Abstract
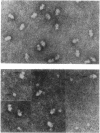
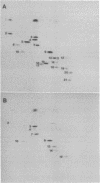
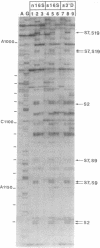
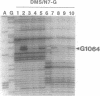
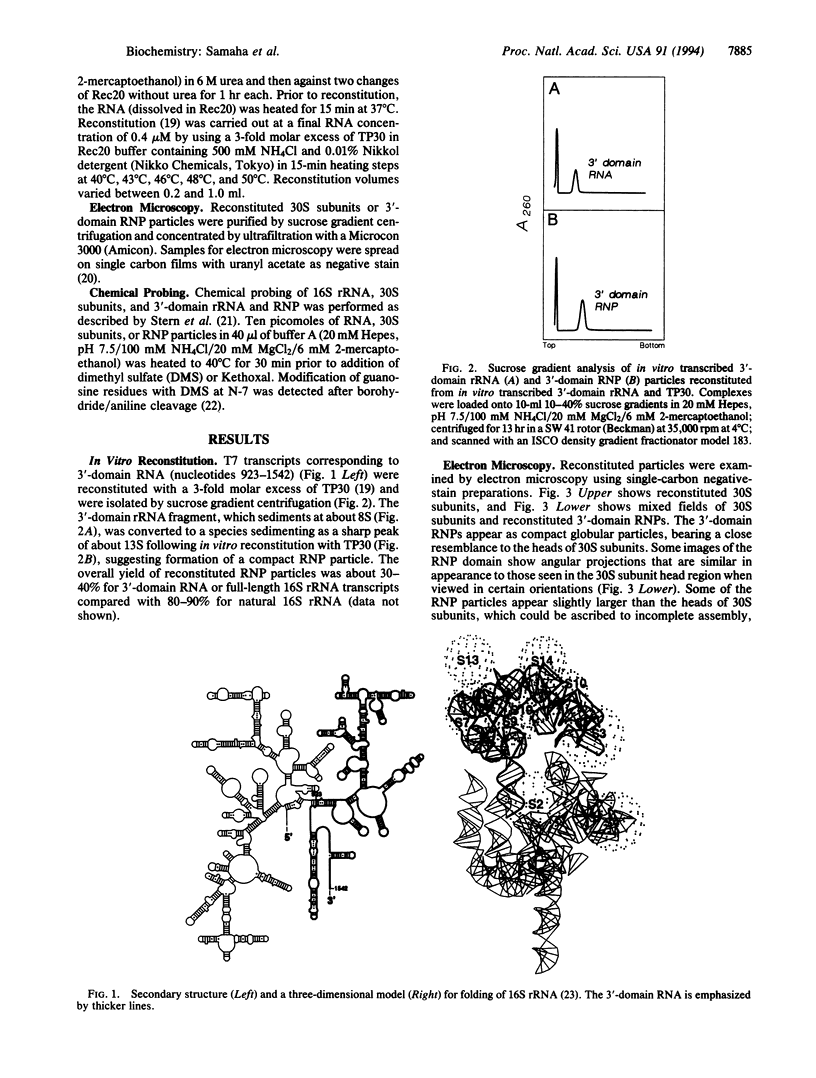
Small (30S) subunits of Escherichia coli ribosomes are composed of 21 proteins and a 1542-nucleotide 16S rRNA, whose secondary structure is divided into three domains. An in vitro transcript of the 3' domain of 16S rRNA (residues 923-1542), assembles efficiently with 30S ribosomal proteins to form a compact ribonucleoprotein (RNP) particle. Isolated particles examined under the electron microscope have a globular appearance, similar in size and shape to the head of the 30S ribosomal subunit. Two-dimensional gel analysis of the particles indicates the presence of proteins S3, S7, S9, S10, S13, S14, and S19 and smaller amounts of S2, all of which have been localized to the head of the 30S subunit by immunoelectron microscopy and neutron diffraction and belong to the S7 assembly family. Interestingly, protein S4, which is believed to interact exclusively with the 5' domain, is also reproducibly found associated with the particles in significant amounts. Chemical probing of the RNA in the assembled particle reveals characteristic cleavage protection patterns, showing that the proteins assemble with the 3'-domain RNA similarly to the way in which they assemble with 16S rRNA, although some of the later steps of assembly appear to be incomplete. These results show that the 3' domain of 16S rRNA can indeed assemble independently of the rest of the 30S subunit into a particle that resembles its structure in the ribosome. In addition, the assembled particles are able to bind spectinomycin with an affinity comparable to that of 30S subunits.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bilgin N., Richter A. A., Ehrenberg M., Dahlberg A. E., Kurland C. G. Ribosomal RNA and protein mutants resistant to spectinomycin. EMBO J. 1990 Mar;9(3):735–739. doi: 10.1002/j.1460-2075.1990.tb08167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen A., Davies J., Ozaki M., Mizushima S. Ribosomal protein conferring sensitivity to the antibiotic spectinomycin in Escherichia coli. Science. 1968 Jul 4;165(3888):85–86. [PubMed] [Google Scholar]
- Capel M. S., Engelman D. M., Freeborn B. R., Kjeldgaard M., Langer J. A., Ramakrishnan V., Schindler D. G., Schneider D. K., Schoenborn B. P., Sillers I. Y. A complete mapping of the proteins in the small ribosomal subunit of Escherichia coli. Science. 1987 Dec 4;238(4832):1403–1406. doi: 10.1126/science.3317832. [DOI] [PubMed] [Google Scholar]
- Dragon F., Brakier-Gingras L. Interaction of Escherichia coli ribosomal protein S7 with 16S rRNA. Nucleic Acids Res. 1993 Mar 11;21(5):1199–1203. doi: 10.1093/nar/21.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyl D., Böck A., Isono K. An improved method for two-dimensional gel-electrophoresis: analysis of mutationally altered ribosomal proteins of Escherichia coli. Mol Gen Genet. 1981;181(3):309–312. doi: 10.1007/BF00425603. [DOI] [PubMed] [Google Scholar]
- Gurevich V. V., Pokrovskaya I. D., Obukhova T. A., Zozulya S. A. Preparative in vitro mRNA synthesis using SP6 and T7 RNA polymerases. Anal Biochem. 1991 Jun;195(2):207–213. doi: 10.1016/0003-2697(91)90318-n. [DOI] [PubMed] [Google Scholar]
- Held W. A., Ballou B., Mizushima S., Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974 May 25;249(10):3103–3111. [PubMed] [Google Scholar]
- Krzyzosiak W., Denman R., Nurse K., Hellmann W., Boublik M., Gehrke C. W., Agris P. F., Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987 Apr 21;26(8):2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A., Pendergast M., Kahan L., Nomura M. Localization of Escherichia coli ribosomal proteins S4 and S14 by electron microscopy of antibody-labeled subunits. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4688–4692. doi: 10.1073/pnas.71.12.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J Mol Biol. 1990 Jan 5;211(1):135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Morgan J., Brimacombe R. A series of specific ribonucleoprotein fragments from the 30-S subparticle of Escherichia coli ribosomes. Eur J Biochem. 1972 Sep 25;29(3):542–552. doi: 10.1111/j.1432-1033.1972.tb02020.x. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Nomura M., Traub P., Guthrie C., Nashimoto H. The assembly of ribosomes. J Cell Physiol. 1969 Oct;74(2 Suppl):241+–241+. doi: 10.1002/jcp.1040740428. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Changchien L. M., Craven G. R., Noller H. F. Probing the assembly of the 3' major domain of 16 S ribosomal RNA. Quaternary interactions involving ribosomal proteins S7, S9 and S19. J Mol Biol. 1988 Mar 20;200(2):309–319. doi: 10.1016/0022-2836(88)90243-4. [DOI] [PubMed] [Google Scholar]
- Powers T., Noller H. F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991 Aug;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Stern S., Changchien L. M., Noller H. F. Probing the assembly of the 3' major domain of 16 S rRNA. Interactions involving ribosomal proteins S2, S3, S10, S13 and S14. J Mol Biol. 1988 Jun 20;201(4):697–716. doi: 10.1016/0022-2836(88)90468-8. [DOI] [PubMed] [Google Scholar]
- Rave N., Crkvenjakov R., Boedtker H. Identification of procollagen mRNAs transferred to diazobenzyloxymethyl paper from formaldehyde agarose gels. Nucleic Acids Res. 1979 Aug 10;6(11):3559–3567. doi: 10.1093/nar/6.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman M. G., Liljas A. Letter: Recognition of structural domains in globular proteins. J Mol Biol. 1974 May 5;85(1):177–181. doi: 10.1016/0022-2836(74)90136-3. [DOI] [PubMed] [Google Scholar]
- Siegmann M., Thomas G. Separation of multiple phosphorylated forms of 40 S ribosomal protein S6 by two-dimensional polyacrylamide gel electrophoresis. Methods Enzymol. 1987;146:362–369. doi: 10.1016/s0076-6879(87)46037-0. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Morgan E. A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984 Jun 11;12(11):4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Changchien L. M., Craven G. R., Noller H. F. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J Mol Biol. 1988 Mar 20;200(2):291–299. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989 May 19;244(4906):783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Weitzmann C. J., Cunningham P. R., Nurse K., Ofengand J. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 1993 Jan;7(1):177–180. doi: 10.1096/fasebj.7.1.7916699. [DOI] [PubMed] [Google Scholar]
- Yuki A., Brimacombe R. Nucleotide sequences of Escherichia coli 16-S RNA associated with ribosomal proteins S7, S9, S10, S14 and S19. Eur J Biochem. 1975 Aug 1;56(1):23–34. doi: 10.1111/j.1432-1033.1975.tb02203.x. [DOI] [PubMed] [Google Scholar]