Abstract
Background
Brucellosis is one of the most common zoonotic diseases worldwide. It can cause acute febrile illness in human and is a major health problem. Studies in human brucellosis in Malaysia is limited and so far no genotyping studies has been done on Brucella isolates. The aim of the study was to determine the genetic diversity among Brucella species isolated from human brucellosis, obtained over a 6-year period (2009–2014).
Methods
In this study, the genotypic characteristics of 43 human Brucella melitensis isolates were analysed using multiple-locus variable-number tandem-repeat analysis (MLVA) which consisted of eight minisatellite loci (panel 1) and eight microsatellite loci; panels 2A (3 microsatellite loci) and panel 2B (5 microsatellite loci). Two human Brucella suis isolates were also investigated using the MLVA assay.
Results
Using panel 1 (MLVA8), two genotypes namely genotype 43 and 44 were obtained from the 43 B. melitensis isolates. Using the combination of panels 1 and 2A loci (MLVA11), two genotypes were obtained while using the complete panels 1, 2A and 2B, nine genotypes were obtained. The polymorphisms in using the complete panels (MLVA16) were observed in three loci from panel 2B, which showed a diversity index higher than 0.17. All B. melitensis isolates were closely related to the East Mediterranean group. For B. suis isolates, only genotype 6 and genotype 33 were obtained using panel 1 and MLVA11 respectively.
Conclusion
In conclusion, the results of the present study showed a low genetic diversity among B. melitensis and B. suis isolates from human patients. Based on the MLVA16 assay, B. melitensis belonging to the East Mediterranean group is responsible for the vast majority of Brucella infections in our Malaysian patients. To our knowledge, this is the first genotyping study of human Brucella isolates in Malaysia.
Keywords: Brucellosis, MLVA, Genotype
Background
Brucellosis, a disease caused by the bacteria of genus Brucella is a re-emerging zoonosis worldwide. This disease is highly prevalent in many regions, particularly in Central Asia, the Middle East, the Mediterranean region, Africa and Latin America [1]. Brucellosis pose a major health concern in humans with initial clinical symptoms reported as fever, myalgia, malaise and may later develop into chronic disease affecting multiple organs [2]. In the livestock industry, brucellosis cause significant reproductive losses in animals and subsequently causes great economic losses [3]. Infection of Brucella sp. from animals to humans can occur by consumption of infected unpasteurised milk and dairy goods, inhalation of infected aerosolized particles and through direct contact with infected animal parts [4].
Currently eleven species are recognized in the Brucella genus [5–9], four of which are known to infect humans. They are Brucella melitensis, Brucella abortus, Brucella suis and Brucella canis [4]. In addition, Brucella inopinata was recovered from a human patient [8]. Bacterial typing has seen a shift towards molecular identification using multiple-locus variable-number tandem repeat analysis (MLVA) as tools which is widely used in the study of Brucella species and other bacterial species of medical importance [10–13]. Based on the approach of high mutation rate in tandem DNA repeats loci, this important methodology has proven its discriminatory power in the study of Brucella species with high homogeneity and the ability to distinguish isolates of various geographic origins [10, 11]. MLVA typing has been useful in outbreak and epidemiological trace-back investigations and in confirming laboratory or foodborne acquired infections [14–18].
Malaysia, a small country located in Southeast Asia has reported cases of animal brucellosis but the percentage is lower compared to its neighbouring countries [19]. The presence of brucellosis in the Malaysian livestock population has occurred for many decades although the seroprevalence is relatively low [20]. In a recent study conducted from year 2000 to 2009, the seroprevalence of brucellosis among goats was detected throughout Malaysia but at generally low seroprevalences. The seroprevalence of brucellosis among goats was 0.91 % and among farms was 7.09 % [21]. The seroprevalence of bovine brucellosis among cattle population in Malaysia was 21.8 % [22]. No current data on the seroprevalence of porcine brucellosis is available except for data from 1955–56 which reported low titres of brucellosis among tested pig sera [20].
In Malaysia, occupations associated with animals or animal related materials such as livestock workers, veterinarians and laboratory workers are most likely to be infected by brucellosis [23, 24]. Human brucellosis cases in Malaysia also occurred through consumption of unpasteurized goat milk [25]. In a study among suspected human brucellosis cases, the seropositivity of brucellosis was only 5.4 % which suggests that the seroprevalence of brucellosis among individuals who have contact with infected animals in Malaysia is low [26]. According to Ministry of Health Malaysia, less than five cases were reported in government hospitals between the year 2000 and 2008 [27]. From year 2009 onwards, human brucellosis cases continued to be reported and most were sporadic cases. However, in the year 2011, an outbreak occurred in Penang, a state in the northern part of Malaysia with more than 30 cases being reported [28]. There are limited studies on human brucellosis in Malaysia and to date no genotyping studies on the human Brucella isolates have been reported in Malaysia.
In this study, the MLVA assay was used to investigate the genetic diversity of both outbreak and sporadic human Brucella isolates collected between the year 2009 and 2014 to determine the genotype of isolates causing human brucellosis in Malaysia. To the best of our knowledge, this is the first genotyping study conducted in Malaysia for human Brucella isolates.
Methods
Ethics statement
This study was carried out with the approval of Medical Research & Ethics Committee, Ministry of Health Malaysia in accordance to the International Conference of Harmonization Good Clinical Practice Guidelines (KKM/NIHSEC/08/0804/P11-666).
Brucella isolates and DNA preparation
A total of 45 isolates from human brucellosis cases in the year 2009–2014 were investigated. All samples were taken as part of standard care. Thirty-five isolates were outbreak cases occurring in the year 2011 while ten isolates were sporadic cases occurring between 2009 and 2014. All isolates were identified at the genus level based on classical identification tests such as Gram staining, positive oxidase and catalase tests, urease activity, dye sensitivity, agglutination with monospecific antisera for A and M antigens (Animal Health Veterinary Laboratories Agency, Weybridge, UK) and hydrogen sulfide production as described [29]. Forty – three isolates were identified as Brucella melitensis and two isolates were identified as Brucella suis. Total genomic DNA from these isolates were extracted using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) and the DNA concentrations were measured using NanoDrop Spectrophotometer (Nanodrop Technologies, Wilmington, USA). Brucella melitensis 16 M was used as the control strain.
Polymerase chain reaction (PCR) amplification and MLVA
The sixteen sets of primers used in this MLVA assay were based on previously described methods [10, 11]. The 16 loci are categorized in three panels namely panels 1, 2A and 2B. Panel 1 composed of eight minisatellite loci (Bruce06, Bruce08, Bruce11, Bruce12, Bruce42, Bruce43, Bruce45, Bruce55) are used for species identification. Panel 1 also provides subspecies classification. The remaining three microsatellite loci in panel 2A (Bruce18, Bruce19, Bruce21) and five microsatellite loci in panel 2B (Bruce04, Bruce07, Bruce09, Bruce16, Bruce30) are used for further subspecies differentiation. All forward primers for each primer set except for Bruce07 and Bruce19 were synthesized with fluorescence dyes (FAM, HEX, or TAMRA). For Bruce07 and Bruce19, the primers were synthesized with either FAM or HEX as the reverse primer.
Monoplex PCR was performed in a total volume of 50 μl containing 10 ng of DNA, 0.3 μM of each primers and 25 μl of My Taq Mix, 2x (Bioline, London, UK). Amplifications were performed using an Eppendorf Mastercycler (Eppendorf AG, Hamburg, Germany) based on PCR conditions as previously reported [10]. The amplified products were diluted between 1:50 to 1:300 with sterile deionised water. Denaturation of the samples were performed individually in microamp plates with a total volume of 10 μl containing 8.7 μl HI-DI formamide (Applied Biosystems, Lincoln City, USA), 0.3 μl GeneScan 1200 LIZ size standard (Applied Biosystems, Lincoln City, USA) and 1 μl of diluted amplified products at 95 °C for 5 min using an Eppendorf Mastercycler (Eppendorf AG, Hamburg, Germany). The denatured products were further resolved by capillary electrophoresis on an ABI 3730xl DNA analyzer (Applied Biosystems, Lincoln City, USA). Fragment sizes obtained were then analyzed using the GeneMapper software version 4.0 (Applied Biosystems, Lincoln City, USA).
MLVA data analysis
The copy number for each locus was managed as a character dataset using BioNumerics version 6.0 (Applied-Maths, Belgium). The clustering analysis was based on the categorical coefficient and Unweighted Pair Group Method Using Arithmetic Averages (UPGMA) method. The genotypes identified were compared with the isolates in the Brucella2012 MLVA database [30]. The genetic diversity and confidence interval of each locus were calculated using the Hunter-Gaston Diversity Indices (HGDI) via an online tool V-DICE available at the HPA website [31].
Results
Genotyping of human Brucella melitensis isolates using MLVA16 assay
The 43 B. melitensis isolates were shown to have genetic similarity coefficient ranging from 75.5 to 100.0 % (Fig. 1). Thirty-four isolates (cluster A2) shared the same MLVA16 genotype. Cluster A1 comprising two isolates differs at one locus (one repeat unit difference at the highly variable panel 2B locus Bruce04). The genetic similarity coefficient between cluster A1 and A2 is 93.8 %. These isolates were part of the outbreak strains that occurred in Penang in year 2011. Interestingly, isolate BRC7/11 which was from a patient from the state of Kedah, was shown to have 100.0 % similarity coefficient with the outbreak strains. This case was previously thought to be a sporadic case. The remaining isolates were from sporadic cases with similarity coefficient ranging from 75.5 to 87.5 %.
Fig. 1.
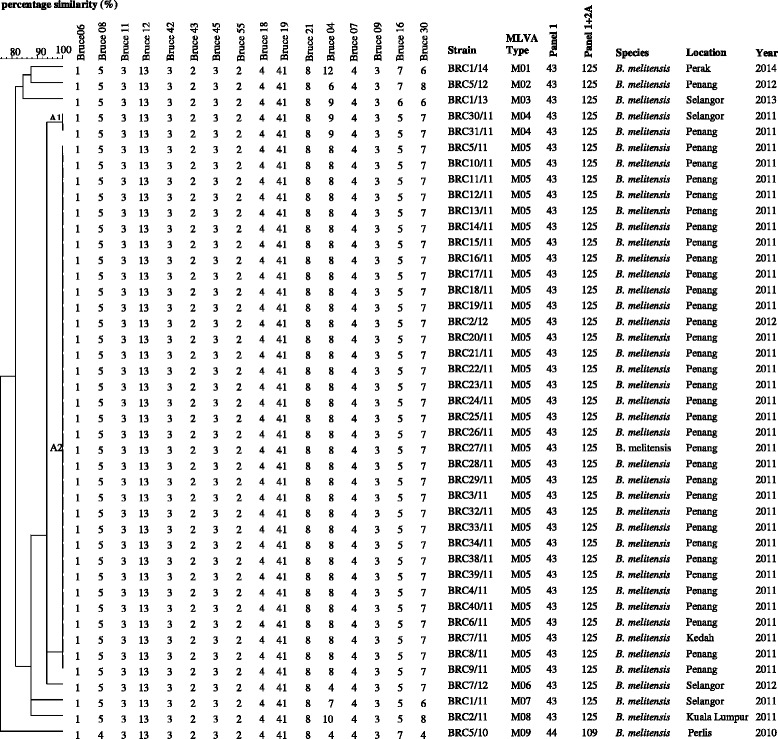
Dendrogram for 43 Brucella melitensis isolates from cases of human brucellosis based on MLVA16 assay. Data indicated in the dendrogram: Strain name in the laboratory (Strain), complete panels (MLVA16) genotypes in this article (MLVA type), genotypes corresponding to each isolate in the Brucella2012 database (panel 1, panel 1 + 2A), species, state where the case was reported (Location) and year of isolation (Year)
Using panel 1, two genotypes were observed namely genotype 43 (42 isolates) and genotype 44 (one isolate). The genotypes 43 and 44 belonged to the East Mediterranean group [11]. Further analysis with panel 1 and 2A revealed two genotypes, where the majority of the isolates were genotype 125 (42 isolates) followed by genotype 109 (1 isolate). Using the complete MLVA assay (panels 1, 2A and 2B), nine genotypes were obtained namely MLVA type M01 to MLVA type M09.
The increase in number of genotypes when using MLVA16 was the result of the high polymorphism displayed in panel 2B where Bruce04, Bruce16 and Bruce30 showed the highest discriminatory markers, with a diversity index of > 0.17 harbouring 3, 4 and 7 alleles respectively (Table 1). Among the three markers, Bruce04 shows the highest polymorphism. Bruce07 and Bruce09 with relatively small alleles (4 and 3 repeat units respectively) do not vary in the present collection. All panel 1 and panel 2A showed no diversity except Bruce08 of panel 1 which showed slight diversity, harbouring 2 alleles.
Table 1.
Number of alleles and HGDI values of 43 isolates
| Locus | Number of alleles | Number of repeats | HGDIa | Confidence of Interval |
|---|---|---|---|---|
| Panel 1 | ||||
| Bruce 06 | 1 | 6 | 0.000 | 0.000 - 0.151 |
| Bruce 08 | 2 | 4, 5 | 0.047 | 0.000 - 0.132 |
| Bruce 11 | 1 | 3 | 0.000 | 0.000 – 0.151 |
| Bruce 12 | 1 | 13 | 0.000 | 0.000 – 0.151 |
| Bruce 42 | 1 | 42 | 0.000 | 0.000 – 0.151 |
| Bruce 43 | 1 | 43 | 0.000 | 0.000 – 0.151 |
| Bruce 45 | 1 | 3 | 0.000 | 0.000 – 0.151 |
| Bruce 55 | 1 | 2 | 0.000 | 0.000 – 0.151 |
| Panel 2A | ||||
| Bruce 18 | 1 | 4 | 0.000 | 0.000 – 0.151 |
| Bruce 19 | 1 | 41 | 0.000 | 0.000 – 0.151 |
| Bruce 21 | 1 | 8 | 0.000 | 0.000 – 0.151 |
| Panel 2B | ||||
| Bruce 04 | 7 | 4, 6, 7, 8, 9, 10, 12 | 0.374 | 0.193 – 0.556 |
| Bruce 07 | 1 | 4 | 0.000 | 0.000 – 0.151 |
| Bruce 09 | 1 | 3 | 0.000 | 0.000 – 0.151 |
| Bruce 16 | 3 | 5, 6, 7 | 0.176 | 0.029 – 0.324 |
| Bruce 30 | 4 | 4, 6, 7, 8 | 0.258 | 0.091 – 0.425 |
aHunter –Gaston Diversity Index
Genotyping of human Brucella suis isolates using MLVA16 assay
Two B. suis isolates from two sporadic cases showed 81.3 % in similarity (Fig. 2). Both B. suis isolates belonged to genotype 6 with isolate BRC3/09 showing closest relation to the B. suis biovar 1 group. The other B. suis isolate (BRC1/12) was not categorized in any group. MLVA11 shows isolate BRC3/09 and BRC1/12 belonged to genotype 33. Using the complete MLVA16 assay, two genotypes were obtained from the two B. suis isolates with the numbering of MLVA type M10 to MLVA type M11.
Fig. 2.

Dendrogram for two Brucella suis isolates from cases of human brucellosis based on MLVA16 assay. Data indicated in the dendrogram: Strain name in the laboratory (Strain), complete panels (MLVA16) genotypes in this article (MLVA type), genotypes corresponding to each isolate in the Brucella2012 database (panel 1, panel 1 + 2A), species, state where the case was reported (Location) and year of isolation (Year)
Discussion
This is the first human brucellosis investigated using MLVA as the genotyping tools on 45 human Brucella isolates in Malaysia. Human brucellosis cases in this study were reported in seven states namely Perlis, Perak, Penang, Kedah (northern region), Selangor, Kuala Lumpur (federal territory) and Negeri Sembilan (central region). All the states were located in Peninsular Malaysia (Fig. 3). No human brucellosis cases were reported in the states of Sabah and Sarawak (East Malaysia). In our study, Penang had the highest reported cases of infection in humans by B. melitensis followed by the state of Selangor.
Fig. 3.
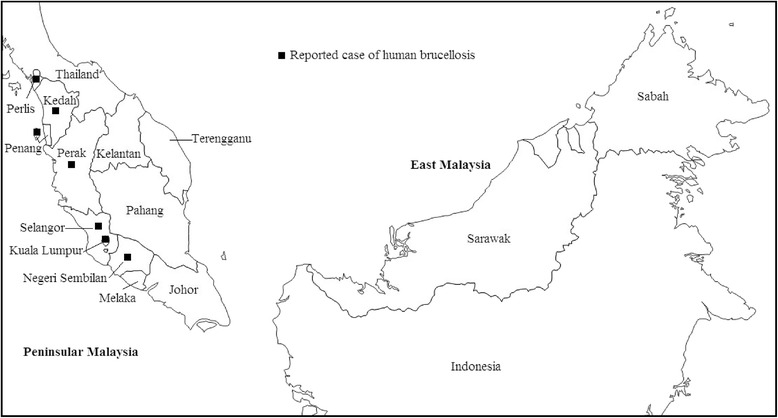
Map of Malaysia showing states with reported case of human brucellosis (adapted from Google maps [35])
Our findings most likely could be supported by a serosurveillance study conducted from 2000 to 2009 showing prevalence and distribution of zoonotic B. melitensis in goats which is higher in Peninsular Malaysia as compared to East Malaysia [21]. From the same study, it was also found that infection of B. melitensis in goats is widespread among farms, affecting all 13 states and federal territories in Malaysia. The state of Perlis had the highest seroprevalence followed by Penang and Melaka. A recent study confirmed the presence of brucellosis in livestock by the isolation of B. melitensis in seropositive goats in Peninsular Malaysia [32]. All the findings from these animal brucellosis studies are of importance in the epidemiology of human brucellosis in Malaysia.
Several studies have shown that B. melitensis is the most isolated Brucella species in human cases [16, 18]. This is in agreement with our findings as B. melitensis accounts for most of the cases reported in this study. Our data reveal that human brucellosis in Malaysia is highly-related to caprine infection as suggested by the clinical history of the patients.
Isolate BRC2/12 from a sporadic case reported in year 2012 shared similar MLVA profile as the 33 outbreak cases occurred in year 2011. This sporadic case was reported in the state of Penang and clinical history showed the patient consumed unpasteurised goat milk and the case is unrelated to the outbreak cases. Interestingly, in the outbreak cases, it was discovered that 33 patients consumed unpasteurised goat milk sourced from the same farm. All 33 isolates showed identical MLVA profiles (MLVA type M05) with good correlation with the epidemiological data. The MLVA assay also revealed that isolate BRC7/11 reported in Kedah, a state situated in the northern region was linked to the 32 cases reported in Penang indicating an outbreak from a common source of infection. However two of the isolates (BRC30/11 and BRC31/11) which were reported during similar outbreak period showed slight difference from the 33 outbreak isolates.
Both isolates BRC30/11 and BRC31/11 differed from the 33 isolates (MLVA type M05) at the bruce04 microsatellite locus from panel 2B by a difference of one repeat unit. Clinical history of isolate BRC30/11 indicated that the patient consumed unpasteurised goat milk during the outbreak period. Clustering analysis showed that isolates BRC30/11 and BRC31/11 were 93.8 % related to the 33 isolates of MLVA type M05, indicating that they are closely related. The two isolates did not share the same MLVA type which could be the result of mutation events in the course of outbreak. A study in Turkey also noted the same finding where two of the isolates yielded different MLVA profile from the outbreak isolates [18].
High polymorphism is associated with some of the most variable loci in panel 2B (Bruce 04, Bruce16 and Bruce30) as observed in this study. The three highly variable loci of panel 2B were also observed in Turkish and Chinese isolates [16, 18]. MLVA16 assay has also been shown to have the ability to differentiate a new exposure from a relapse case in the same patient [11]. In this study, isolates BRC26/11 and BRC33/11 from the outbreak were isolated from the same patient before treatment and after two months of completion of treatment respectively. Both isolates showed identical MLVA profile (MLVA type M05) implying that the patient most likely had a relapsed infection with the same strain of B. melitensis. To confirm this, whole genome sequencing analysis may provide a more definitive answer.
Isolate BRC5/10 was grouped in panel 1 genotype 44 and no clinical history was given for the patient who hails from Perlis, a state situated in the northern region of Malaysia. The panel 1 of MLVA16 assay indicated the isolate belonging to genotype 44 (1-4-3-13-3-2-3-2) clustered in the Brucella melitensis of East Mediterranean group. The panel 1 genotype 44 has been reported in Turkey and also present in Israel [11, 18].
B. melitensis has previously been clustered into three major lineages comprising the East Mediterranean, West Mediterranean and American group [11]. B. abortus and B. suis have no prior groupings. Our results suggest that the B. melitensis isolates in Malaysia most likely form a homogenous group belonging to the East Mediterranean group. The data also showed that human Brucella isolates in Malaysia comprised of panel 1 genotypes 6, 43 and 44. Genotype 43 was also observed in other parts of the world including Turkey and China [16, 18]. Panel 1 genotypes 49 and 51 of Brucella melitensis in the Western Mediterranean group which are common in Italy [11] were not detected in this study.
B. suis especially of biovars 1 and 3 are the causative agent of swine brucellosis in human and are commonly sporadic [4]. In our study, B. suis infection also occurred among human patients. Both were sporadic cases reported in Penang and Negeri Sembilan. Isolate BRC1/12 originated from a single patient who works as a palm estate farmer and the case was reported in Penang. Even though he was not occupationally-involved in animals, he may have some contact with a close family member who was working in a pig slaughter farm. When queried upon the Brucella2012 database, the genotype did not cluster to any particular group and the isolate belonged to panel 1 genotype 6. Another isolate which was typed as B. suis was BRC3/09 which was identified in a patient in the state of Negeri Sembilan in the central region of Peninsular Malaysia. Isolate BRC3/09 also belonged to panel 1 genotype 6. Panel 1 genotyping identified the profile of both isolates as genotype 6 (2-3-6-11-3-1-5-2) which is closely related to B. suis biovar 1 group. The two B. suis isolates yielded MLVA11 genotype 33.
The presence of brucellosis in Malaysia was first documented in animals with the isolation of Brucella abortus in 1950 [33], Brucella suis in 1960s [33] and B. melitensis in 1994 [23]. Subsequently, brucellosis was detected in humans in 1980 by the isolation of B. suis biotype 3 [34] and in 1994 by the isolation of B. melitensis [23]. The detection of B. melitensis and B. suis among Malaysian population decades ago still affect the current population today as shown in our data. One of the reasons of the continuous brucellosis infection in human population in Malaysia could be due to the animal reservoir. It is suspected that the annual mass importation of goats from endemic countries and importation of various breeding stocks of swines from Europe into Malaysia that causes the persistence of brucellosis in the animal population [19–21].
Our present study consists of 45 human isolates only. It is expected that more local isolates will be continuously studied to determine presence of new genotypes in the country, subsequently allowing for the genotyping results made accessible for querying and comparison to other Brucella isolates from wider geographical origins. Combined with new methodologies of whole genome sequencing, data obtained from analyses of MLVA11 genotype 125 from our present collection and from different parts of the world will be an important approach to the molecular epidemiological studies of brucella throughout the country as well as globally.
Conclusions
In conclusion, the results of the present study showed a low genetic diversity among B. melitensis and B. suis isolates from human patients. The genotypes found in Malaysia comprise mainly of panel 1 genotypes 43 and 44, grouped in B. melitensis of East Mediterranean while panel 1 genotype 6 belonged to the B. suis biovar 1 group. Panel 1 genotype 43 made up the most common genotype circulating in this country as observed in both outbreak and sporadic isolates. All the three panel 1 genotypes were confined to Peninsular Malaysia only since no cases of human brucellosis have been reported in Sabah and Sarawak (East Malaysia).
Acknowledgements
We would like to thank the Director General of Health Malaysia for permission to publish this article. This study was supported by the Ministry of Health Malaysia Grant JPP_IMR 11–035. We also thank Siti Hawa Hamzah for providing the materials for this study.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TBY performed the overall work and drafted the manuscript. NA managed the project and reviewed the manuscript. RH and AMN performed bacterial identification of the isolates. TKL provided the analysis tools, analyzed data and improving the manuscript. KXP provided technical help of analysis tools, analysis and interpretation of cluster analysis. JMZ prepared the DNA samples. All authors read and approved the final manuscript.
Contributor Information
Bee Yong Tay, Email: tayby@imr.gov.my.
Norazah Ahmad, Email: norazah@imr.gov.my.
Rohaidah Hashim, Email: rohaidah@imr.gov.my.
Jama’ayah Mohamed Zahidi, Email: jama@imr.gov.my.
Kwai Lin Thong, Email: thongkl@um.edu.my.
Xiu Pei Koh, Email: xp_527@yahoo.com.
Azura Mohd Noor, Email: azura.imr@gmail.com.
References
- 1.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. The new global map of human brucellosis. Lancet Infect Dis. 2006;6:91–9. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- 2.Pappas G, Akritidis N, Bosilkovski M, Tsianos E. Brucellosis. N Engl J Med. 2005;352:2325–36. doi: 10.1056/NEJMra050570. [DOI] [PubMed] [Google Scholar]
- 3.Bamaiyi PH, Abd-Razak NS, Zainal MA. Seroprevalence and economic impact of eradicating zoonotic brucellosis in Malaysia: a case study of Melaka state of Malaysia. Veterinary World. 2012;5:398–4. doi: 10.5455/vetworld.2012.398-404. [DOI] [Google Scholar]
- 4.Corbel MJ. Epidemiology. In: Corbel MJ, Elberg SS, Cosivi O, editors. Brucellosis in humans and animals. Geneva: WHO Press; 2006. pp. 13–21. [Google Scholar]
- 5.Osterman B, Moriyón I. International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of Brucella: minutes of the meeting. Int J Syst Evol Microbiol. 2006;56:1173–5. doi: 10.1099/ijs.0.64349-0. [DOI] [Google Scholar]
- 6.Foster G, Osterman BS, Godfroid J, Jacques I, Cloeckaert A. Brucella ceti sp. nov. and Brucella pinnipedialis sp. nov. for Brucella strains with cetaceans and seals as their preferred hosts. Int J Syst Evol Microbiol. 2007;57:2688–93. doi: 10.1099/ijs.0.65269-0. [DOI] [PubMed] [Google Scholar]
- 7.Scholz HC, Hubalek Z, Sedláček I, Vergnaud G, Tomaso H, Al Dahouk S, et al. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int J Syst Evol Microbiol. 2008;58:375–82. doi: 10.1099/ijs.0.65356-0. [DOI] [PubMed] [Google Scholar]
- 8.Scholz HC, Nöckler K, Göllner C, Bahn P, Vergnaud G, Tomaso H, et al. Brucella inopinata sp. nov., isolated from a breast implant infection. Int J Syst Evol Microbiol. 2010;60:801–8. doi: 10.1099/ijs.0.011148-0. [DOI] [PubMed] [Google Scholar]
- 9.Whatmore AM, Davison N, Cloeckaert A, Al Dahouk S, Zygmunt MS, Brew SD, et al. Brucella papionis sp. nov., isolated from baboons (Papio spp.). Int J Syst Evol Microbiol. 2014; doi:10.1099/ijs.0.065482-0 [DOI] [PMC free article] [PubMed]
- 10.Le Flèche P, Jacques I, Grayon M, Al Dahouk S, Bouchon P, Denoeud F, et al. Evaluation and selection of tandem repeat loci for a Brucella MLVA typing assay. BMC Microbiol. 2006;6:9. doi: 10.1186/1471-2180-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al Dahouk S, Le Flèche P, Nöckler P, Jacques I, Grayon M, Scholz HC, et al. Evaluation of Brucella MLVA typing for human brucellosis. J Microbiol Methods. 2007;69:137–45. doi: 10.1016/j.mimet.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 12.van Belkum A. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA) FEMS Immunol Med Microbiol. 2007;49:22–7. doi: 10.1111/j.1574-695X.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 13.Lindstedt BA, Torpdahl M, Vergnaud G, Le Hello S, Weill FX, Tietze E, et al. Use of multilocus variable-number tandem repeat analysis (MLVA) in eight European countries. Euro Surveill. 2013;18(4):pii=20385. doi: 10.2807/ese.18.04.20385-en. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira AC, Chambel L, Tenreiro T, Cardoso R, Flor L, Dias IT, et al. MLVA16 typing of Portuguese human and animal Brucella melitensis and Brucella abortus isolates. PLoS One. 2012;7:e42514. doi: 10.1371/journal.pone.0042514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garofolo G, Di Giannatale E, De Massis F, Zilli K, Ancora M, Cammà C, et al. Investigating genetic diversity of Brucella abortus and Brucella melitensis in Italy with MLVA-16. Infect Genet Evol. 2013;19:59–70. doi: 10.1016/j.meegid.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Fan MG, Chen JD, Mi JC, Yu RP, Zhao HY, et al. MLVA genotyping of Chinese human Brucella melitensis biovar 1, 2 and 3 isolates. BMC Microbiol. 2011;11:256–7. doi: 10.1186/1471-2180-11-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucero NE, Tenenbaum M, Jacob NR, Escobar GI, Groussaud P, Whatmore AM. Application of variable number of tandem repeats typing to describe familial outbreaks of brucellosis in Argentina. J Med Microbiol. 2010;59:648–52. doi: 10.1099/jmm.0.017525-0. [DOI] [PubMed] [Google Scholar]
- 18.Kiliç S, Ivanov IN, Durmaz R, Bayraktar MR, Ayaşlioğlu E, Uyanik MH, et al. Multiple-locus variable-number tandem-repeat analysis genotyping of human brucella isolates from Turkey. J Clin Microbiol. 2011;49:3276–83. doi: 10.1128/JCM.02538-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bamaiyi PH, Hassan L, Khairani-Bejo S, Zainal AM. Updates on brucellosis in Malaysia and Southeast Asia. Malaysian J Vet Res. 2014;5:71–82. [Google Scholar]
- 20.Bahaman AR, Joseph PG, Siti KB. A review of the Epidemiology and Control of Brucellosis in Malaysia. Jurnal Veterinar Malaysia. 2007;19:1–6. [Google Scholar]
- 21.Bamaiyi PH, Hassan L, Khairani-Bejo S, ZainalAbidin M, Ramlan M, Adzhar A, et al. The prevalence and distribution of Brucella melitensis in goats in Malaysia from 2000 to 2009. Prev Vet Med. 2015; doi:10.1016/j.prevetmed.2015.02.001 [DOI] [PubMed]
- 22.Anka MS, Hassan L, Adzhar A, Khairani-Bejo S, Mohamad RB, Zainal MA. Bovine brucellosis trends in Malaysia between 2000 and 2008. BMC Vet Res. 2013;18(9):230. doi: 10.1186/1746-6148-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kementerian Pertanian dan Industri Asas Tani Malaysia. Penyakit Melitensis. In: No Dokumentasi: PVM 1(5):1/2010. Protokol Veterinar Malaysia. 2015. http://www.dvs.gov.my/documents/10157/5c0aa392-708d-4f94-bdcc-80ff39647523. Accessed 18 February 2015.
- 24.Hartady T, Saad MZ, Khairani Bejo S, Salisi MS. Clinical human brucellosis in Malaysia: a case report. Asian Pac J Trop Dis. 2014;4:150–3. doi: 10.1016/S2222-1808(14)60332-7. [DOI] [Google Scholar]
- 25.National Poison Centre: Raw goat milk sicken boys. http://www.prn.usm.my/poison_news.php?news_id=6116. (2015). Accessed 23 February 2015.
- 26.Jama’ayah MZ, Heu JY, Norazah A. Seroprevalance of brucellosis among suspected cases in Malaysia. Malays J Pathol. 2011;33:31–4. [PubMed] [Google Scholar]
- 27.Ministry of Health Malaysia. Guidelines for the Diagnosis, Management, Prevention and Control of Human Brucellosis in Malaysia. 2014. http://www.moh.gov.my/images/gallery/Garispanduan/Human_Brucellosis.pdf. Accessed 22 July 2014.
- 28.Mardiyah Z, Laina M, Naziehah MD, Tosiah A: Brucella spp. di dalam susu kambing. http://los.moh.gov.my/ebook/laporantahunan2011/files/assets/basic-html/page162.html (2013). Accessed May 2013.
- 29.Corbel MJ, Banai M. Genus I. Brucella. In: Brenner DJ, Krieg NR, Garrity GM, Staley JT, Boone DR, Vos PD, Goodfellow M, Rainey FA, Schleifer KH, editors. Bergey’s Manual of Systematic Bacteriology. New York: Springer; 2005. pp. 370–386. [Google Scholar]
- 30.Brucella2012 MLVA database. http://mlva.u-psud.fr. 2014. Accessed 25 July 2014.
- 31.VNTR Diversity and Confidence Extractor. 2014. http://www.hpa-bioinformatics.org.uk/cgi-bin/DICI/DICI.pl. Accessed 25 July 2014.
- 32.Bamaiyi PH, Hassan L, Khairani-Bejo S, Zainal Abidin M, Ramlan M, Krishnan N, et al. Isolation and molecular characterization of Brucella melitensis from seropositive goats in Peninsula Malaysia. Trop Biomed. 2012;29:513–8. [PubMed] [Google Scholar]
- 33.Kementerian Pertanian dan Industri Asas Tani Malaysia. Penyakit Brusela suis dalam babi. In: No Dokumentasi: PVM 1(6):1/2011. Protokol Veterinar Malaysia. 2015. http://www.dvs.gov.my/documents/10157/27f59410-3ab9-4244-8596-f12b95b3420a. Accessed 10 March 2015.
- 34.Heng NH, Joseph PG. Eradication of brucellosis in cattle in Malaysia and its public health significance. Kajian Veterinar. 1986;18:255–60. [Google Scholar]
- 35.Daniel Dalet. Malaysia. 2015. http://d-maps.com/carte.php?num_car=5391&lang=en. Accessed 3 March 2015.


