Abstract
Since its relatively recent discovery, RNA interference (RNAi) has emerged as a potent, specific, and ubiquitous means of gene regulation. Through a number of pathways that are conserved from yeast to humans, small non-coding RNAs direct molecular machinery to silence gene expression. In this review, we focus on mechanisms and structures that govern RNA silencing in higher organisms. In addition to highlighting recent advances, parallels and differences between RNAi pathways are discussed. Together, the studies reviewed herein reveal the versatility and programmability of RNA-induced Silencing Complexes (RISCs) and emphasize the importance of both upstream biogenesis and downstream silencing factors.
Discovery and Biological Perspectives of RNA interference
RNAi was first described by Fire and Mello in the 1990s when, in an attempt to use anti-sense RNA to down-regulate gene expression, they observed that double-stranded RNA (dsRNA) was more potent than sense or anti-sense RNA alone [1]. This seminal work boasted robust and specific gene knock down in addition to coining the term “RNA interference” (RNAi). Shortly beforehand, the discovery of individual regulatory RNAs in C. elegans hinted that small, non-coding RNAs might be a pervasive means of gene regulation in higher organisms [2, 3]. In the following decade, with the mechanistic insight of Fire and Mello’s work in hand, this idea was confirmed and it is now accepted that well-over 1,000 small RNAs are encoded in the human genome that may regulate over 60% of our genes [4, 5]. It also became apparent that several seemingly disconnected phenomena are variations of RNAi-type pathways including co-suppression in plants, DNA elimination in Tetrahymena, and quelling in Neurospora [6]. The prevalence of RNAi is impressive and its importance is underscored by the fact that RNAi dysfunction is associated with numerous diseases and disorders including neurological maladies, cancers, and infertility.
Shortly after the initial description of RNAi phenomenology, a burst of genetic, biochemical, biophysical, and bioinformatic efforts laid a strong foundation for identifying the molecular pathways, players, and parameters that govern silencing via RNAi. Work from numerous groups defined the molecular apparatus of RNAi as RISC (RNA-induced Silencing Complex), a ribonucleoprotein complex minimally comprised of a small single-stranded RNA (~20-31 nucleotides) and an Argonaute protein which serves as the effector molecule (reviewed in [7]). In this configuration, the loaded “guide” RNA acts as a specificity determinant that directs Argonaute and any other associated machinery to the target. The guide-loaded Argonaute platform underlies every example in the expansive array of known RNAi pathways in eukaryotes regardless of the source of the guide (structured loci, transposons, viral, etc.), the machinery used to generate it, or the target RNA. The specific manner in which the target is silenced differs, but the essence of all of these pathways is conserved.
In most animals, there are three principal modes of RNA interference: the micro- (mi-), small interfering (si-), and Piwi-interacting (pi-) RNA pathways. These differ most notably in the cellular source of guide RNA precursors (biogenesis phase) (Box 1) and the mechanism of target silencing (effector phase) (Box 2).
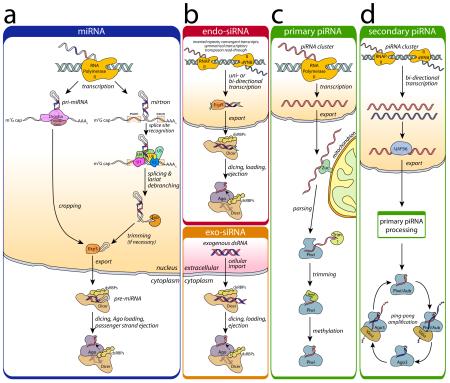
Box 1. Overview of Eukaryotic Small RNA Biogenesis.
(a) Canonical miRNA precursors are derived from transcripts with internal hairpins termed pri-miRNAs (grey; complementary regions in red/blue) [123, 124]. Cropping of pri-miRNAs by the microprocessor complex, consisting of Drosha and Pasha/DGCR8 (pink), results in pre-miRNAs. In a minor, alternative mirtron pathway, precursors are excised from protein-coding transcripts during splicing [125, 126]. pre-miRNAs are exported to the cytoplasm via Exportin-5 (orange) for endonucleolytic processing by Dicer (peach); target recognition is aided by dsRNA binding proteins (dsRBPs) (yellow). As the dsRNA is loaded into Argonaute (Ago, purple) one strand is discarded (the “passenger” strand) (reviewed in [127]). The remaining guide strand directs Ago to its target via complementary base pairing. Some specific miRNAs are processed in a Dicer-independent pathway [128, 129]. (b) siRNAs typically derive from exogenous sources (i.e. viral infection or chemical synthesis) (exo-siRNA, bottom), but endogenous siRNAs (endo-siRNAs, top) (reviewed in [130]) can also originate from transcription of hairpins, convergent transcription or transposon transcriptional read-through. In the cytoplasm, the biogenesis of siRNA-loaded RISCs follows the same processing as shown in (a). (c, d) In the germline-specific piRNA pathway, most piRNA precursors originate from piRNA clusters found in pericentromeric heterochromatin that may be uni- or bidirectionally transcribed [41]. The primary transcripts are exported to the cytoplasm to be parsed by the endonuclease Zucchini (Zuc, green) [44, 45, 47] and possibly other factors. The parsed transcripts are loaded in a Dicer-independent manner into a Piwi-clade Ago protein (blue), exonucleolytically trimmed (green) [57], and 2′-O-methylated at their 3′ end (yellow circle) by Hen1 (green) [58, 59] (c). In the secondary piRNA pathway, transcripts arising from bidirectional transcription are exported from the nucleus with the aid of factors such as UAP56 (blue) and directed to the primary piRNA processing machinery. An important adaptation is the utilization of a signal amplification loop, the ping-pong cycle [41, 107], in which pairs of Piwi proteins work in concert to adaptively boost the number of loaded piRNAs. In insects, primary piRNAs are loaded into either Piwi or Aubergine (Aub). Piwi/Aub piRISCs slice the target transcript, the remnants of which are loaded into Ago3 with the assistance of an “amplifier” complex containing Vasa [131] and then matured. In turn, Ago3 cleaves primary transcripts that can be loaded into Piwi/Aub, thus amplifying the silencing signal.
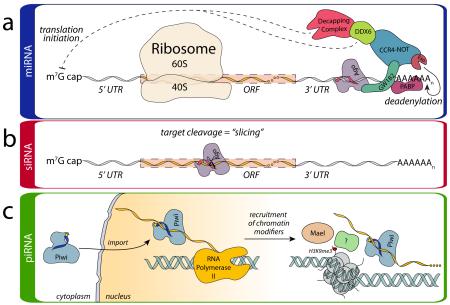
Box 2. Overview of Effector Step Mechanisms.
(a) mi- and si-RNA-induced Silencing Complexes (RISCs) affect silencing either through mRNA endonucleolytic cleavage, termed “slicing” [23, 67] or slicing-independent mechanisms (reviewed in [99]). In the miRNA pathway, RISC comprised of Ago loaded with the guide RNA localizes to target mRNAs through imperfect base pairing. Once bound, Ago (purple) recruits GW182 (teal), which mediates interactions with additional silencing machinery. The interaction of GW182 with PABP (fuchsia) is believed to interfere with PABP stimulation of translation initiation [73-75]. Alternatively, as a result of PABP displacement, mRNA is made more sensitive to deadenylation, leading to destabilization [72]. GW182 also binds the CCR4-NOT complex (blue) that in turn recruits deadenylases (Exo) (orange) that destabilize the transcript. In addition CCR4-NOT interacts with DDX6 (green), which functions as a translational repressor and recruits the decapping complex (red). (b) In contrast to the miRNA pathway, siRNA loaded RISC serves as an endonuclease. siRNA guides recognize their target with perfect or nearly-perfect base complementarity, thus favoring silencing through slicing (scissors) of the target (b).In all of these cases, this regulation is termed post-transcriptional gene silencing (PTGS) as it acts directly on transcribed mRNA substrates. (c) While the slicing activity of Piwi proteins is requisite for the ping-pong cycle discussed in Box 1, it is believed that the majority of piRNA silencing activity arises from slicer-independent, transcriptional gene silencing (TGS). In contrast to PTGS mechanisms, mature piRISC is imported into the nucleus and localizes to its target co-transcriptionally. Recognition of the target is believed to recruit silencing factors, including Maelstrom (beige) and chromatin modifiers that promote deposition of histone 3 K9 trimethylation (H3K9me3) marks (red dots), which alter the chromatin landscape and impart silencing [132].
Many recent reviews have provided excellent, contemporary descriptions of individual RNAi pathways [8-11]. Here, we aim to synthesize the latest findings across fields focusing on the mechanisms and molecular structures of RNAi. Insights from yeast, worms, and plants (all of which have a rich RNAi literature themselves) are included when broadly applicable; however, a full treatment is outside the scope of this review. Moreover, while the molecular players for each phase are introduced in Boxes 1 and 2, the majority of this review focuses on the molecular mechanisms that govern each of their activities.
The RNAi Biogenesis Machinery
Most of the small RNA biogenesis machines that have been characterized are, perhaps unsurprisingly, nucleases. This is true for both the mi/siRNA and the piRNA pathways; however, the biological logic of processing and the cascade of nucleases involved are quite different. In addition to the mechanisms described below, the activities of biogenesis molecules are themselves regulated (reviewed in [12]), providing additional layers to silencing control.
Microprocessor
The first step of processing for miRNAs occurs in the nucleus and is performed by the microprocessor, a heterodimeric complex comprised of the proteins Drosha and DGCR8/Pasha. The function of this complex is to crop long, hairpinned, pri-miRNAs into ~70 nt pre-miRNA products for export (reviewed in [13]). Following transcription, dsRNA substrates are recognized through double-stranded RNA-binding domains (dsRBDs) similar to those found in many proteins, including TRBP (Fig. 1a). Although both microprocessor components harbor dsRBDs that are indispensible in vivo [14], biochemical experiments suggest that substrate recognition is accomplished by DGCR8 [ref. 15]. Drosha provides the endonucleolytic activity of the complex with each of Drosha’s RNase III domains being responsible for cleaving one strand of the pri-miRNA [16, 17].
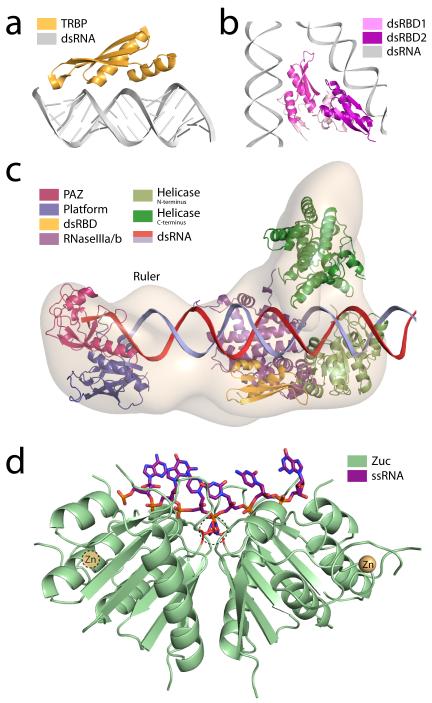
Figure 1.
Strategies for RNA recognition and processing a) Human TRBP (gold), the quintessential double-strand RNA binding protein (dsRBP), is comprised of three double-strand RNA binding domains (dsRBDs) connected by linkers. Each dsRBD binds one face of the double helix (grey) and is situated over the major groove with additional interactions along the adjacent minor grooves. The stability of this complex is almost entirely driven by direct and water-mediated hydrogen-bonding interactions (PDBs: 3ADL [119] and 1DI2 [120]). b) The microprocessor complex (PDB: 2YT4 [20]) responsible for processing pri-miRNAs is comprised of the nucleases Drosha and DGCR8/Pasha. The crystal structure of the dsRBDs of human DGCR8 (pink/purple) suggests it recognizes dsRNA in the same manner as TRBP. A-form RNA (grey) was modeled on each dsRBD based on the structure of TRBP in the presence of dsRNA. c) Crystal structures of mammalian Dicer fragments or homologs (helicase domains from P. furiosis, RNaseIII domain from Giardia) are positioned within the electron microscopic map of human Dicer (grey) based on structures from lower organisms and electron microscopic labeling [31]. The PAZ domain (magenta) recognizes the 3′ 2 nucleotide RNA overhang. Opposing RNaseIII domains (purple) perform duplex cleavage. The cleavage product’s length is measured based on the size of the ruler domain between the PAZ and RNaseIII domains. The helicase domain (green) has been implicated in processivity. d) The endonuclease Zucchini (green) (PDB: 4GGJ [44]) recognizes the phosphate backbone of single-stranded (purple; modeled) piRNA precursor substrates via a narrow, positively-charged groove. This positions the scissile phosphate between two opposing active-site histidines (center dashed circle). Additionally, each monomer binds a single zinc atom through a CCCH zinc finger (beige).
To date, several efforts have yielded biophysical and structural data on portions of the microprocessor complex [18-20]. The structure of Drosha’s single dsRBD has been determined by NMR and modeled in complex with dsRNA [18]. While there are nuances to the Drosha dsRBD structure that differentiate it from other dsRBDs, its structure is largely canonical and adopts the usual αβββα fold observed for other dsRBDs. Similarly for DGCR8, crystal structures of the dimerization domain and the tandem dsRBDs (dubbed the DGCR8 core) have been determined [19, 20] (Fig. 1b). While the structures of the individual domains are not particularly informative, the relative positioning of the tandem dsRBDs is noteworthy. Modeling dsRNA onto each domain, three assembly schemes can be envisioned. The first possibility is that in vivo the dsRBD of Drosha and at least one dsRBD of DGCR8 cooperate to effectively recognize dsRNA substrates. Alternatively, each dsRBD of DGCR8 could bind a single pri-miRNA simultaneously. This seems unlikely, as it would result in severe bending of the substrate, however, FRET studies have provided some evidence for this option [20]. Finally, larger assemblies could be generated by reciprocal binding of distinct pri-miRNAs to individual dsRBDs of the microprocessor. Electron tomographic studies of pri-miRNAs in complex with DGCR8 have produced images of ~400 kDa pri-miRNA:DGCR8 complexes that suggest such an arrangement [21]. Given that many miRNA precursors are transcribed in clusters, this last model may in fact be a biological means to improve the efficiency of pre-miRNA formation.
Dicer
After nuclear export, pre-miRNAs are processed by a Dicer family enzyme [22]. This results in a 21-25 nt mature dsRNA that is competent for RISC loading [23]. Like Drosha, the endonucleolytic cleavage by Dicers relies on two RNase III domains with each domain being responsible for cleaving one strand of the duplex [24]. Moreover, most of these enzymes conduct activity by forming an intramolecular pseudodimer between two tandem RNase III domains [25]. In these cases, RNA recognition is accomplished primarily through a PAZ domain that binds the 3′ end of the pre-miRNA [26-28] with a preference for 2 nt overhangs [29], though other parts of the molecule—including the C-terminal dsRBD and platform—also contribute to substrate recognition. Ultimately, Dicers couple the functionality of their RNase III domains to distal RNA recognition domains like PAZ. The physical distance between these two (determined by the size of the intervening Platform domain and connector helix) ultimately dictates the size of the nucleolytic products, allowing Dicer to act as a molecular ruler [30]. Structural work on Giardia intestinalis Dicer revealed the overall architecture of this enzyme, the relative positions of the PAZ and RNase III domains, and the mechanics that underlie pre-miRNA measurement [30].
More recently, cryo-electron microscopy studies of human Dicer showed a somewhat similar arrangement and suggested that the helicase domain found in higher eukaryotes functions to channel incoming dsRNA substrates toward the PAZ/RNase III surface [31] (Fig. 1c). The relative orientation of the PAZ and RNase III domains differs between Giardia and human structures, partially explaining the difference in product sizes between the two enzymes (25 versus 21 nt, respectively).
While a high-resolution structure of a full-length metazoan Dicer remains to be determined, several individual and tandem domains of animal Dicers have been solved [32-34]. The crystal structure of the RNase IIIb and dsRBD domains demonstrated the importance of a conserved lysine residue in 5′ cleavage product stabilization. Very recently, a suite of crystal structures of the Platform-PAZ domains bound to various substrates revealed a dual-pocket architecture capable of stabilizing the 2 nt 3′ overhang as well as reorienting the dsRNA after cleavage to assist in RISC loading [32] (Fig. 1c). Additional recognition of the 5′ end by the Platform-PAZ region allows for measurement and stabilization at both ends of the substrate, resulting in increased processing efficiency [35].
Other components of micro and siRNA biogenesis
Pre-miRNA dicing and Ago loading can be facilitated by associated dsRNA binding proteins (dsRBPs). These dsRBPs typically contain two or three individual dsRBDs that mediate interactions with A-form dsRNA [36]. Interestingly, many dsRBPs have refashioned their C-terminal dsRBD for protein-protein interactions suggesting that these proteins may direct mature dsRNA substrates from Dicer to Ago [37]. Delineating the precise function and mechanism of dsRBPs has proven challenging due to the number of dsRBPs found across species and the variability of their interactions. Nonetheless, some preliminary studies on these proteins have demonstrated their involvement in dicing, stabilization of the RISC-loading complex (RLC), and strand selection. In flies, Dicer-1 or Dicer-2 bind the dsRBP Loquacious (Loqs) to process endogenous small RNA precursors. A second dsRBP, R2D2, cooperates with Dicer-2 to process exogenous siRNAs (reviewed in [38]). In mammals, two dsRBPs, TRBP and PACT, have been shown to interact with Dicer and modulate its substrate specificity [39].
Alongside these biochemical studies, biophysical and structural efforts on dsRBPs have been successful in establishing the means by which the authentic dsRBDs recognize dsRNA [40]. In the case of the human dsRBP TRBP, structures of the first two dsRBDs domains, particularly the structure of the second domain bound to dsRNA, have shown that these RBDs exemplify the canonical dsRBD fold, binding the dsRNA through both major and minor groove interactions along the helical surfaces of the protein (Fig. 1a).
piRNA biogenesis
Biogenesis of piRNAs is markedly different from that of micro- and siRNAs. First and foremost, these are famously Dicer-independent. piRNA precursors are produced in the nucleus as long, single-stranded transcripts, each of which contains many individual elements that can be processed into mature small RNAs. The loci that give rise to these transcripts are termed piRNA clusters, and they serve as a catalog for what will ultimately become piRNA guide strands [41, 42].
Much of what we know about piRNA biogenesis mechanisms has been derived using the Drosophila melanogaster model system and only some of which has clear parallels in mammals. After transcription, primary (presumably intact) transcripts are exported from the nucleus, then localized to the nuage – a perinuclear/perimitochondrial cytoplasmic region enriched in many piRNA-related molecules [43]. While a complete list of the molecular factors that are responsible for this trafficking remains to be determined, some factors, like UAP56, bear similarities to other RNA-export mechanisms [43]. After export, cluster transcripts undergo parsing by the endonuclease Zucchini (Zuc) [44, 45] and possibly other enzymes.
As has been the case for many piRNA components, Zuc was first identified as a silencing factor through genetic screening [46]. The function of Zuc as a nuclease in piRNA biogenesis was then established by biochemical and structural studies [44, 45, 47]. In contrast to the previously discussed RNA nucleases, Zuc does not employ a canonical RNase fold. Rather, Zuc belongs to the HKD family of phosphodiesterases, named for its conserved His-Lys-Asp residues in and near the active site [48]. The HKD motif has been appropriated by phospholipases and nucleases alike, performing hydrolysis in a cation-independent fashion. The structures of Zuc from both mouse and fly revealed a long, positively-charged groove running through the active site, which unlike Dicer, can only accommodate a single-stranded substrate [44, 45] (Fig. 1d). After cleavage, the parsed products retain 5′ phosphate and 3′ hydroxyl chemistry, and can presumably be loaded into Piwi.
Recent genome-wide screens by multiple groups identified a large number of piRNA pathway components [49-51], and it is presumed that the current model of piRNA silencing is rather incomplete. Nonetheless, two proteins in Drosophila, Rhino and Cutoff, were shown to be necessary for the production of dual-strand piRNA cluster transcripts [52, 53]. Very recently, it was determined that Rhino (an HP1a homolog) recognizes piRNA clusters through chromatin interactions. In addition, nuclear Piwi mediates Rhino localization to cluster transcripts [54, 55]. Rhino, in turn, interacts indirectly with Cutoff through the protein Deadlock. This is reminiscent of transcriptional gene silencing in S. pombe, with dual recognition of loci by Ago1 and chromodomain protein Chp1 in the RNA-induced transcriptional silencing (RITS) complex [56]. Ultimately, the current model proposes that Cutoff protects cluster transcripts from transcriptionally-coupled pre-mRNA processing (such as polyA-tailing and splicing) [54, 55] consequently favoring these transcripts to undergo piRNA rather than mRNA processing.
Two additional primary processing factors are known: Trimmer and HEN1. After parsing and 5′ end formation, immature piRNAs that require 3′ end processing are loaded into Piwi. While the mechanism of 3′ end formation remains unknown, work in a silkworm-derived cell line demonstrated the presence of a nuclease activity that trims precursor transcripts to mature piRNA length [57]. This “Trimmer” acts in a Mg2+-dependent, 3′ to 5′ exonucleolytic fashion. Unfortunately, the molecular identity of this protein remains elusive, hampering further mechanistic and structural efforts. Finally, coupled to trimming activity is the 2′-O-methylation at the 3′ end of piRNAs. This activity is carried out by the S-adenosyl-methionine dependent methyltransferase HEN1 [58, 59] (Box 1). The structure of Arabadopsis thaliana HEN1 bound to a 22 nt RNA duplex revealed a novel Mg2+-dependent methylation mechanism [60]. It is presumed that methylation by HEN1 works similarly in animals, however, significant differences in the RNA recognition mechanism is likely as the substrates in plants are diced, dsRNA duplexes.
RNAi Effector Machinery
Argonaute family proteins serve as the programmable, central effector molecules across all RNAi pathways. When loaded with a single-stranded guide RNA, Argonautes form functional RISCs, which can be directed to their target by base pairing interactions and impart silencing through a variety of direct or indirect mechanisms (reviewed in [61, 62]). Currently, this family is broken down into three clades in animals, each with slightly different properties: Ago, Piwi, and Wago (worm-specific Argonautes) [63]. Due to their relevance across species, this review will solely focus on effector step mechanisms of the Ago and Piwi clades.
Argonaute
Biologically, the role of Argonaute (Ago) silencing is widespread, not only in terms of the number of different targets silenced [4, 5], but also in the clever means by which this system has been appropriated for specific tasks. RISCs are used for regulating gene expression networks, development, proliferation, metabolism, and DNA damage response (reviewed in [64]), highlighting the versatility and programmability of this system. It was recently reported that miRNA-Ago binary complexes could in fact be very long-lived (>3 weeks) and still maintain their silencing potency [65]. This suggests that miRNA-Ago complexes may serve as a form of cellular memory, cataloging gene silencing instructions during times of quiescence, and responding quickly to stimuli [65]. In correspondence with the stability that Agos confer upon loaded small RNAs, Ago itself is destabilized when miRNA levels decline [83]. Taken together, this symbiotic relationship between protein and RNA allow for silencing responses to be finely tuned both temporally and in intensity.
Once loaded, miRISCs ultimately affect post-transcriptional gene silencing (PTGS). However, the mechanism by which silencing occurs depends on specific qualities of the RNA, protein, and the other associated components in RISC. With respect to the RNA guide, siRNAs characteristically base pair perfectly with targets, promoting slicing [23, 67]. While miRNA guides are typically complementary with their target along the seed sequence (nt 2-8) they base pair imperfectly along the remainder of the guide sequence [68]. As a result, miRISCs remain engaged with their target strands and induce silencing by translational repression and/or target deadenylation/destabilization mechanisms [69-71] (Box 2); however, the relative contributions of these non-slicing mechanisms remains hotly debated [72-75]. In addition to guide RNA considerations, different Ago subfamily members also serve as molecular determinants for silencing output. In Drosophila, the division of labor between the two Agos is clear: DmAgo1 acts as a miRNA-guided silencer and DmAgo2 responds to siRNAs. In humans, of the four Ago subfamily members (hAgo1-4), only hAgo2 has slicing activity and the mouse version of this protein is the only one that results in embryonic lethality upon knockout [76-78]. On the other hand, all appear to participate in miRNA-mediated silencing.
In organisms from bacteria to humans, Argonaute proteins maintain a standard architecture comprised of four predominant domains: N-terminal, PAZ, MID, and PIWI (Fig. 2a) [78]. These domains have been ascribed biological functions for guide RNA recognition and catalysis (reviewed in [61]). The 5′ phosphate of the guide strand is held firmly in place by the MID domain with some important contributions from the PIWI domain. Additionally, interactions between an ordered loop of the MID domain and the first base of the guide favor interactions with uridine, explaining the 1U bias observed from small RNA sequencing of RISCs. The so-called nucleotide specificity loop varies in non-animal Argonautes and consequently modulates the nucleotide bias at the first position [79]. Following the path of the guide strand through the core of the structure, there are abundant protein interactions with the RNA, almost exclusively with the sugar-phosphate backbone. Interactions with ribose 2′-OH groups provides RNA specificity, however base-specific interactions remain restricted to the 5′ nucleotide. The central part of the RNA is disordered in all of the available eukaryotic structures. This region would be occupied with target strand mismatches. Finally, the 3′ end of the RNA is bound by the PAZ domain, with most of the relevant interactions occurring between the final phosphate and sugar in a sequence non-specific manner [27, 80, 81].
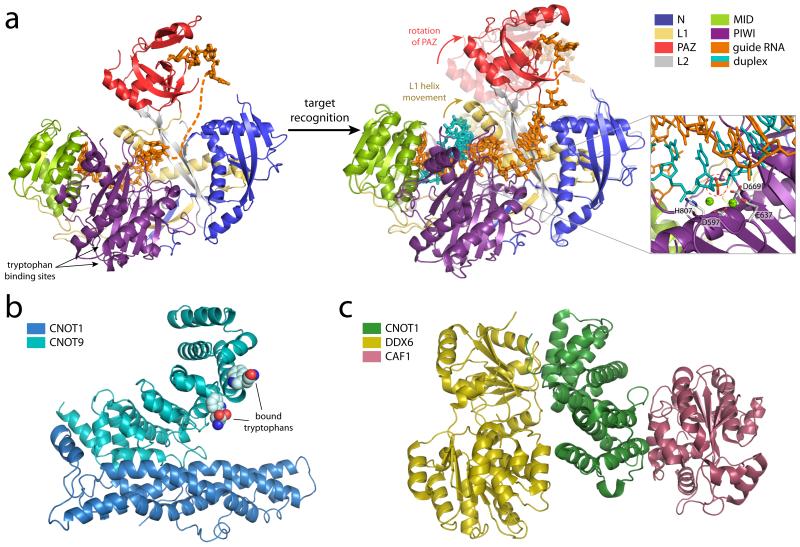
Figure 2.
Structures and modes of effector step silencing a) The overall structure of human Argonuate2 (composite of PDBs: 4F3T [90] and 4OLB [91]) bound to an RNA guide colored by domain: N-terminal (N) (blue), linker 1 (L1) (grey), PAZ (red), linker 2 (L2) (yellow), MID (green), and PIWI (purple). Upon target recognition (PDBs 4F3T (faded, prerecognition) to 4W5O [93] (solid, postrecognition), substantial conformational changes in the PAZ domain and a helix in the L1 linker are observed (arrows), the latter being essential for duplex accommodation. Additional ordering of the guide RNA also occurs upon target binding. A model of target slicing, based on the structure of RNase H bound to a DNA–RNA hybrid (PDB 1ZBI [121]) and focusing on the active site is shown in the inset. The catalytic tetrad (D597, E637, D669 and H807) coordinates two magnesium ions and mediates the cleavage of the target (scissile phosphate circled in red). When directing silencing without slicing, Ago2 interacts with GW182 family proteins through tryptophan binding sites in the PIWI domain, indicated by arrows. GW182, in turn, recruits downstream silencing machinery. (b) The structure of the binding domains of CNOT1 (blue) and CNOT9 (teal), two CCR4–NOT subunits, in the presence of tryptophan (PDB 4CRV [102]) support that CNOT9 can be directly recruited to GW182 family proteins rich in glycine and tryptophan residues. (c) The MIF4G domain of the CCR4–NOT subunit CNOT1 (green) interacts directly with the DEAD-box helicase DDX6 (yellow; PDB 4CT4 [103]) in addition to the deadenylase CAF1 (burgundy; PDB 4GMJ [122]). Together, these interactions provide a platform to link Ago targets to deadenylation, translational repression and the decapping machinery via GW182.
With regard to catalysis, the PIWI domain contains the enzyme’s DEDH active site, which requires Mg2+ for activity [78, 82, 83] (Fig. 2a). In human Agos, the presence of these catalytic residues is necessary but not sufficient for slicing (demonstrated by the fact that both hAgo2 and hAgo3 contain all four requisite residues while hAgo2 is the only active slicer). Recent biochemical and structural studies of hAgos have identified determinants of Argonaute slicer activity that extend beyond the active site. These are most notably in an adjacent loop to the active site and in the N domain [84-88]. Moreover, the activity of Agos can be modulated through post-translational modifications including proline hydroxylation that increases slicing activity, sumoylation that increases protein stability, ADP-ribosylation that relieves both slicing and translation repression, and phosphorylation that can either enhance or inhibit silencing efficacy (reviewed in [89]).
Holistically, Ago presents an extended binding interface for the guide RNA that spans all four protein domains [82, 90, 91]. With the ability to reproducibly purify unbound hAgos [84, 90] and load them with specific guides [84, 90], it became apparent that this binding confers stability to not only to the guide [65, 90, 91, 92] but also Ago itself [90]. Moreover, this binding holds the guide RNA in a conformation that is consistent in all observed structures; this is particularly evident and critical in the seed region [61]. Here, the bases point outwardly from a narrow RNA-binding groove, poised to recognize target molecules. From the structures of hAgo2 bound to guide RNA, it seems that the MIDPIWI lobe will not require significant conformational changes to accommodate target binding. It has been suggested that linkers adjacent to the PAZ domain would, however, involve rearrangement. The PAZ domain has already been shown to be the most mobile domain and releases the guide upon target binding (reviewed in [61]).
Very recently, a series of structures of hAgo2 in complex with both guide and various short target RNAs provided further insight regarding the mechanisms of target recognition [93]. This work illustrated that mismatches between the guide and target could not be tolerated in the seed region due to close packing of the RNA duplex with adjacent regions of hAgo2. Upon target binding, significant conformational changes of the protein’s PAZ domain and a helix in the L1 linker were observed, the latter of which must shift position in order to accommodate the forming duplex. This also stabilizes the guide RNA structure and “irons out” the kink between nucleotides 6 and 7 found in the RNA path of the guide-only structure. Together, the structures suggest a stepwise model of target recognition in which the guide strand first recognizes putative targets solely with seed sequence nucleotides 2-5. Subsequent conformational changes allow for validation of the target using nucleotides 6-7, followed by nucleotides 8 and/or 13-16 [93]. Additional structures are still needed, however, to better explain the atomic underpinnings of slicing and product ejection as the formation of more extended duplexes will require additional conformational rearrangements.
In the case of Ago silencing via non-slicer mechanisms, GW182-family proteins, named for their enrichment in glycine and tryptophan residues, are recruited to RISC (reviewed in [105]) through interactions with the PIWI domain [91, 95] (Fig. 2a). These factors are required for effective miRNA silencing in animals where they serve as key mediators between Agos and downstream RNA turnover/translational repression factors [96, 98] (Box 2). After binding RISC, these proteins localize to P bodies—cytoplasmic foci that are associated with mRNA decapping, degradation, and translational repression (reviewed in [94, 99]). GW182-Ago interactions promote target degradation via the sequestration of stabilizing poly(A)-binding proteins [100, 101]. Moreover, GW182 proteins interact with the CNOT9 subunit of the CCR4-NOT complex via W-motifs (distinct from GW-motifs), thus recruiting deadenylase activity that catalyzes removal of the poly(A) tail and consequently promotes mRNA degradation [96-98] (Fig. 2b). In addition, CCR4-NOT interacts via its CNOT1 subunit with the DEAD-box helicase DDX6 which functions as a translational repressor and activator of decapping [102, 103] (Fig 2c) (Box 2). Finally, post-translational modification of Argonautes by numerous factors can also affect silencing activity by influencing small RNA binding or protein stability (reviewed in [104]), thus adding another layer of regulation, particularly to miRNA silencing.
Piwis
After primary biogenesis, piRNA loaded RNA-induced silencing complexes (piRISCs) are loaded with 26-31 nt guides that are not only typically longer than si- or miRNAs, but are also 2′-O-methylated at their 3′ end. Our current understanding of the molecular mechanisms of Piwi proteins is primarily based on numerous genetic screens, biochemical assays, and comparisons to their Ago counterparts. The existing structural data on eukaryotic Piwis focuses on individual domains, specifically the 3′-binding PAZ domain and 5′-binding MID domain [105, 106], both of which show extensive interactions with the ends of the RNA guide. As is the case for Ago, the 5′ phosphate is bound tightly by the MID-PIWI lobe and the identity of the 5′ nucleotide is “read” by the nucleotide specificity loop [105]. Also similar to Agos, the opposite end of the guide RNA is recognized by the PAZ domain. A key difference, however, is the accommodation of the 2′-O-methyl at the 3′ end by a preformed hydrophobic pocket in the PAZ domain [106]. It remains to be determined how Piwi proteins are physically able to bind larger RNAs than their Ago cousins. One possibility is that changes in the orientation of the PAZ domain will allow for longer guides to be accommodated. Alternatively, inherent flexibility of the PAZ domain itself may be sufficient for the binding of the larger substrates in the context of Piwi loading.
Work in Drosophila has demonstrated that piRISCs can act in two fashions. As a slicer, Piwi can cleave targets [107, 108]. Subsequently, the RNA products of this reaction may be loaded into the complementary Piwi-clade protein Ago3 for ping-pong amplification [107] (Box 1). Piwi also appears to initiate transcriptional gene silencing (TGS) by shuttling to the nucleus, recognizing transposon transcripts, and recruiting DNA [109, 110] and/or chromatin modifiers, [111, 112] which repress transcription of transposon loci (Box 2). This link between RNAi and heterochromatin formation is highly reminiscent of RNA induced transcriptional silencing (RITS) in budding yeast. However, the slicing activity of Piwi is dispensable for silencing these loci, emphasizing that the predominant mode of piRNA silencing is indeed through TGS [113].
Other factors in effector step silencing
In addition to RISC, numerous additional factors are necessary for effective silencing. During miRISC loading, for instance, the endonucease complex C3PO binds and degrades pre-RISC passenger strands thereby facilitating the maturation of RISC [114]. Strikingly, crystal structures of C3PO revealed an ovoid architecture with the nucleolytic residues along the inner cavity of the complex [115]. The means by which ssRNA gains access to this interior surface is still being explored, however, the leading speculation is that C3PO is a dynamic complex and that partial disassembly could expose the interior and provide an entry point for the RNA.
The piRNA effector step also relies on numerous downstream molecules that help to localize Piwi to its target and estabilsh silencing. Post-translational methylation of Piwi arginine residues near the protein’s N-terminus allow Piwi to be recognized by a suite of Tudor domain-containing proteins. These Tudor proteins are believed to serve as localization and scaffolding molecules which tether piRNA components together (reviewed in [116]). Alternatively, if Piwi is translocated to the nucleus, factors including Maelstrom and chromatin modifiers are recruited to genomic loci to establish hetrochromatic silencing marks (Box 2). Other effector proteins, including asterix/Gtsf1 show a pronounced effect on transposon silencing while leaving piRNA levels unchanged [51, 117, 118]. The precise mechanisms by which these factors are recruited and enforce TGS are being investigated.
Outlook
From the initial discoveries of RNA interference to the more recent biochemical and structural findings, the biology of small RNA silencing continues to impress us with its versatility, programmability, and potency. Ongoing work to reveal the underpinnings of target recognition, the recruitment of downstream silencing factors, and the high-resolution structures of many key players promises exciting results yet to come.
ACKNOWLEDGEMENTS
The field of RNA interference has been evolving rapidly and is filled with many excellent works. We apologize that we were unable to cite all of these due to space limitations. We thank Christopher Hammell and Christopher Faehnle for critical reading of the manuscript and advice. We also thank Ian MacRae for assistance with Figure 1c. L.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92. doi: 10.1101/gr.082701.108. 2612969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68. doi: 10.1093/nar/gkt1181. 3965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci U S A. 2000;97:11650. doi: 10.1073/pnas.200217597. 17255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94. doi: 10.1038/nrg2504. 2724769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzzardo PM, Muerdter F, Hannon GJ. The piRNA pathway in flies: highlights and future directions. Curr Opin Genet Dev. 2013;23:44. doi: 10.1016/j.gde.2012.12.003. 3621807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 11.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51. doi: 10.3109/10409238.2012.738643. 3557704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 14.Bellemer C, Bortolin-Cavaille ML, Schmidt U, Jensen SM, Kjems J, Bertrand E, Cavaille J. Microprocessor dynamics and interactions at endogenous imprinted C19MC microRNA genes. J Cell Sci. 2012;125:2709. doi: 10.1242/jcs.100354. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 16.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Mueller GA, Miller MT, Derose EF, Ghosh M, London RE, Hall TM. Solution structure of the Drosha double-stranded RNA-binding domain. Silence. 2010;1:2. doi: 10.1186/1758-907X-1-2. 2836000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senturia R, Faller M, Yin S, Loo JA, Cascio D, Sawaya MR, Hwang D, Clubb RT, Guo F. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci. 2010;19:1354. doi: 10.1002/pro.414. 2974827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. Crystal structure of human DGCR8 core. Nat Struct Mol Biol. 2007;14:847. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 21.Faller M, Toso D, Matsunaga M, Atanasov I, Senturia R, Chen Y, Zhou ZH, Guo F. DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures. Rna. 2010;16:1570. doi: 10.1261/rna.2111310. 2905756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 23.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 25.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Ma E, Zhou K, Kidwell MA, Doudna JA. Coordinated activities of human dicer domains in regulatory RNA processing. J Mol Biol. 2012;422:466. doi: 10.1016/j.jmb.2012.06.009. 3461841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 29.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. Rna. 2005;11:674. doi: 10.1261/rna.7272305. 1370754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 31.Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nat Struct Mol Biol. 2012;19:436. doi: 10.1038/nsmb.2268. 3319852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y, Simanshu DK, Ma JB, Park JE, Heo I, Kim VN, Patel DJ. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol Cell. 2014;53:606. doi: 10.1016/j.molcel.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Z, Lee JK, Tjhen R, Stroud RM, James TL. Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proc Natl Acad Sci U S A. 2008;105:2391. doi: 10.1073/pnas.0711506105. 2268147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J Mol Biol. 2007;374:106. doi: 10.1016/j.jmb.2007.08.069. [DOI] [PubMed] [Google Scholar]
- 35.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doyle M, Jantsch MF. New and old roles of the double-stranded RNA-binding domain. J Struct Biol. 2002;140:147. doi: 10.1016/s1047-8477(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 37.Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16:1148. doi: 10.1038/nsmb.1673. 2845538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19. doi: 10.1038/nrg2916. 3703915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HY, Zhou K, Smith AM, Noland CL, Doudna JA. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013;41:6568. doi: 10.1093/nar/gkt361. 3711433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamashita S, Nagata T, Kawazoe M, Takemoto C, Kigawa T, Guntert P, Kobayashi N, Terada T, Shirouzu M, Wakiyama M, Muto Y, Yokoyama S. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Sci. 2011;20:118. doi: 10.1002/pro.543. 3047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 42.Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA. Production of artificial piRNAs in flies and mice. Rna. 2012;18:42. doi: 10.1261/rna.029769.111. 3261743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, Weng Z, Theurkauf WE. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871. doi: 10.1016/j.cell.2012.09.040. 3499805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279. doi: 10.1038/nature11502. 3493678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, Ishitani R, Siomi H, Siomi MC, Nureki O. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 46.Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119. doi: 10.1093/genetics/129.4.1119. 1204776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010;24:2499. doi: 10.1101/gad.1968110. 2975926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selvy PE, Lavieri RR, Lindsley CW, Brown HA. Phospholipase D: enzymology, functionality, and chemical modulation. Chem Rev. 2011;111:6064. doi: 10.1021/cr200296t. 3233269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Czech B, Preall JB, McGinn J, Hannon GJ. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell. 2013;50:749. doi: 10.1016/j.molcel.2013.04.007. 3724427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. The genetic makeup of the Drosophila piRNA pathway. Mol Cell. 2013;50:762. doi: 10.1016/j.molcel.2013.04.031. 3679447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell. 2013;50:736. doi: 10.1016/j.molcel.2013.04.006. 3724422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pane A, Jiang P, Zhao DY, Singh M, Schupbach T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. Embo J. 2011;30:4601. doi: 10.1038/emboj.2011.334. 3243597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, Seitz H, Zamore PD, Weng Z, Theurkauf WE. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137. doi: 10.1016/j.cell.2009.07.014. 2770713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157:1353. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 56.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 57.Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 58.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 59.Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603. doi: 10.1101/gad.1563607. 1899469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang Y, Ji L, Huang Q, Vassylyev DG, Chen X, Ma JB. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature. 2009;461:823. doi: 10.1038/nature08433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuhn CD, Joshua-Tor L. Eukaryotic Argonautes come into focus. Trends Biochem Sci. 2013;38:263. doi: 10.1016/j.tibs.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 62.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 63.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 64.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425:3582. doi: 10.1016/j.jmb.2013.03.007. 3757117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olejniczak SH, La Rocca G, Gruber JJ, Thompson CB. Long-lived microRNA-Argonaute complexes in quiescent cells can be activated to regulate mitogenic responses. Proc Natl Acad Sci U S A. 2013;110:157. doi: 10.1073/pnas.1219958110. 3538211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smibert P, Yang JS, Azzam G, Liu JL, Lai EC. Homeostatic control of Argonaute stability by microRNA availability. Nat Struct Mol Biol. 2013;20:789. doi: 10.1038/nsmb.2606. 3702675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 68.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 69.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 70.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 71.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034. doi: 10.1073/pnas.0510928103. 1449641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835. doi: 10.1038/nature09267. 2990499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233. doi: 10.1126/science.1215704. 3547538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716. doi: 10.1038/embor.2012.82. 3410385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237. doi: 10.1126/science.1215691. 3971879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 77.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 78.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 79.Frank F, Hauver J, Sonenberg N, Nagar B. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. Embo J. 2012;31:3588. doi: 10.1038/emboj.2012.204. 3433783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 81.Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 82.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368. doi: 10.1038/nature11211. 3853139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 84.Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua-Tor L. The making of a slicer: activation of human Argonaute-1. Cell Rep. 2013;3:1901. doi: 10.1016/j.celrep.2013.05.033. 3769929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hauptmann J, Dueck A, Harlander S, Pfaff J, Merkl R, Meister G. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat Struct Mol Biol. 2013;20:814. doi: 10.1038/nsmb.2577. [DOI] [PubMed] [Google Scholar]
- 86.Hauptmann J, Kater L, Loffler P, Merkl R, Meister G. Generation of catalytic human Ago4 identifies structural elements important for RNA cleavage. Rna. 2014;20:1532. doi: 10.1261/rna.045203.114. 4174435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, Patel DJ. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 2013;3:1893. doi: 10.1016/j.celrep.2013.06.010. 3757560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schurmann N, Trabuco LG, Bender C, Russell RB, Grimm D. Molecular dissection of human Argonaute proteins by DNA shuffling. Nat Struct Mol Biol. 2013;20:818. doi: 10.1038/nsmb.2607. [DOI] [PubMed] [Google Scholar]
- 89.Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100. doi: 10.1016/j.cell.2012.05.017. 3464090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037. doi: 10.1126/science.1221551. 3521581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winter J, Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 93.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Gene regulation. Structural basis for microRNA targeting. Science. 2014;346:608. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Braun JE, Huntzinger E, Izaurralde E. The role of GW182 proteins in miRNA-mediated gene silencing. Adv Exp Med Biol. 2013;768:147. doi: 10.1007/978-1-4614-5107-5_9. [DOI] [PubMed] [Google Scholar]
- 95.Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EK. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. Rna. 2009;15:804. doi: 10.1261/rna.1229409. 2673069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 97.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218. doi: 10.1038/nsmb.2166. 3885283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 99.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 100.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol. 2010;17:238. doi: 10.1038/nsmb.1768. 2920127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zekri L, Kuzuoglu-Ozturk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. Embo J. 2013;32:1052. doi: 10.1038/emboj.2013.44. 3616289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014;54:737. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 103.Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 2014;54:751. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 104.Johnston M, Hutvagner G. Posttranslational modification of Argonautes and their role in small RNA-mediated gene regulation. Silence. 2011;2:5. doi: 10.1186/1758-907X-2-5. 3199228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cora E, Pandey RR, Xiol J, Taylor J, Sachidanandam R, McCarthy AA, Pillai RS. The MID-PIWI module of Piwi proteins specifies nucleotide- and strand-biases of piRNAs. Rna. 2014;20:773. doi: 10.1261/rna.044701.114. 4024632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tian Y, Simanshu DK, Ma JB, Patel DJ. Structural basis for piRNA 2′-O-methylated 3′-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc Natl Acad Sci U S A. 2011;108:903. doi: 10.1073/pnas.1017762108. 3024652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 108.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214. doi: 10.1101/gad.1454806. 1553205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785. doi: 10.1016/j.molcel.2008.09.003. 2730041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aravin AA, Bourc’his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970. doi: 10.1101/gad.1669408. 2732394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430. doi: 10.1093/nar/gkm576. 2018648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci U S A. 2011;108:21164. doi: 10.1073/pnas.1107892109. 3248523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Darricarrere N, Liu N, Watanabe T, Lin H. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proc Natl Acad Sci U S A. 2013;110:1297. doi: 10.1073/pnas.1213283110. 3557079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750. doi: 10.1126/science.1176325. 2855623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:650. doi: 10.1038/nsmb.2032. 3109212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 117.Donertas D, Sienski G, Brennecke J. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013;27:1693. doi: 10.1101/gad.221150.113. 3744727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ohtani H, Iwasaki YW, Shibuya A, Siomi H, Siomi MC, Saito K. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 2013;27:1656. doi: 10.1101/gad.221515.113. 3744724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure. 2010;18:594. doi: 10.1016/j.str.2010.02.006. 3119452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. Embo J. 1998;17:7505. doi: 10.1093/emboj/17.24.7505. 1171094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 122.Petit AP, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res. 2012;40:11058. doi: 10.1093/nar/gks883. 3510486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A. 2007;104:17719. doi: 10.1073/pnas.0703890104. 2077053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844. doi: 10.1016/j.cell.2013.01.031. 3707628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328. doi: 10.1016/j.molcel.2007.09.028. 2763384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83. doi: 10.1038/nature05983. 2475599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35:368. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 128.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, Wolfe SA, Giraldez AJ. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694. doi: 10.1126/science.1190809. 3093307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584. doi: 10.1038/nature09092. 2995450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642. doi: 10.1016/j.cell.2009.01.035. 2675692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Coute Y, Conn S, Kadlec J, Sachidanandam R, Kaksonen M, Cusack S, Ephrussi A, Pillai RS. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157:1698. doi: 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 132.Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964. doi: 10.1016/j.cell.2012.10.040. 3504300. [DOI] [PMC free article] [PubMed] [Google Scholar]