dwell time for free ubiquitin chains or K64cyclinB carrying a single ubiquitin chain plateaued at four to five ubiquitins, the dwell time for multi-lysine substrates, such as cyclinB and securin, increased continually as more ubiquitins were added. In a conventional competition assay, the Ki (≈Kd=koff/kon) of a tetraubiquitin chain is 170nM(5). The Kd value measured by the SM method is (210±60)nM, given an estimated kon = 8.5×105/M/sec(30). A K6A mutation on ubiquitin weakens the interaction of ubiquitylated substrates with the proteasome(29, 31). Consistent with this, the dwell time of UbK6A-securin on the proteasome was shorter than that of wtUb-securin in the SM assay (Fig. S16). The interaction between substrate backbone and the proteasome appears to be minimal, because the dwell time for substrates conjugated with a single ubiquitin was very short (dwell time < 300ms)(Fig. 2B).
The dwell time obtained from SM measurements includes both a period for the initial interaction with proteasome and, for productive interactions leading to degradation, a period for translocation and degradation. To assess the contribution of each step to the observed dwell time, we blocked enzymatically active sites of the proteasome with inhibitors that act downstream of the initial binding events (Fig. S17). Neither ubiquitin-aldehyde, which inhibits deubiquitylation by Uch37, nor epoxomicin, which inhibits substrate proteolysis, changed the dwell time. Using 1,10-phenanthroline to inhibit Rpn11 activity and proteasomal degradation also caused little change in the dwell time (Fig. S18–19). Therefore, dwell time primarily reports on the initial binding event between the ubiquitylated substrate and the proteasome.
Cooperative and stochastic features of proteasome-substrate interactions
Single-molecule studies can provide insights into mechanisms of the initial proteasome-substrate interaction, an important step for deconstructing the specificity of degradation. Only two proteasomal subunits, Rpn10 and Rpn13, contain ubiquitin-binding domains: Rpn10 has two UIM domains, and Rpn13 has one Pru domain (4). Together, they could maximally engage 3 ubiquitins at once. If a substrate molecule simultaneously bound to the three domains, this would constitute a type of cooperative or avidity binding process (Fig. 2C). Binding could also be enhanced by a “stochastic” mechanism, in which larger numbers of ubiquitins enhance the affinity of binding by increasing local ubiquitin concentrations, without requiring simultaneous interactions with different receptor proteins. These two mechanisms can be distinguished by dwell time analysis, because a cooperative mechanism primarily affects the enthalpic component of the free energy of binding, whereas a stochastic mechanism should change only the entropic contribution. Kinetically, the cooperative mechanism should result in an exponential increase of dwell time with the number of conjugated ubiquitins, but a stochastic mechanism would be characterized by a linear increase, reflecting mass action (Fig. 2C)(32).
For both cyclinB-NT and securin, the dwell time initially increased exponentially as a function of the number of conjugated ubiquitins, up to a total of 3 (Fig. 2D), consistent with a cooperative mechanism (Fig. 2C). In contrast, further ubiquitylation (from 4 to 9) led to a linear increase in dwell time, consistent with a stochastic mechanism (Fig. 2C,D). From the slope of the binding curve on a semi-log plot, we calculated the mean free energy of binding per bound ubiquitin to the proteasome: 0.92kcal/mol for securin and 0.97kcal/mol for cyclinB, suggesting weak interactions. The cooperative behavior at low ubiquitin stoichiometry and the linear behavior at higher levels provide keys to understanding how ubiquitin configurations are discriminated.
The role of the ubiquitin chain structure in proteasome-substrate interactions
Ubiquitin chains interact more strongly with the proteasome than do ubiquitin monomers(5). Most proteasomal substrates require conjugation of ubiquitin chains for degradation. As we showed, multi-monoubiquitylated securin (or geminin) was degraded much more slowly than securin with ubiquitin chains (Fig. 1C,B). The simplest explanation would be that this difference is due to weaker interaction of the multi-monoubiquitylated forms with the proteasome. To test this explanation, we measured the interaction between Ub0K-conjugated securin and the proteasome, and compared it to securin linked to wild-type ubiquitin. Surprisingly, Ub0K-conjugated securin and wtUb-conjugated securin showed almost identical dwell times on the proteasome, when substrates having the same total number of ubiquitins were compared (Fig. 3A). This result was not caused by surface-immobilization of proteasome, because it also occurred in binding assays performed in bulk, measuring interactions with the ubiquitin receptor Rpn10 on the proteasome (Fig. S20). Likewise, cyclinB, which can be degraded without forming ubiquitin chains, had a similar binding affinity whether conjugated with wtUb or Ub0K (Fig. 3B).
Figure 3. Interaction with the proteasome is mainly determined by the total number of ubiquitins on a substrate, insensitive to Ub-chain structures.
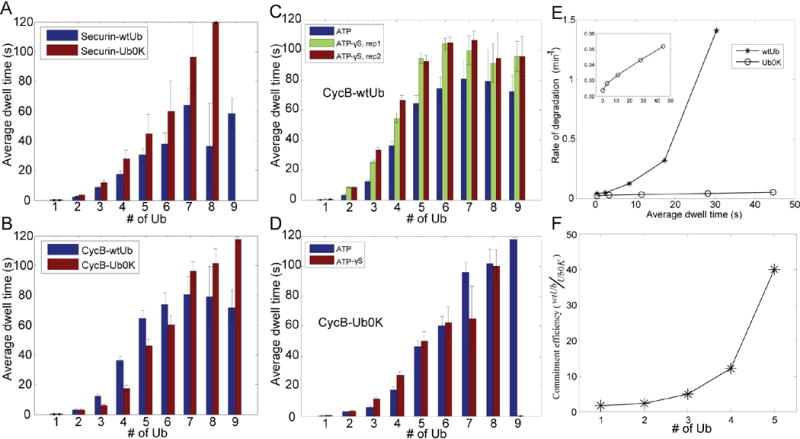
A–B. Securin or cyclinB-NT was ubiquitylated by APC with either wtUb or Ub0K, and tested for interaction with the proteasome as in Fig. 2B. C–D. Interaction of wtUb- or Ub0K-conjugated cyclinB with the proteasome in the presence of ATP or ATP-γS. ‘rep1’ & ‘rep2’ are two experimental replicates. The data for ATP-proteasome is plotted for comparison as is identical to B. CyclinB-ATP data in C is identical to that in B. E. Degradation rate and proteasomal dwell time relationship, for wtUb- and Ub0K-conjugated securin. Degradation rates were measured in the quantitative degradation assay (Fig. 1C), and dwell time on the proteasome was measured using the SM method. Inset shows the Ub0K result on a smaller Y-axis. F. the ratio of “commitment efficiency” for securin-wtUb over securin-Ub0K, as a function of total conjugated ubiquitins. “Commitment efficiency” is defined as the degradation rate divided by the dwell time.
Although degradation of most substrates appears to be much more efficient if chains are formed, our results show that the presence of chains does not alter the affinity of substrate binding to the proteasome. Instead affinity is primarily determined by the total number of ubiquitins on a substrate molecule, irrespective of their configurations except in the special case of single-chain substrates. Nevertheless, the increase in degradation rate when chains are present implies that ubiquitin configuration has an important role in the fate of the substrate after its binding to the proteasome. Therefore, we used SM methods to examine how the presence of ubiquitin chains affects various steps in the degradation process, in order to understand how ubiquitin chains stimulate substrate degradation.
During the process of degradation, the proteasomal subunits in the 19S regulatory particle alternate among different ATPase-driven conformational states(33–35). To test whether the recognition of ubiquitin chains may require a particular proteasome conformation, we locked the proteasome in the ATP-bound state with the non-hydrolyzable analog ATP-γS (Fig. S18), and studied the proteasome’s interaction with ubiquitylated substrates. There was a consistent increase of ~25% in dwell time for binding of wtUb-conjugated cyclinB to the ATP-γS proteasome, as compared to proteasomes in ATP buffer. This increase in affinity was not observed when cyclinB was conjugated with Ub0K(Fig. 3C,D). As a more sensitive metric for this effect, 29% of the binding events between ATP-γS proteasome and wtUb-conjugated cyclinB lasted longer than 10 seconds, whereas only 10% of binding interactions had such persistence under the three control conditions(Fig. S21). Another substrate, securin, gave similar results (Fig. S22). Proteasomes in buffer containing ADP showed weaker interactions with ubiquitylated cyclinB than those in ATP-containing buffer and the binding of wtUb-conjugated cyclinB to ADP-proteasomes was not different from that of Ub0K-conjugated cyclinB (Fig. S23). We thus conclude that the discrimination of different configurations of ubiquitin on a substrate is enhanced in the ATP-bound state of the proteasome, and therefore, may result from rearrangement of the dynamic conformations of the proteasomal subunits. This discrimination was observed as a small binding enhancement, which by itself is unlikely to explain the generally large degradation-rate enhancement for Ub-chain containing substrates. Perhaps the rearrangement of proteasomal subunits may expose or activate a hidden ubiquitin chain receptor that contributes to substrate discrimination. Two alternative mechanisms for the enhancement of protein degradation in the presence of ubiquitin chains: allosteric opening of the gate of 20S complex and stimulation of proteasomal ATPase activity(36–38), were found in control studies to have little effect, when tested with ubiquitylated Ube2S as the substrate(Fig. S24–25), and therefore unlikely to explain the much larger effect of ubiquitin chains in promoting degradation(Fig. 1B,C).
The role of ubiquitin chains in initiating substrate translocation
If binding strength were the only determinant for degradation, the degradation rate should be proportional to the dwell time on the proteasome, as predicted by a probabilistic model of chemical reactions. For Ub0K-conjugated securin, this appeared to be the case over most of the range of ubiquitylation (Fig. 3E), where the rate versus dwell time was linear. By contrast, the degradation rate of wtUb-conjugated securin increased superlinearly with dwell time(Fig. 3E), suggesting that ubiquitin chains, in addition to providing interaction with the proteasome, may also increase the efficiency of commitment of a substrate to degradation once bound to the proteasome(i.e. a kcat effect) (Fig. 3F). Remarkably, at high modification levels, wtUb was up to 40-fold more efficient in promoting securin degradation than Ub0K, per unit dwell time.
In addition to binding kinetics, SM traces can provide critical mechanistic information about events that follow binding, such as substrate translocation and degradation by the proteasome, in the form of the kinetics of ubiquitin removal from the substrate. We observed two different types of signals after binding of substrate to the proteasome: structureless, in which there was a complete loss of the ubiquitin within 200ms (the sampling rate of the camera), and stepped, in which the ubiquitin signal decreased in several discrete steps with very short intervals (Fig. 4A). These effects were not attributable to photobleaching or the limited speed of camera detection (Fig. S26–27).
Figure 4. Ubiquitin chains on substrates promote translocation initiation.
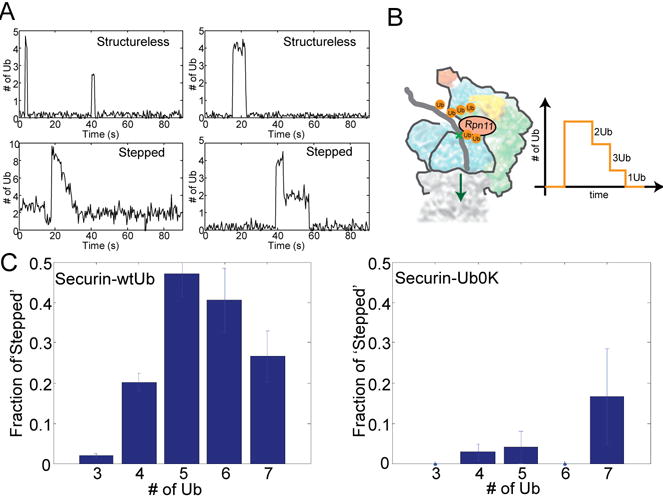
A. Examples of ‘structureless’ and ‘stepped’ traces from a SM experiment on wtUb-conjugated securin. ‘Stepped’ traces are due to progressive deubiquitylation by Rpn11 as illustrated in B. C. The fraction of ‘stepped’ traces, as a function of Ub#, for wtUb-(N=663) or Ub0K-conjugated(N=133) securin. Errorbars represent standard deviation of the mean.
Although the likely explanation for structureless events is dissociation of substrate from the proteasome before ubiquitin removal, the stepped decrease of ubiquitin signal could represent a series of deubiquitylation events, catalyzed by Rpn11, the only effective DUB in our samples (Fig. 4B). In support of this interpretation, inhibitors of Rpn11 activity, such as 1,10-phenanthroline and ATP-γS, almost completely eliminated the stepped decreases (Fig. S28). When cyclinB-NT was conjugated with multiple diubiquitin chains, we observed with each step the removal of two ubiquitins rather than one (Fig. S29). Thus, the proposal that Rpn11 cleaves ubiquitin chains en bloc(23) is strongly supported at the SM level. Accordingly, we did not observe the stepped loss of ubiquitin signals for ubiquitylated K64cyclinB, a single-lysine substrate that carries only one ubiquitin chain(Fig. S27). Presumably, if all ubiquitins on a multi-lysine substrate happened to form a single chain, only structureless events would be observed. However, such cases should be rare for APC substrates, because the E2 UbcH10 forms long ubiquitin chains with low efficiency. Rather, it acts broadly to increase the number of sites of mono- or di- ubiquitylation on multiple lysines(3).
Kinetic studies also reveal the sequence of events in degradation. Before the very processive phase of deubiquitylation, there was always a delay. The distribution of delay time was not exponential, as would be expected for a single-step reaction. Instead the distribution corresponded closely to a Γ-distribution with a modal delay of 2 seconds (Fig. S30), suggesting intermediate rate-limiting steps between initial binding and the onset of deubiquitylation.
Rpn11 sits at the substrate entry port, where it has a critical role in removing ubiquitin chains on translocating peptides(6, 39–42). Translocation may signal irreversible engagement of the substrate into the degradation process. This tight coupling between deubiquitylation and translocation was supported by the absence of deubiquitylated but non-degraded substrates in our bulk assays (Fig. S1). We therefore used the processive deubiquitylation mediated by Rpn11 as a kinetic indicator for substrate translocation. To test if the substrate was cleaved by the proteasome contemporaneously with deubiquitylation, we labeled both ends of cyclinB-NT with Dylight550. We observed a ‘stepped’ decrease of fluorescence on cyclinB that was sensitive to proteasome inhibition by MG132 and was coincident with the deubiquitylation, suggesting co-translocational activity of Rpn11 (Fig. S31; S3). We compared the fraction of stepped events among all binding events, for wtUb-conjugated securin and Ub0K-conjugated securin to assess their relative probability of undergoing deubiquitylation and translocation steps. At increased amounts of conjugated ubiquitin, this probability increased dramatically for securin conjugated with wt-ubiquitin capable of forming chains. By contrast, it remained low if Ub0K, which precludes chain formation, was used (Fig. 4C). This difference was large enough to account for the 12-fold degradation-rate disparity between wtUb-conjugated and Ub0K-conjugated securin. Multi-monoubiquitylated securin was not intrinsically refractory to deubiquitylation, which still occurred processively in the few cases of stepped events on multi-monoubiquitylated securin (Fig. S32). Thus ubiquitin chains on substrates appear to specifically promote initiation of translocation and degradation.
Discussion
The multiple lysines on a substrate and on ubiquitin itself generate a large number of possible ubiquitin configurations. These configurations represent a “ubiquitin code” of unknown degeneracy that must be read by the proteasome and converted into a rate of substrate degradation. We have approached this problem with two methods: construction of substrates with defined ubiquitin configurations and single-molecule techniques. We found that tetraubiquitin chains are not essential for rapid proteasomal degradation of APC substrates, which would explain why a tetraubiquitin receptor on proteasome has not been found. In fact, ubiquitin chains on cyclinB, and possibly other APC substrates, are typically short(3) and multiple ubiquitylatable lysine residues are a common feature of these substrates. A distributed array of short ubiquitin chains appears to be a superior and perhaps an optimal signal for proteasomal degradation; this conclusion could probably extend to substrates of other E3 ligases. Although a single ubiquitin chain may be sufficient for degrading certain substrates, such as Sic1 mutants and IκB(16, 43), increasing the number of ubiquitylated lysine residues of the canonical single-chain substrate, β-galactosidase, greatly accelerates its degradation(44). Similarly wt-cyclinB was degraded faster than mutants having fewer lysines at the same total amount of ubiquitylation (Fig. 1F, Ub=7~8). Besides K48 chains, the APC also establishes K11 and K63 linkages on substrates(3). We found that K48-chains promoted more efficient degradation than K11, K27 and K63 chains (Fig. S6).
By studying cyclinB mutants, we found that proximity of the first ubiquitylated lysine to the N-terminus was associated with faster degradation (Fig. 1E), suggesting that degradation rate is sensitive to the position of ubiquitylated lysine residues. There was a correspondence between long dwell times and elevated rates of degradation (Fig. S33A), suggesting that ubiquitin-chain position could control the rate of degradation, at least partially, through controlling affinity with the proteasome. Single-lysine mutants of cyclinB with a ubiquitin chain at different positions had indistinguishable binding kinetics to the proteasome, suggesting that mono-chain and multi-chain substrates may interact with the proteasome by different mechanisms (Fig. S33B). For wildtype substrates, our current method of constructing defined ubiquitin configurations does not specify the chain positions; the results are understood as a populational average of all actual combinations of positions, of which the vast majority can promote efficient degradation.
Comparison of the Kd values for tetraubiquitin chains measured by our SM methods with those from bulk assays suggests that surface immobilization of the proteasome is unlikely to distort kinetic rate constants. In addition, ubiquitylated securin took on average 10 seconds to complete translocation and possibly the degradation process on surface-bound proteasome (Fig. S34). This result is consistent with time for degrading similar-size proteins, such as DHFR and Sic1, measured in bulk assays(45), suggesting the surface-bound proteasome is unimpaired for unbiased kinetic studies.
The rate of degradation is determined by both binding (i.e. Kd-effect) and post-binding (i.e. kcat-effect) events on the proteasome. For APC substrates, multiple diubiquitin chains were more efficient degradation signals than tetraubiquitin chains, given the same total number of conjugated ubiquitins (Fig. 1B–D). The explanation for this distinction may be their different binding strength with the proteasome. Using SM methods, we observed weaker binding if ubiquitins were assembled into a single chain on K64cyclinB when compared to that for wt-cyclinB, which had short and distributive chain configurations (Fig. 2B). High resolution cryo-EM structures of the proteasome are consistent with the potential effectiveness of multiple short ubiquitin chains. Since proteasomal ubiquitin receptors Rpn10 and Rpn13 are distant from each other (6, 7), distributive configurations of ubiquitins on a substrate molecule might promote use of more ubiquitin receptors on the proteasome; a single ubiquitin chain might also be less effective due to steric constraint. Furthermore, Rpn10 and Rpn13 may not be the only ubiquitin receptors on proteasome since budding yeast can tolerate mutations of both(46). Additional receptors or shuttle factors for the proteasome are also thought to contribute to the binding of ubiquitylated proteins(47, 48)(see below).
The SM binding measurements suggest a model whereby a substrate molecule samples multiple modes of binding during its interaction with the proteasome (Fig. 5). Evidence for such mechanism comes from measurement of the dwell time as a function of ubiquitylation levels on the substrate. Beyond the ubiquitin-binding capacity of the proteasome, limited most likely to 3~4 ubiquitins by available ubiquitin receptors, a further increase of binding affinity relies on an increase in the local ubiquitin concentration on the substrate – the stochastic interaction. This stochastic mechanism stabilizes the bound state by increasing its entropy, or the number of microscopic states, since entropy is proportional to the logarithm of the number of these states. In this system, an increase in relevant entropy may occur if the substrate molecule can explore multiple conformations on the proteasome through intramolecular diffusion, while remaining associated with the proteasome (Fig. 5). Such dynamic sampling should also increase the likelihood that the peptide terminus would be captured by the substrate entry port on the ATPases, thereby facilitating initiation of translocation. A cooperative process is implied by the exponential increase of dwell time as a function of the number of conjugated ubiquitins, while a stochastic process is implied by a linear increase (Fig. 2C,D). A similar, biphasic binding relationship (1/Ki~Ub number) was in fact suggested in an early publication using competition assays, though the interpretation was different(5) (Fig. S35). An exponential increase of dwell time involving greater than three simultaneous interactions would further increase the discrimination of ubiquitin levels over a linear increase. Why then is the process no longer exponential after four ubiquitins? Cooperative mechanisms tend to promote tight binding, which has potential risks for the cell. If a highly ubiquitylated substrate could not be degraded by the proteasome, it would stably block the proteasome. Such an inhibition by stably binding complexes has been proposed to underlie the accumulation of ubiquitylated intermediates in various neurodegenerative diseases(49). A linear increase in affinity at high ubiquitin stoichiometry, though less discriminating, is also less prone to form unproductive, inhibitory substrate-proteasome complexes.
Figure 5. An integrated model for the degradation of ubiquitylated substrates by the 26S proteasome.
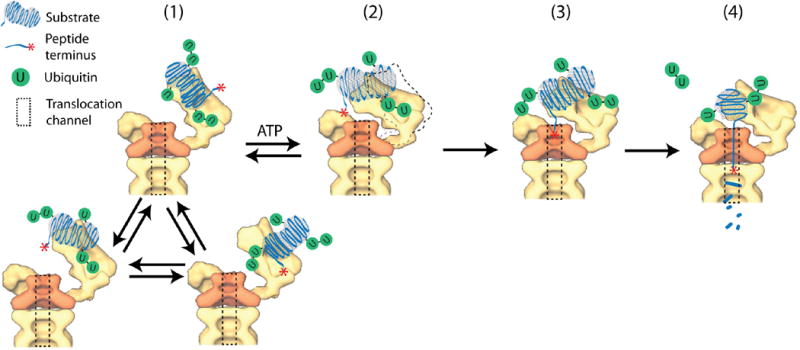
Polyubiquitylated substrates could simultaneously interact with multiple ubiquitin receptors on the proteasome. While remaining bound, a substrate molecule explores multiple configurations on the proteasome through intramolecular diffusion (1). Rearrangements of proteasomal subunits, fueled by the ATPases in response to binding and hydrolysis of ATP, expose or activate a deeper ubiquitin chain receptor, and facilitate the transfer of ubiquitylated substrates from initial binding to a deeper engagement (2,3). In this engagement, the substrate or its terminus is closer to the substrate entry port, which expedites translocation initiation (3) and ensuing degradation (4).
Although binding to the proteasome is a prerequisite for degradation, it does not in itself determine the rate of degradation. For example, wtUb-conjugated securin and Ub0K-conjugated securin bind equally tightly, but the former is degraded much faster than the latter. The specificity of degradation must also reflect post-binding events. The SM analysis of Rpn11-dependent deubiquitylation indicates that the chain structure of ubiquitin promotes the initiation of translocation, a requirement for degradation. This effect of Ub-chains also applies to cyclinB, a special substrate that can be degraded even without ubiquitin chains. We observed a shorter delay between binding and the initial deubiquitylation event for wtUb-conjugated cyclinB than Ub0K conjugates, consistent with the translocation-promoting activity of ubiquitin chains (Fig. S36).
Most substrate-proteasome encounters do not lead to degradation, especially for substrates with a low number of conjugated ubiquitins (Fig. 4C). Even for highly ubiquitylated substrates, binding events sometimes lasted for tens of seconds without leading to degradation. Thus there may be a latent state of the proteasome, where heterogeneous ubiquitin chain conformations might affect deubiquitylation(50), or, perhaps more likely, might affect the orientation of a bound substrate, placing the translocation-initiating element far away from the substrate entry port. In this context the presence of a short flexible domain at a substrate’s terminus should substantially accelerate its rate of degradation(20, 51). Therefore, engagement of the translocation-initiating element by force-generating pore loops of the proteasomal ATPases, which are reached via the substrate entry port, may generally be a rate-limiting step in degradation. Translocation initiation has been proposed to underlie ‘commitment’, a hypothetical point where substrates are irreversibly destined to degradation(39). We would argue that translocation initiation, sensitive to the configuration of ubiquitin groups on the substrate, is either the commitment step or is closely coupled to it.
To understand how ubiquitin chains promote translocation initiation, we propose a model based on our experimental observations (Fig. 5). Conformational changes of the proteasome, driven by the ATPases quickly transiting through different nucleotide-bound states(33, 40), may activate a ubiquitin-chain receptor(s) that participates in substrate recognition(52). Candidates for such a receptor include ATPase Rpt5, which can be crosslinked to bound ubiquitin chains(53). The same result can also be explained by rearrangement of ubiquitin receptors (Rpn10&Rpn13) into a higher-affinity state for ubiquitin chains. It would make sense if the additional ubiquitin-chain receptor were closer to the substrate entry port than Rpn10 or Rpn13, to facilitate translocation initiation by engaging the substrate into a ‘deeper’ conformation after the initial interaction involving mainly Rpn10 and Rpn13 (Fig. 5). Such an intermediate step is indicated by the delay before deubiquitylation or translocation (Fig. S30). Consistently, Rpt5 is very close to the substrate entry port; and conformational changes induced by ATP-γS dramatically reduce the distance between Rpn10 and Rpt4/5, indicating a possible direct transfer of substrates from initial binding to deeper engagement(33, 54)(Fig. S37).
In summary, we have shown that there is no simple length threshold for ubiquitin chains for degradation by the proteasome. Rather there are at least two requirements, a minimal number of ubiquitins to result in tight binding, and a certain number or length of chains to promote translocation into the axial channel. The ultimate rate of degradation is likely set by ubiquitin stoichiometry, chain configuration, and properties of the substrate that affect not only the capacity to be ubiquitylated and the configuration of chains but also the orientation of the chains and translocation-initiating elements once bound to the proteasome.
Supplementary Material
Acknowledgments
We thank L. B. and W. M. for commenting on the manuscript. We thank the Nikon imaging center at Harvard medical school for providing instruments. Y. L. is supported by a Damon Runyon research fellowship and a Lallage Feazel Wall Fellow. This work was supported by in part by National Institutes of Health Grant GM43601 (to D. F.) and GM66492 (to R.W.K.)
Footnotes
References and notes
- 1.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004 Nov 29;1695:55. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997 Dec;11:1245. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 3.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006 Jul;8:700. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 4.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000 Jan 4;19:94. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander GC, et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012 Feb 9;482:186. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakata E, et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2012 Jan 31;109:1479. doi: 10.1073/pnas.1119394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimova NV, et al. APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat Cell Biol. 2012 Feb;14:168. doi: 10.1038/ncb2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciechanover A, Stanhill A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim Biophys Acta. 2013 Jul 18; doi: 10.1016/j.bbamcr.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Asher G, Reuven N, Shaul Y. 20S proteasomes and protein degradation “by default”. Bioessays. 2006 Aug;28:844. doi: 10.1002/bies.20447. [DOI] [PubMed] [Google Scholar]
- 11.Shabek N, et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol Cell. 2012 Oct 12;48:87. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Barford D. Structure, function and mechanism of the anaphase promoting complex (APC/C) Q Rev Biophys. 2011 May;44:153. doi: 10.1017/S0033583510000259. [DOI] [PubMed] [Google Scholar]
- 13.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006 Jan 13;124:89. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 14.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996 Dec 15;10:3081. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 15.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010 Jan;17:86. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petroski MD, Deshaies RJ. Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol Cell. 2003 Jun;11:1435. doi: 10.1016/s1097-2765(03)00221-1. [DOI] [PubMed] [Google Scholar]
- 17.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011 Oct 21;44:325. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011 May 19;473:337. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 19.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002 Sep;4:725. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 20.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nature structural & molecular biology. 2004 Sep;11:830. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 21.Lasker K, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proceedings of the National Academy of Sciences of the United States of America. 2012 Jan 31;109:1380. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011 May;10:R110 003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002 Sep 26;419:403. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 24.Finley D, et al. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Molecular and cellular biology. 1994 Aug;14:5501. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee BH, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010 Sep 9;467:179. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006 Oct 6;127:99. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 27.Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat Cell Biol. 2011 Mar;13:223. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Wang W, Kirschner MW. Specificity of the anaphase promoting complex: a single-molecule study. Science. 2014 doi: 10.1126/science.1248737. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. The Journal of biological chemistry. 2001 Aug 10;276:30483. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 30.See note in the supplement: “Estimating the on-rate of tetraubiquitin chains with immobilized 26S proteasome in SM experiments”.
- 31.Shang F, et al. Lys6-modified ubiquitin inhibits ubiquitin-dependent protein degradation. The Journal of biological chemistry. 2005 May 27;280:20365. doi: 10.1074/jbc.M414356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.See note in the supplement: “Background information on the stochastic binding mechanism”.
- 33.Sledz P, et al. Structure of the 26S proteasome with ATP-gammaS bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proceedings of the National Academy of Sciences of the United States of America. 2013 Apr 30;110:7264. doi: 10.1073/pnas.1305782110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beckwith R, Estrin E, Worden EJ, Martin A. Reconstitution of the 26S proteasome reveals functional asymmetries in its AAA+ unfoldase. Nature structural & molecular biology. 2013 Oct;20:1164. doi: 10.1038/nsmb.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011 Feb 18;144:526. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell. 2009 Dec 11;36:794. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peth A, Kukushkin N, Bosse M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37 homologs. The Journal of biological chemistry. 2013 Mar 15;288:7781. doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bech-Otschir D, et al. Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nature structural & molecular biology. 2009 Feb;16:219. doi: 10.1038/nsmb.1547. [DOI] [PubMed] [Google Scholar]
- 39.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002 Oct 18;298:611. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 40.Matyskiela ME, Lander GC, Martin A. Conformational switching of the 26S proteasome enables substrate degradation. Nature structural & molecular biology. 2013 Jul;20:781. doi: 10.1038/nsmb.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Worden EJ, Padovani C, Martin A. Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nature structural & molecular biology. 2014 Mar;21:220. doi: 10.1038/nsmb.2771. [DOI] [PubMed] [Google Scholar]
- 42.Pathare GR, et al. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proceedings of the National Academy of Sciences of the United States of America. 2014 Feb 25;111:2984. doi: 10.1073/pnas.1400546111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annual review of immunology. 2000;18:621. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki T, Varshavsky A. Degradation signals in the lysine-asparagine sequence space. EMBO J. 1999 Nov 1;18:6017. doi: 10.1093/emboj/18.21.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peth A, Nathan JA, Goldberg AL. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. The Journal of biological chemistry. 2013 Oct 4;288:29215. doi: 10.1074/jbc.M113.482570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008 May 22;453:481. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. The Journal of biological chemistry. 2004 Jun 25;279:26817. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 48.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004 Jul 9;118:99. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 49.Imamura S, Yabu T, Yamashita M. Protective role of cell division cycle 48 (CDC48 protein against neurodegeneration via ubiquitin-proteasome system dysfunction during zebrafish development. The Journal of biological chemistry. 2012 Jun 29;287:23047. doi: 10.1074/jbc.M111.332882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye Y, et al. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012 Dec 13;492:266. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun. 2011;2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell. 2010 Nov 24;40:671. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002 Apr 18;416:763. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 54.Forster F, Unverdorben P, Sledz P, Baumeister W. Unveiling the long-held secrets of the 26S proteasome. Structure. 2013 Sep 3;21:1551. doi: 10.1016/j.str.2013.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


