This article describes the different ways in which protected health information can be erroneously released in research images, various approaches to image de-identification, and a standards-based approach to image de-identification that is in use by the Cancer Imaging Archive and supported by the National Cancer Institute.
Abstract
Online public repositories for sharing research data allow investigators to validate existing research or perform secondary research without the expense of collecting new data. Patient data made publicly available through such repositories may constitute a breach of personally identifiable information if not properly de-identified. Imaging data are especially at risk because some intricacies of the Digital Imaging and Communications in Medicine (DICOM) format are not widely understood by researchers. If imaging data still containing protected health information (PHI) were released through a public repository, a number of different parties could be held liable, including the original researcher who collected and submitted the data, the original researcher’s institution, and the organization managing the repository. To minimize these risks through proper de-identification of image data, one must understand what PHI exists and where that PHI resides, and one must have the tools to remove PHI without compromising the scientific integrity of the data. DICOM public elements are defined by the DICOM Standard. Modality vendors use private elements to encode acquisition parameters that are not yet defined by the DICOM Standard, or the vendor may not have updated an existing software product after DICOM defined new public elements. Because private elements are not standardized, a common de-identification practice is to delete all private elements, removing scientifically useful data as well as PHI. Researchers and publishers of imaging data can use the tools and process described in this article to de-identify DICOM images according to current best practices.
©RSNA, 2015
SA-CME LEARNING OBJECTIVES
After completing this journal-based SA-CME activity, participants will be able to:
■ List issues involved in de-identifying DICOM images for use in research trials.
■ Explain why images contain private elements not defined by the DICOM Standard.
■ Describe a mechanism for safely retaining some private elements when de-identifying a set of images.
Introduction
A researcher or organization involved in an imaging-based study may be required to export imaging data to satisfy collection and measurement requirements or to publish the original data at the conclusion of the study. In the first case, imaging data are exported to one or a limited number of contract organizations to perform analyses, while in the second case, the imaging data are published for consumption by other researchers or interested parties. Government regulations in most countries (the Health Insurance Portability and Accountability Act [HIPAA] in the United States, the Convention for the Protection of Individuals with Regard to the Automatic Processing of Personal Data in Europe [1]) prohibit the release of protected health information (PHI) but do not specify the mechanisms or software needed to adhere to the regulations. In the first case, an organization might be allowed to export data containing PHI if it had a business relationship with the contract organization; however, those details differ for each study and each imaging center and its institutional review board (IRB). Even with a business associate agreement (2) in place, the IRB may prohibit the export of PHI. In the case of public consumption, the regulations are quite clear and do prohibit the release of PHI.
Although the staff at the imaging center that originally collected the data may be aware of the regulations, they may not have the technical expertise to properly de-identify image data before the files are exported or released for publication. The organization that manages the research study may provide software to de-identify the imaging data, but the imaging center will still have the legal responsibility for images that are exported or shared.
The Digital Imaging and Communications in Medicine (DICOM) standard (3) is the global convention used by manufacturers to define and store diagnostic imaging data. DICOM images are encoded as a set of elements; public elements are defined by the DICOM standard, and private elements are defined on an individual basis by each manufacturer. Each public or private element in an image file has a unique hexadecimal tag (eg, 0010 0020) and the data defined for that tag (eg, “Patient ID”). The DICOM standard defines hundreds of elements to encode items ranging from “Patient Name” to “Slice Thickness” to magnetic resonance (MR)–specific parameters such as “Echo Time.” Modality manufacturers use private elements to encode acquisition parameters that are not yet defined by the DICOM standard or that they consider proprietary. Modality manufacturers also define and include private elements that contain PHI. These PHI private elements can be as obvious as the name of a patient and as subtle as an identifier string that could be tracked back to a patient by someone with access to the departmental image archive.
A DICOM conformance statement is a document published by a manufacturer that contains technical information concerning data exchange with a specific type of device (eg, an imaging unit, workstation, printer, image archive) (4,5). The conformance statement provides the mechanism for a manufacturer to publish the set of private elements that are stored in the DICOM files created by an imaging system. Manufacturers do not document and publish all of their private elements. It is simple to extract the data encoded in private elements in images, but one might not know if an acquisition parameter describes an MR sequence or an identifier for the patient without confirmation from the conformance statement or directly from the manufacturer.
The software system used to de-identify DICOM images should also meet these two conflicting requirements: (a) The system must not allow any PHI to be included in exported data, and (b) the system must retain all data (public and private elements) that describe the acquisition. These include physical parameters for individual images (eg, MR parameters), as well as other parameters such as the series description or the time point in the study for this acquisition (year 1, year 2, etc).
There are a number of technical challenges to creating a system that will satisfy these requirements.
1. DICOM standard elements with well-defined semantics are abused at the time of collection. Some elements are encoded as text strings and are taken from technologist input at the console. Instead of using the field for its intended purpose (eg, “Image Comment”), the technologist may enter PHI if allowed by local convention. There are comment fields defined in DICOM that are intended to contain information concerning individual images or possibly the imaging study as a whole. We have found that some sites do use these fields for useful comments that have scientific value, whereas other sites use the same fields to record the name of the referring physician (which constitutes PHI). With no defined practice across imaging sites and collections, free-text fields such as these require scrutiny to remove PHI but retain useful information.
2. Imaging system vendors use private elements to encode acquisition parameters not yet documented by the DICOM standard. They also use private elements to record study or demographic information to support legacy data structures. Only a small set of vendor private elements are listed in the DICOM standard; the 2013 version of the standard lists a total of 84 private elements from six vendors.
3. DICOM sequences provide a mechanism to nest data elements at different levels in DICOM objects. PHI may be encoded at these lower levels and can be missed in a simplistic approach that scans only the first level of elements in a DICOM object.
4. Manufacturers do document some but might not document all private elements in their DICOM conformance statements.
The de-identification system developer may not have sufficient knowledge about a particular imaging modality to know that it is important to search for and retain certain acquisition parameters that might be recorded only in a private element. For example, some MR diffusion parameters may be available only in private elements. A specific collection site might have the expertise to identify such parameters in private elements for their imaging data, but it would be difficult to find an expert in all imaging technologies who would be able to identify the acquisition parameters that might be encoded in private elements and to locate those elements.
5. Image providers or others involved in the original image submission remove information from the images that identifies the vendor model and software version. We can normally identify the vendor directly from the private elements, but it is difficult to locate the proper conformance statement without the scanner model and software version.
6. The users and managers of the de-identification system may not be able to discuss the collection of images with the original imaging center. Even if one locates the appropriate staff members, it may be difficult to determine if private elements were actually recorded with the original DICOM images if they were not used as part of the original interpretation or analysis.
7. Missing acquisition parameters might not be noticed until months or years after the images have been stored in a centralized repository.
These parameters are noticed quickly during an ongoing study that requires measurements reliant on private elements. However, for data stored in a centralized repository for secondary research, the end users are disassociated from the data submitters. There are likely no formal communication channels in this case, and the time elapsed between acquisition and consumption makes it difficult to know how the images were originally recorded and where the loss may have occurred.
8. Sites may include screen captures with PHI or billing documents with DICOM data. Topograms for computed tomographic (CT) data and ultrasonographic (US) images may have patient demographic information burned into the pixel data.
The system used to perform the de-identification includes a number of steps to try to satisfy the two requirements while managing the other complexities. One common step is to delete all private elements. This provides maximum security for elimination of PHI in private elements, but this practice might blindly remove scientifically essential data in violation of the second requirement listed previously in this section. Another common step is to delete all comment and similar free-text fields without further review. Again, this practice removes another source of PHI, but also might remove useful information such as the presence of contrast agent in a series of images.
It is extremely difficult to eradicate all PHI from DICOM images with automated software while at the same time retaining all useful information. It is not always clear what constitutes data that would be useful in the future versus a string that might contain an identifier for a patient.
What Is the Cancer Imaging Archive?
The Cancer Imaging Archive (TCIA) (6,7) is a central repository, funded by the National Cancer Institute (NCI), of high-value image collections useful for research. The NCI interacts with imaging centers and sites that collect cancer imaging data involving DICOM images. The de-identification methods described in this article are derived from hands-on experience in making TCIA image collections available to the public. However, these methods could be applied by anyone who has a need to de-identify DICOM image data without degrading the images’ research value. A number of other image databases currently exist that might be able to adopt these strategies (8).
A complete overview of the image submission and de-identification process has been described by Clark et al (6). A high-level summary of the steps for submission and publication of DICOM objects includes the following: (a) The imaging center or collection site uses software to remap patient identifiers from their local scheme to anonymized identifiers and then transfers the resulting images to TCIA; (b) TCIA staff uses automated software and performs human review to ensure that all PHI has been removed from submitted data; and (c) de-identified images are published through the archive software and made available to researchers.
More details about the de-identification process and software used are provided later in this article. We were able to design a standards-based solution that we believe eliminates PHI in the published data in accordance with guidelines from our IRB. Furthermore, we have gained valuable experience in implementing the process with collection sites, communicating with collection sites and end users, and reading through numerous DICOM conformance statements from acquisition system manufacturers. This experience is reflected in the list of conflicting requirements listed previously in this article.
In TCIA’s 3-plus years of operation, we have de-identified and published over 80,000 DICOM imaging studies from 40 collections submitted by 40 sites. Some of the sites submit images for multiple collections; one collection may contain data from multiple sites obtained with acquisition units from different manufacturers or different software versions for the same manufacturer. The majority of the imaging studies are from CT, MR imaging, or positron emission tomography (PET) systems. A smaller number of studies contain data from computed radiography, digital radiography, mammography, or nuclear medicine systems.
TCIA is a centralized repository of de-identified images released for secondary research. Publication of the data obtained from the de-identified images deposited in TCIA required that our system remove all PHI to satisfy U.S. HIPAA regulations and local IRB policies.
Using the DICOM Standard to Drive De-identification
Part 15 of the DICOM standard includes Annex E: Attribute Confidentiality Profiles (9). This annex provides a number of definitions and recommendations concerning de-identification for different uses. The foundation of this annex is a defined set of actions that are to be applied to each element in a DICOM object. Table 1 contains the coded actions and description of each action.
Table 1:
DICOM Action Codes for Confidentiality
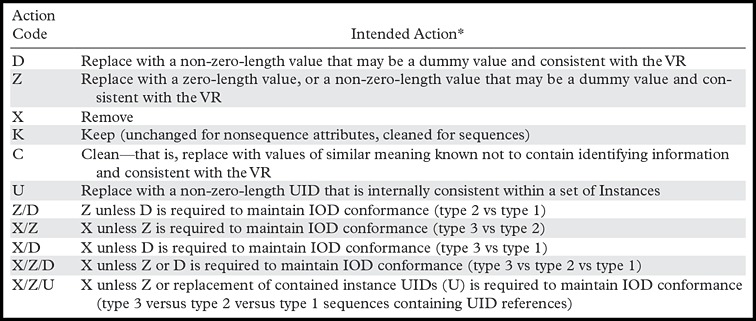
Source.—Reference 9.
Note.—IOD = information object definition, UID = unique identifier, VR = value representation.
*Type 1 data elements must be included and are mandatory elements. Type 2 elements must be included and are mandatory; however, it is permissible that if a value for a type 2 element is unknown, it can be encoded with zero value length and no value. Type 3 elements are optional data elements.
The Basic Application Level Confidentiality Profile defines a baseline set of requirements for de-identification in terms of the action codes listed in Table 1. A number of options are defined that can be applied in addition to this profile. DICOM provides these optional levels of de-identification to support different usage requirements. For example, exchanging research images within the same department might not require the same level of de-identification as publishing images to a global access repository. Table 2 is a modified extract from table E.1-1 in part 15 of the DICOM standard. Each row describes the action to be applied for a specific DICOM element. The columns in the table refer to different profiles and options with different levels of confidentiality; the entries in the columns indicate the appropriate action for the DICOM element.
Table 2:
Extract from DICOM Application Level Confidentiality Profile Attributes
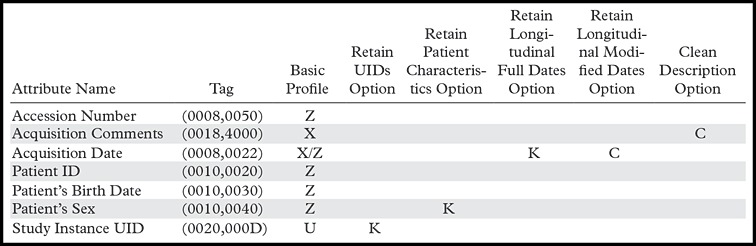
Note.—Adapted and reprinted, with permission, from reference 9.
The DICOM standard does not describe how to select or combine profiles and options. The goal of TCIA is to retain the scientifically useful information in the images while removing all PHI. These requirements mean that we cannot take the simplest approach, which would include the following: (a) Delete all private elements, and (b) delete or clean all standard elements that could possibly have PHI without review.
For the TCIA publication process, we have chosen the “Basic Application Level Confidentiality Profile” with the following options: (a) clean pixel data, (b) clean graphics, (c) clean descriptors, (d) retain longitudinal temporal information with modified dates, (e) retain patient characteristics, (f) retain device identity, and (g) retain safe private tags.
Many of the options provide explicit instructions on whether to keep or delete elements. However, to safely implement options that include instructions to clean the contents of an element, we must review those elements on an individual basis to ensure PHI is removed or cleaned while retaining any scientifically useful information. The DICOM standard also provides a minimal set of instructions on how to retain safe private tags. Currently, only a small subset of elements known to be safe is mentioned in the DICOM standard.
Conversely, we have explicitly chosen to not implement these options: (a) clean recognizable visual features (we do not obscure facial features), (b) clean structured content (we only accept DICOM structured report objects that are fully de-identified by the submitting organization), and (c) retain longitudinal temporal information with full dates.
The DICOM standard provides detailed guidance on mechanisms for de-identifying images, but it does not provide a blanket approach that will work for all cases. The staff at each organization responsible for de-identifying data must understand the level of confidentiality required and select proper methods for de-identification. The use of options that advise the cleaning of free-text data requires special attention, and there is minimal guidance on dealing with private elements.
Solution
Our goal is to release images for public use that contain no embedded PHI (standard or private elements) but contain as much data as possible for future researchers. The De-Identification Knowledge Base is a key component of our system and is described in more detail in the next section. For the discussion of our solution, the De-Identification Knowledge Base contains the following:
Private element definitions that we have determined by reading manufacturers’ DICOM conformance statements.
A list of acquisition modalities from imaging manufacturers that we identify by the combination of manufacturer, modality, model, and software version. These four parameters are used to constitute a signature that identifies a single modality and is used in our processing steps.
Profiles of modalities that are similar devices that share private element definitions. Each modality profile is tied to one manufacturer and one type of modality from that manufacturer (eg, CT or MR imaging).
De-identification scripts that we have defined that have been created on the basis of modality profiles and underlying private element definitions. Each script is tied directly to one modality profile.
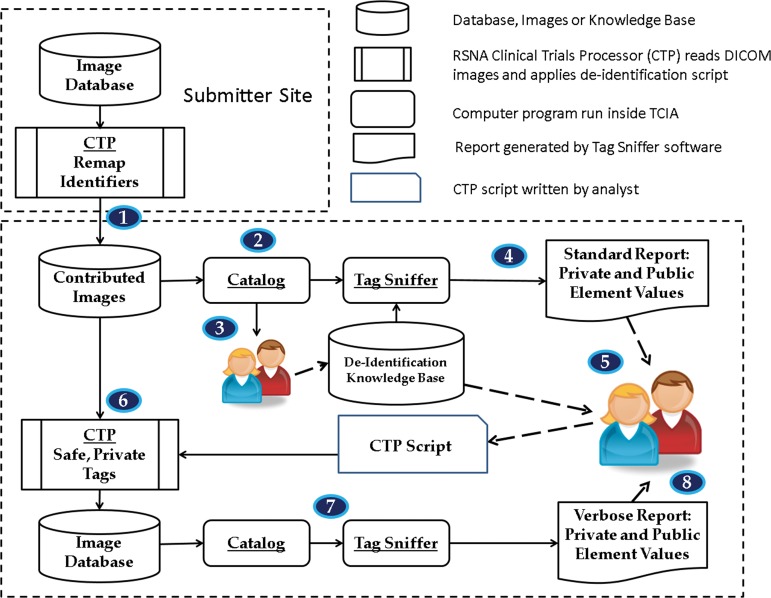
Figure 1 shows the process and applications we use to de-identify images. The knowledge base contains action codes defined for DICOM standard elements in version PS3.15 and action codes we have defined for manufacturer private elements on the basis of their conformance statements.
Figure 1.
TCIA image de-identification process. Flowchart shows image submission and full de-identification steps, with a detailed description in the text. Both the submitting site and receiving site participate in the process. Specialized open-source software is used at the receiving site with reports reviewed by senior analysts to ensure removal of all PHI.
Step 1.—Contributing sites use the Radiological Society of North America (RSNA) Clinical Trial Processor (CTP) (10) and a common script that we provide to de-identify and submit images to our central collection system in accordance with the basic application profile and options mentioned earlier. This common base script uses a lookup table (completed by the contributing site) to map local patient identifiers to anonymized patient identifiers. Only the submitting site ever sees the original identifiers. The images that are transmitted by the CTP with this first script are stored as the contributed images in Figure 1.
Step 2.—A catalog application is used to organize the contributed images by manufacturer, modality, model, and software version. These four parameters are the signature described previously in this section. No images are modified or moved in this step.
Step 3.—A senior analyst reviews the output of the catalog application. If all of the acquisition modalities for these contributed images have been previously encountered, we can skip to the next step. For those modalities that do not have a matching signature in a database, the senior analyst performs the following steps: (a) finds the appropriate conformance statement for this device; (b) updates the knowledge base with the signature and any new private elements that have been found in the conformance statement; and (c) updates the CTP de-identification script stored in the knowledge base per any new private elements. For new modalities, the analyst will update the appropriate CTP script or write a new script. The new or updated script is written back into the knowledge base for future work.
Step 4.—A “tag sniffer” application reads each image in the set of contributed images. This application records unique values of all standard elements found in the images, as well as all private elements found in the images that are also listed in the knowledge base. These values are combined with the action codes in the knowledge base to generate a report identifying elements that might contain PHI.
Step 5.—A senior analyst reviews the tag sniffer standard report and creates a single CTP script that will be used for final de-identification.
Because contributed images from different sites have different characteristics in their standard elements, the senior analyst customizes the CTP script to de-identify those standard elements. For example, the script might remove a physician name from a comment field but leave intact an indication of contrast agent in the same field.
The senior analyst retrieves the appropriate de-identification scripts from the knowledge base and combines those with the custom work mentioned previously in this section. If the work in step 3 was performed properly, the analyst only needs to retrieve CTP scripts for private elements at this step and does not need to alter them.
The output of step 5 is a CTP script that will be applied to this set of contributed images.
Step 6.—We use the CTP script created in step 5 to de-identify the contributed images and to process them for inclusion in our public image database. At our site, we now have a copy of the contributed images as sent by the contributing site and the fully de-identified images that are in our image database.
Step 7.—The catalog and tag sniffer applications are run on the de-identified images in the image database. A more verbose report is generated in this step. We are checking that values that should have been changed (eg, “Study Date”) are changed.
Step 8.—Trained data analysts review the verbose output generated by the tag sniffer. They look for any data that contain PHI. Should any such data be found, the CTP de-identification script will be updated and applied to the images again.
All elements, standard or private, are carefully reviewed at the end of the process to ensure they are free of PHI.
This second-pass review by the data analysts is needed to check the work done in the preceding steps and satisfies the requirements of our IRB.
An important part of the software process involves the configuration of CTP for de-identification. We explicitly configure CTP to discard all private elements unless they are contained in our list of safe elements. Thus, a private element that has not been reviewed by our staff will not inadvertently appear in the DICOM images after they have been processed by our system. This means that we might omit some data if the private element was not listed in a conformance statement, but we do this to ensure that no PHI is allowed to pass through our system.
TCIA uses a system that combines automated software and visual inspection. We have chosen a conservative approach for imaging files that are released for public consumption.
De-identification Knowledge Base
Over time, we are able to build a knowledge base of private elements by reading DICOM conformance statements. As we publish imaging data from more sources, we add to the knowledge base for each different acquisition modality we encounter. Over time, we begin to see similar modalities at different sites and are able to reuse the existing data in the knowledge base.
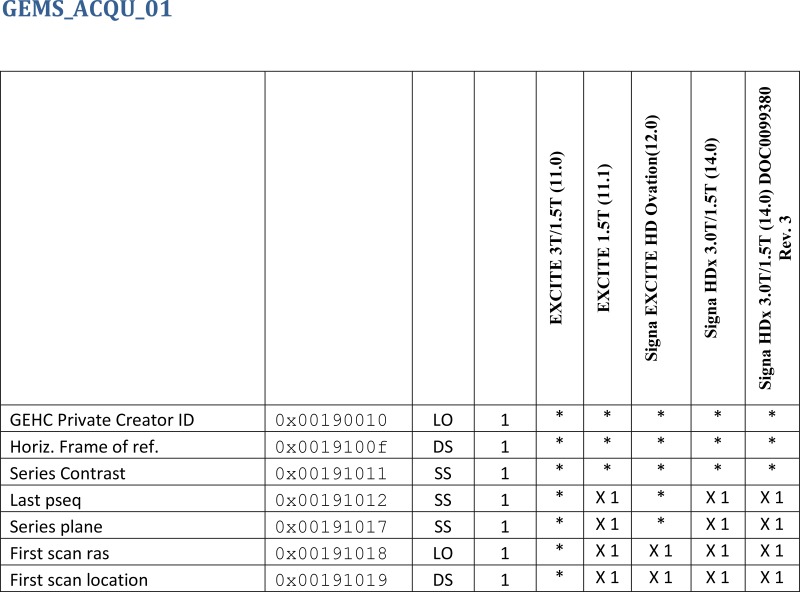
The knowledge base is available on the TCIA Web site’s wiki (11) and as a searchable database. Figure 2 is an extract of a Portable Document Format (PDF) document available on our wiki. It lists a subset of the private elements defined by GE Healthcare for MR imaging modalities.
Figure 2.
Chart shows level of detail in our DICOM knowledge base. Each row represents one private element found in the conformance statement published by a manufacturer. From left to right, the columns contain the name of the element, hexadecimal tag of the element, value representation indicating the type of string or binary value used to encode the element, and value multiplicity defining the number of different values found in a single element. The five rightmost columns contain coded entries indicating that the element is referenced in the conformance statement for this model (*) or is no longer supported (X1). (This chart and others can be downloaded from reference 11.)
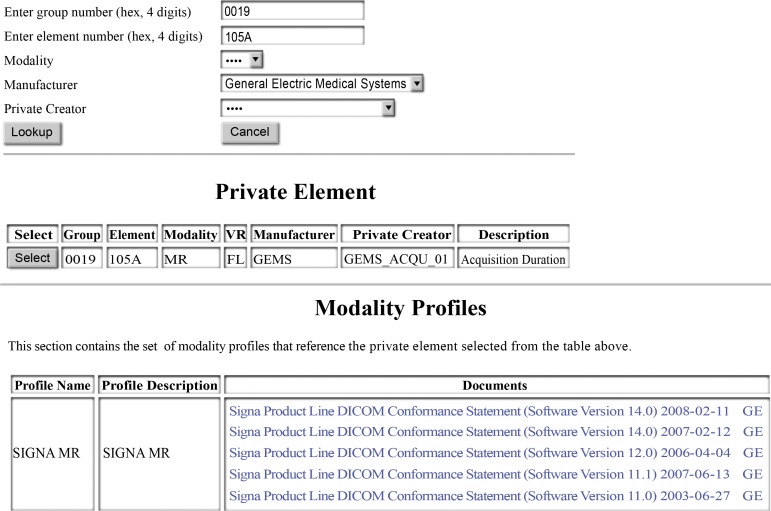
Figure 3 shows a Web-based interface that is available to the public. Researchers who receive images might discover private elements that their software does not understand. The Web-based system will allow researchers to enter some information about the private element (hexadecimal tag, manufacturer, modality, private creator identity) and find all private elements in our database that match the criteria.
Figure 3.
Screenshots show an example of the knowledge base’s Web-based user interface. In addition to internal use, the knowledge base allows external users to search for the definitions of private elements. A researcher who finds a private element in his or her own data can use this resource to understand the data without having to search through conformance statements.
The first search returns a list of all elements that match the query criteria. A user may select any individual element from that list, and the software will make a further search and show a set of documents that are relevant to the private element: (a) DICOM conformance statements, (b) our summary documents (Fig 2) on our wiki, (c) spreadsheets that contain the action codes we have defined for private elements, and (d) CTP de-identification scripts.
An important aspect of our knowledge base of private elements is that it contains only entries that have been identified in DICOM conformance statements published by manufacturers. Given that TCIA is publishing images to a large audience without business agreements with that audience, we took a conservative approach. We only use data from conformance statements and do not rely on documentation or advice from individuals outside of the manufacturer’s organization. We do not attempt to reverse engineer data in private elements. This is in accordance with the procedures that have been approved by our IRB.
What Tools Are Available for the End User?
The De-Identification Knowledge Base contains the data we have obtained by reading DICOM conformance statements. These data are available online with a Web-based user interface through which users can search the database and download all or parts of the information for their own use. The data available to end users include definitions of DICOM private elements that can be searched and filtered by modality and manufacturer and CTP scripts for de-identifying DICOM objects created on the basis of our interpretation of DICOM conformance statements.
The CTP software we use is a standard version that is supported by the RSNA. Researchers are welcome to obtain that software directly from the RSNA Web site. The only local modifications are the use of de-identification scripts that have been derived from our knowledge base. Any user of CTP would perform a similar customization step to define his or her own de-identification rules.
The DICOM tag sniffer software is a reporting system that scans through nested folders of DICOM images that are somehow related. For example, a folder and corresponding subfolders might contain the images submitted by one site for one collection. The DICOM tag sniffer records and generates reports at different levels of detail to allow human review. This open-source software is available on our collaborative software development site (“software forge”) (https://mirgforge.wustl.edu/gf/project/dicomtagsniffer/docman).
The experience gained through this effort is available to other researchers by means of an online knowledge base, scripts that drive the de-identification process, and reporting software that can be used to review imaging data for PHI.
Conclusion
We have implemented what we believe to be a rigorous system to de-identify public collections on the basis of DICOM standard practices and manufacturer conformance statements. This system is targeted to support the public release of DICOM images for TCIA, sponsored by the NCI. We have created open-source tools and a knowledge base of private elements that will help researchers faced with similar tasks.
Presented as an education exhibit at the 2012 RSNA Annual Meeting.
For this journal-based SA-CME activity, the authors S.M.M., K.E.S., J.S.K., J.B.F., and L.R.T have provided disclosures (see “Disclosures of Conflicts of Interest”); all other authors, the editor, and the reviewers have disclosed no relevant relationships.
Funding: The work was supported by the National Cancer Institute (NCI) (contract number HHSN261200800001E); Washington University funding to support TCIA comes from SAIC-F subcontract 10XS220 and Leidos Biomedical Research (contract 14XS007 for the NCI).
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Abbreviations:
- CTP
- Clinical Trial Processor
- DICOM
- Digital Imaging and Communications in Medicine
- IRB
- institutional review board
- NCI
- National Cancer Institute
- PHI
- protected health information
- TCIA
- the Cancer Imaging Archive
Disclosures of Conflicts of Interest.—: J.B.F. Activities related to the present article: employed by Leidos Biomedical Research. Activities not related to the present article: disclosed no relevant relationships. Other activities: disclosed no relevant relationships. J.S.K. Activities related to the present article: employed by Leidos Biomedical Research. Activities not related to the present article: disclosed no relevant relationships. Other activities: disclosed no relevant relationships. K.E.S. Activities related to the present article: institution receives funding from Leidos Biomedical Research through a subcontract with the National Cancer Institute (NCI). Activities not related to the present article: disclosed no relevant relationships. Other activities: disclosed no relevant relationships. L.R.T. Activities related to the present article: institution receives funding from Leidos Biomedical Research through a subcontract with the NCI. Activities not related to the present article: disclosed no relevant relationships. Other activities: disclosed no relevant relationships. S.M.M. Activities related to the present article: institution receives funding from Leidos Biomedical Research through a subcontract with the NCI. Activities not related to the present article: disclosed no relevant relationships. Other activities: disclosed no relevant relationships.
References
- 1.Levin A, Nicholson MJ. Privacy law in the United States, the EU and Canada: the allure of the middle ground. Univ Ott Law Technol J 2005;2(2):357–395. [Google Scholar]
- 2.U.S. Department of Health & Human Services . Business Associate Contracts: Sample Business Associate Agreement Provisions. U.S. Department of Health & Human Services Web site. http://www.hhs.gov/ocr/privacy/hipaa/understanding/coveredentities/contractprov.html. Published January 25, 2013. Accessed May 28, 2014.
- 3.Digital Imaging and Communications in Medicine (DICOM) . NEMA Web site. http://medical.nema.org. Accessed April 13, 2014.
- 4.Prior FW. Specifying DICOM compliance for modality interfaces. RadioGraphics 1993;13(6):1381–1388. [DOI] [PubMed] [Google Scholar]
- 5.Bidgood WD, Jr, Horii SC, Prior FW, Van Syckle DE. Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc 1997;4(3):199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26(6):1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prior F, Clark K, Commean P, et al. TCIA: an information resource to enable open science. Conf Proc IEEE Eng Med Biol Soc 2013;2013:1282–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freymann J, Kirby J. CIP Survey of Biomedical Imaging Archives. National Cancer Institute Web site. https://wiki.nci.nih.gov/display/CIP/CIP+Survey+of+Biomedical+Imaging+Archives. Accessed May 28, 2014.
- 9.DICOM Standards Committee . Digital Imaging and Communications in Medicine (DICOM). Part 15: security and system management profiles (PS 3.15-2011). Rosslyn, Va: National Electrical Manufacturers Association, 2012; 60–92. ftp://medical.nema.org/medical/dicom/2011/11_15pu.pdf. Accessed April 13, 2014. [Google Scholar]
- 10.Medical Imaging Resource Center . The RSNA Clinical Trial Processor. Oak Brook, IL: Radiological Society of North America, 2012. http://mircwiki.rsna.org/index.php?title=CTP-The_RSNA_Clinical_Trial_Processor. Updated January 5, 2015. Accessed May 28, 2014. [Google Scholar]
- 11.Moore S, Prior FW, Tarbox L, et al. De-identification knowledge base. The Cancer Imaging Archive Web site. https://wiki.cancerimagingarchive.net/display/Public/De-identification+Knowledge+Base. Updated December 29, 2014. Accessed March 20, 2012.
- 12.Moore S. Tag Sniffer. FORGE Web site. https://mirgforge.wustl.edu/gf/project/dicomtagsniffer/docman/?subdir=56. Accessed May 28, 2014.