Abstract
We describe a method called META RNA profiling (for “modular early-tagged amplification”) that can quantify a broad panel of microRNAs or mRNAs simultaneously across many samples – and requires far less sequence depth than existing digital profiling technologies. The method assigns quantitative tags during reverse-transcription to permit up-front sample pooling before competitive amplification and deep sequencing. This simple, scalable, and inexpensive approach brings large-scale gene expression studies within more practical reach.
Analysis of gene expression within diverse clinical and research specimens underpins our understanding of cellular physiology and informs our approaches to disease. Discerning meaningful expression patterns within complex biological systems usually requires statistical comparisons in two dimensions: across multiple RNAs and multiple samples. While mature technologies exist for highly parallel analysis in the first dimension, throughput efficiency remains limited in the second dimension.
Genome-wide assessment of RNA expression is possible with techniques such as RNA-Seq1–5, serial analysis of gene expression (SAGE)6, or microarrays7. But because these approaches require multi-step processing of each sample separately, they are not designed to facilitate large-scale sample multiplexing. The accuracy, sensitivity, and broad dynamic range of quantitative reverse-transcription PCR (qRT-PCR) make it the method of choice for measuring targeted RNAs. However, because fluorescence must be monitored in separate reaction volumes, applying a multi-gene qRT-PCR assay to a large number of samples can be costly and laborious.
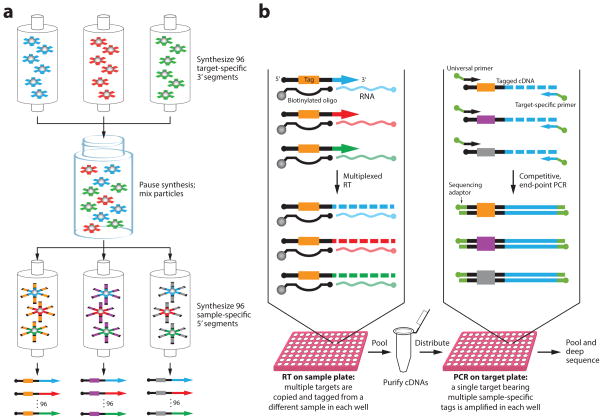
We sought to develop an RNA quantitation strategy that retains the quantification advantages of qRT-PCR while leveraging the simplicity, scalability, and uniformity of pooled sample processing that is afforded by a sequencing-based readout (Fig. 1). Our approach, called modular early-tagged amplification (META) RNA profiling, is composed of three fundamental steps. (i) To enable early parallelization of the workflow, sample-specific counting tags are first assigned to a panel of RNA molecules being targeted within each sample during reverse transcription (RT). Use of a modular primer synthesis scheme ensures that RNAs from different samples are copied to complementary DNAs (cDNAs) in consistent proportions (Fig. 1a; Supplementary Note 1). (ii) Labeled cDNAs from all samples are pooled and purified, and then each cDNA target is separately amplified by competitive, end-point PCR. Because cDNAs bearing tags from multiple samples are co-amplified under identical conditions in the same tube, cross-sample quantitative accuracy is maintained. (iii) Finally, the relative amounts of RNAs in various samples are deduced by enumerating the sample-specific tags associated with each cDNA sequence obtained by massively parallel sequencing of the PCR products.
Figure 1. Schematic of META RNA profiling.
The example depicts measurement of 96 miRNAs from 96 samples. (a) Modular RT primer mixes are synthesized in two stages: 96 partially synthesized 3′ primer segments containing target-specific sequences are pooled prior to redistribution for addition of 96 5′ tag segments that will be used as sample markers. The 96 resulting primer mixes each have distinct tags. Because the second stage of synthesis begins with the same uniform mixture of 3′-segments in each column, the final primer mixes all share similar ratios of target-specific sequences. (b) Each sample first undergoes multiplexed RT using a sample-specific modular primer mix to assign the sample-specific counting tags to cDNAs in proportion to target RNA abundance. Tagged cDNAs from all samples are combined into a single volume and are purified by in-solution hybrid capture using biotin-labeled oligonucleotides complementary to primer-extended sequences. Pooled cDNAs bearing tags from multiple samples are then co-amplified by competitive, singleplex PCRs of each target taken to plateau phase. Counting of tag-target combinations from deep sequenced amplicons reveals the relative abundance of RNAs across all samples.
The method is capable of quantifying either microRNAs (miRNAs) or messenger RNAs (mRNAs). It demands far less mean depth per base than other targeted or whole-transcriptome sequencing methods because separate end-point PCRs serve to roughly equalize total copies of low- and high-abundance RNA species. Thus rare transcripts can be adequately sampled without having to oversample abundant ones. We show that the lowest output mode of an Ion Torrent personal bench-top sequencer (<1,000,000 reads) can be used to rapidly and inexpensively quantify 96 RNAs from 96 samples, so that 96 META PCR reactions provide data equivalent to 9,216 individual qRT-PCR assays (Supplementary Table 1). Analysis of even larger sample sets would further underscore the simplicity of this approach compared to qRT-PCR because the number of reaction tubes scales as the sum – not the product – of the number of RNAs and number of samples being evaluated.
We first tested the performance of META RNA profiling on mixtures of known amounts of synthetic miRNAs. We chose a representative panel of 90 human miRNAs from the miRBase registry8 and added six control RNAs (Supplementary Table 2). Each of these synthetic RNA oligonucleotides was robotically dispensed into 96 separate tubes in varying amounts to achieve final concentrations ranging from 4 to 0.08 nM. We distributed the RNAs in a pattern designed to provide a simple visual assessment of the multiplexing capacity and accuracy of the method; when quantified and plotted on a heat map, the RNA mixtures would reproduce an image of a rose (Supplementary Figs. 1 and 2).
In the first step of META RNA profiling, all 96 targeted RNAs were simultaneously reverse-transcribed in a single well for each sample (Fig. 1b; Supplementary Note 2). Since the ratios of target-specific primer sequences are similar in all reactions (Supplementary Note 1; Supplementary Table 3), the proportions of tagged cDNA copies should faithfully reflect the abundance of RNAs in the respective samples. Upon completion of RT, tagged cDNAs from all 96 samples were pooled into a single tube and were purified by hybridization and capture using biotinylated oligonucleotides (Supplementary Table 4).
The cDNA pool was then distributed into the wells of a 96-well plate for amplification of each target by separate end-point PCRs (taken to plateau phase). Importantly, because all tags associated with a given cDNA species were amplified competitively in a single volume, tag ratios encoding RNA abundance were preserved. The resulting amplicons from all 96 reactions were pooled, gel-purified, and used directly as templates for massively parallel sequencing.
We used an Ion Torrent PGM sequencer with either a low capacity (314) or high capacity (318) chip, yielding an average of 0.42M or 3.48M filtered reads per run, respectively (Supplementary Table 1). Reads were binned based on their target and tag sequences, and heat maps were generated from read counts of all 9,216 bins (Online Methods).
The resulting plots reproduced the intended image of the rose (Supplementary Fig. 1a,b), confirming accurate, highly parallel quantitation of complex synthetic RNA mixtures across a large number of samples. To evaluate the concordance between the amount of synthetic RNA added to a sample and its measured level, we compared the fold-change of known and measured values relative to the mean for each RNA (Supplementary Fig. 1c,d). Regression analysis yielded a slope and R2 of 0.82 and 0.88 for 318 chip data, and 0.89 and 0.84 for 314 chip data, respectively. To then explore the effect of sequence depth on accuracy of measurement, we calculated the Pearson correlation coefficient between known and measured values while varying the total number of reads used (Supplementary Fig. 1e). This analysis showed only modest improvement in accuracy above approximately 500,000 total reads (~54 reads per bin). To assess technical reproducibility, we calculated coefficients of variation (CVs) among three replicate measurements. The median CV for all 9,216 data bins was 19.7%, and CV distributions grouped by RNA are shown (Supplementary Fig. 3).
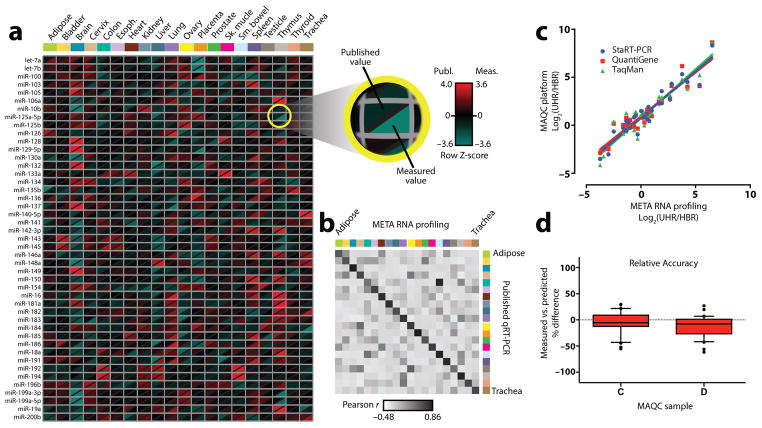
We next tested the performance of the assay on miRNAs derived from 20 normal human tissues and from the NCI-60 panel of cancer cell lines. These sample sets were chosen based on availability of independently published qRT-PCR data9–11 against which our measurements could be validated. Input consisted of 50 ng total RNA from each sample, and resulting read counts were subjected to global mean normalization, mean-centering, and autoscaling as previously described12–14. Results are presented using modified heat maps in which our measurement is compared to the published value in the two halves of a diagonally split pixel (Fig. 2a; Supplementary Figs. 4 and 5a). Concordance between the datasets is evident in the scarcity of pixels having combinations of red and green halves. Analysis of Pearson correlation coefficients showed good agreement between RNA levels measured by META RNA profiling vs. qRT-PCR for a given tissue or cell line (Fig. 2b; Supplementary Fig. 5b; Supplementary Note 3). Comparisons to data from other platforms, including NanoString, RNA-Seq, TaqMan, and several microarray systems15–17 showed good consistency (Supplementary Figs. 6–9). We could also determine absolute rather than relative concentrations by co-amplifying a sample containing known, equimolar amounts of all synthetic miRNAs as a quantitative reference standard (Supplementary Fig. 10). Based on this analysis, we found that the assay was able to measure miRNAs over a concentration range of at least 4–5 orders of magnitude.
Figure 2. Validation with human tissues and reference samples.
(a) A heat map with divided pixels compares levels of miRNAs measured as 3 technical replicates from 20 normal human tissues to published qRT-PCR measurements10. Both data sets were standardized as previously described12–14. Displayed are 45 of 90 measured miRNAs; the full data set is shown in Supplementary Figure 4. (b) Heat map of correlation coefficients of miRNA levels measured by META RNA profiling vs. qRT-PCR from the same tissue (diagonal) or between different tissues (off-diagonal). Color scheme and order of tissues is the same as in a. (c) Pair-wise correlation of fold-difference of mRNA levels in MAQC reference samples as measured by META RNA profiling (in quadruplicate) vs. three other platforms. 30 mRNAs common to all platforms were tested. Linear regression fits are shown. UHR = Universal Human Reference RNA; HBR = Human Brain Reference RNA. (d) Box plot of relative accuracy (for the same 30 genes), defined as the % difference between measured levels of an mRNA in MAQC samples C and D compared to levels predicted based on measurements of samples A and B18. The RA score for a gene is ΔC = (C−C′)/C′ and ΔD = (D−D′)/D′, where C and D are measured levels of the gene, and C′ and D′ are predicted levels. Predicted levels were calculated as C′ = 0.75A + 0.25B and D′ = 0.25A + 0.75B. Horizontal line = median; box = interquartile range; whiskers = 10th – 90th percentile; dots = outliers.
Adapting the method to quantify mRNAs was straightforward; modifications are detailed in Online Methods and Supplementary Table 5. To provide a validation benchmark, we targeted 30 genes whose expression was measured at consistent levels using three distinct quantitative platforms as part of the MicroArray Quality Control (MAQC) consortium project18. Assays were performed in quadruplicate using 100 ng of total RNA from the four MAQC reference samples, which consisted of (A) Stratagene Universal Human RNA, (B) Ambion Human Brain RNA, and mixtures of these two samples at ratios of (C) 3:1 and (D) 1:3. To evaluate the correlation of fold-change measurements between our assay and each of the three quantitative MAQC platforms, pairwise regression analyses were performed of fold-differences between samples A and B (Fig. 2c). For the common set of 30 genes, the respective slope and R2 for META RNA profiling versus TaqMan were 1.02 and 0.89; versus StaRT-PCR, 0.97 and 0.91; and versus QuantiGene, 0.92 and 0.88. As previously described18, 19, since samples C and D are composed of defined ratios of samples A and B, the relative accuracy (RA) of the assay could be assessed by comparing observed expression levels for C and D to predicted levels calculated from measurements of A and B. Box-plots of RA scores for the panel of 30 mRNAs show that values are distributed closely around zero (Fig. 2d).
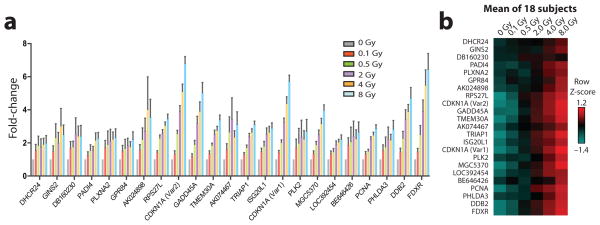
Finally, to test META RNA profiling on clinical samples, we measured radiation-induced gene expression changes in human blood. This has been proposed as an approach to estimate the dose of total-body radiation exposure following a large-scale nuclear disaster20, 21; but optimization of sample throughput would be needed to enable triage of thousands of potentially exposed individuals. To explore the feasibility of using META RNA profiling for this purpose, we developed an assay to quantify expression changes in a panel of 23 previously identified radiation-responsive transcripts21. We used this assay to perform parallel analysis of 108 ex vivo irradiated blood samples from 18 individuals (six dose levels each). Input consisted of 400 ng of total RNA derived from peripheral blood mononuclear cells that were isolated 24 hours after irradiation of whole blood. As expected, a dose-dependent increase in expression was observed for all genes in the panel when the signal was averaged across all 18 individuals (Fig. 3). The expression pattern for each individual also exhibited good consistency with this overall trend (Supplementary Fig. 11).
Figure 3. High-throughput measurement of radiation exposure in human blood.
Expression level changes in a panel of previously identified radiation-responsive genes were measured 24 hours after ex vivo irradiation of 108 blood samples from 18 individuals. All samples were processed and measured in parallel in two replicate META RNA profiling experiments. (a) Mean fold-induction of gene expression at various radiation doses, relative to a mock-irradiated sample. Error bars indicate SEM. (b) Heat map of standardized gene expression values at different doses averaged over 18 subjects, each of whose values are shown separately in Supplementary Figure 11. Mean centering and autoscaling were performed separately across samples from each subject.
Up-front sample parallelization confers several advantages over approaches that combine samples just prior to sequencing. Workflow is greatly simplified, obviating the need for microfluidic devices or automation. Pooled processing at all post-RT steps should reduce quantitative variability across samples. By carrying PCR of each target to completion, sequence depth gets evenly distributed across all targets rather than being mostly consumed by abundant transcripts. Thus, per-sample cost, which is tied to sequence depth, is minimized. Comparisons to existing technologies are further discussed in Supplementary Note 4.
In practice, we are able to quantify 96 RNAs from 96 samples in 2–3 days for ~$1000 with an Ion Torrent 314 chip. The one-time cost of synthesizing primers, which can be amortized over many runs, is ~$2000–5000 depending on the number of targets and tags. The method is readily adaptable to different sequencing platforms, it can be extended to analyze various functional RNA classes, and it requires minimal computational infrastructure and expertise. By removing many of the practical barriers to large-scale sample multiplexing, we anticipate that META RNA profiling will facilitate studies with the statistical power to resolve subtle physiologic and pathologic intricacies of gene regulation.
Online Methods
Modular synthesis of RT primer mixes
A two-stage modular oligonucleotide synthesis strategy was employed to create mixtures of primers, with each mixture having a distinct sample-specific barcode in the 5′-segment and uniform proportions of multiple target-specific sequences in the 3′-segment (Fig. 1a). First, several target-specific 3′-segments were made on separate oligonucleotide synthesis columns. Synthesis was carried out using standard phosphoramidite chemistry in the 3′ to 5′ direction on 40 nanomole polystyrene support columns (Prime Synthesis, Aston, PA) using a Dr. Oligo 192 automated synthesizer. The synthesis was paused after oligomerization of the 3′-segments was complete, and partially synthesized oligonucleotides were left on the polystyrene supports in the protected state with the dimethoxytrityl (DMT) group still on.
Argon gas was blown through the columns to dry the polystyrene supports, and then the columns were cut open and the polystyrene powder was poured into a common glass vial. The particles were suspended in a 2:1 to 3:1 mixture of dichloromethane: acetonitrile that was titrated to make the polystyrene neutrally buoyant. The slurry was constantly agitated to ensure uniform mixing while a pipette was used to dispense equal volumes of the slurry into fresh synthesis columns (with the bottom frit in place). The columns were then flushed with acetonitrile, allowing all polystyrene particles to settle to the bottom. After the acetonitrile had fully drained out by gravity, the top frits were put in place to secure the powder into the columns. One column was made for each sample-specific barcode.
The new columns were placed back on the automated synthesizer for continuation of synthesis. A distinct barcode sequence (Supplementary Table 6) was assigned to each column for incorporation into the 5′-segment of the primer mix. Barcodes were designed to be eight nucleotides in length, with each barcode differing from all other barcodes in the set at a minimum of two positions (to minimize the probability of misclassification caused by sequencer errors). A universal PCR primer binding sequence was also added to the 5′-segment of each oligonucleotide mixture. The synthesizer was programmed with an additional “dummy base” at the 3′-terminus to account for the partially synthesized oligonucleotides already present on the polystyrene supports.
Upon completion of the second stage of the modular synthesis, the oligonucleotide mixtures were cleaved from the polystyrene supports with the DMT group left on. Each mixture was subjected to rapid deprotection followed by purification on a separate Glen-Pak DNA reverse-phase cartridge (Glen Research, Sterling, VA). The cartridge selectively retained the hydrophobic DMT group at the 5′-end of the completed oligonucleotides, enriching for full-length products. The DMT group was removed upon completion of purification. The purified oligonucleotide mixtures were then dried and re-suspended in 10 mM Tris (pH 7.6) to create 10x working stocks. Sequences of miRNA and mRNA modular primer segments are listed in Supplementary Tables 3, 5, and 8.
Preparation of synthetic RNA samples
RNA oligonucleotides comprised of 90 microRNA and 6 control RNA sequences (Supplementary Table 2) were synthesized at a 40 nmole scale with 2′-deprotection and purification at the Yale Keck oligonucleotide synthesis core facility. A Tecan Freedom Evo 200 robotic liquid handler was programmed to dispense pre-defined amounts of each RNA into the wells of a 96-well plate to achieve final concentrations ranging from 4 to 0.08 nM in a pattern designed to produce the rose image shown in Supplementary Fig. 1 on a heat map. The RNAs were dissolved in a buffer containing 10mM Tris (pH 7.6), 0.1 mM EDTA, and 300 ng/mL poly-A carrier RNA (Qiagen) in RNAse-free water. The synthetic RNA solutions were stored at −80°C until needed for RT.
Tissue and cell line RNA samples
Total RNA samples derived from the NCI-60 cell lines were obtained from Dr. Susan Holbeck at the Developmental Therapeutics Program of the National Cancer Institute. The First Choice Human Total RNA Survey Panel (Ambion) was used as the source of total RNA from 20 normal human tissues. MAQC reference samples consisted of the Stratagene Universal Human Reference RNA (composed of total RNA from 10 human cell lines), and the Ambion First Choice Human Brain Reference RNA.
RNA from irradiated blood samples
Peripheral blood was collected in tubes containing sodium citrate after obtaining informed consent from 18 healthy volunteers under approval of the Human Investigation Committee at Yale University. Blood was divided into 2 mL aliquots and subjected to 0, 0.1, 0.5, 2, 4, or 8 Gy of X-irradiation at a dose rate of 1.79 Gy per minute within 1 hour of blood draw. Blood was then incubated for 24 hours at 37°C after addition of an equal volume of RPMI 1640 medium containing 10% fetal bovine serum, as previously described21. Peripheral blood mononuclear cells were isolated using ficoll gradient centrifugation, and total RNA was prepared from these cells using an RNeasy Mini Kit (Qiagen).
Processing of miRNA samples
In the first step of META RNA profiling, multiple RNA targets were reverse-transcribed in a single tube for each sample. The RT primer mix used for a given sample had a sample-specific tag in the 5′-segment, and consistent ratios of multiple target-specific primer sequences in the 3′-segment (Supplementary Table 3). Primers were designed to hybridize to 6 nucleotides at the 3′-end of the short miRNA (and control RNA) targets. A 5′-biotin labeled oligonucleotide was annealed to adjacent complementary common primer sequences to stabilize the short RNA-primer heteroduplex by extending base stacking (Supplementary Table 3)22.
Each reverse transcription cocktail consisted of 5 μM tagged primer mix (~50 nM of each target-specific primer), 7.5 μM biotin-labeled oligonucleotide, 1 × RT buffer, 3 mM MgCl2, 250 μM each dNTP, 5 mM dithiothreitol (DTT), 30 ng/μL carrier RNA (Qiagen), template RNA, and 5 units/μL Multiscribe reverse transcriptase (Life Technologies) in RNAse-free water. Each RT was carried out in a final volume of 10 μL. Prior to addition of template RNA, DTT, and reverse transcriptase, the biotin-labeled oligonucleotide was annealed to the primer mix by heating the cocktail to 95°C for 2 minutes and then cooling to room temperature. The final assembled RT cocktail was subjected to 40 cycles of 16°C for 2 minutes, 42°C for 1 minute, and 50°C for 1 second. Reactions were terminated by heating to 65°C for 20 minutes and adding EDTA at a final concentration of 10 mM. Products of all separate RT reactions were then combined into a single volume.
Pooled cDNAs were purified by capture of the complementary biotin-labeled oligonucleotide using high capacity streptavidin-coated agarose resin (Thermo Scientific) (5μL resin slurry added per 10 μL RT reaction). Resin particles were kept suspended in the solution by slowly turning the tubes end-over-end at room temperature for at least 2 hours to promote biotin binding. Particles were then washed in buffer containing 10 mM Tris pH 7.6 and 50 mM NaCl. cDNAs were released from the resin-bound oligos into a fresh volume of the same buffer (twice the volume of resin slurry) by heat-denaturation at 95°C for 2 minutes. To remove un-extended RT primers, a second round of selective annealing, capture, washing, and elution was performed using a mix of biotin-labeled oligonucleotides complementary to primer-extended sequences (100 nM each; Supplementary Table 4).
The purified cDNA pool was distributed into 96 separate tubes for singleplex endpoint PCR of each cDNA target. Because all sample-specific tags associated with a given target underwent competitive amplification in a single reaction volume, the tag proportions were maintained. The primer pair used in each PCR consisted of a universal forward primer and a distinct target-specific reverse primer as depicted in Fig. 1b (Supplementary Table 4). Sequencing adaptors were incorporated into the 5′-ends of the primers to enable direct sequencing of the PCR products. Each PCR cocktail consisted of a 10 μL volume of 1x AccuPrime PCR Buffer I (which included dNTPs and MgCl2), 100 nM universal forward primer, 100 nM target-specific reverse primer, 2 μL pooled cDNA template, and 0.2 μL AccuPrime Taq DNA polymerase (Invitrogen). Mineral oil was added to minimize evaporation. Thermal cycling parameters were 94°C for 2 minutes, 60°C for 30 seconds, 72°C for 20 seconds, followed by 40 cycles of 94°C for 20 seconds, 65°C for 30 seconds, and 72°C for 20 seconds. A final extension step was performed at 72°C for 2 minutes followed by cooling to 4°C and addition of EDTA (10 mM final) to terminate polymerase activity.
All PCR volumes were combined, and a 20 μL aliquot of the pooled reaction products was purified on a 2% low-melting point agarose gel. DNA was extracted from the excised gel slice using a QIAquick Gel Extraction Kit (Qiagen). Concentration was estimated using a Bioanalyzer 2100 (Agilent) and adjusted to levels recommended for Ion Torrent emulsion PCR.
Processing of mRNA samples
The overall scheme for processing of mRNA samples was the same as that described above for miRNA samples, with a few notable modifications. Because mRNAs were much larger than miRNAs, we were able to design primers to amplify ~100 nucleotide target regions. Accordingly, longer gene-specific RT primers could be used (Supplementary Tables 5 and 8). This enabled RT to be performed at higher temperature with a thermostable polymerase without requiring a complementary biotinylated oligonucleotide to enhance stability via extended base stacking. Each RT reaction was carried out in a 10 μL volume consisting of tagged primer mix (~50 nM each target-specific primer), 1 × First-Strand buffer, 500 μM each dNTP, 5 mM DTT, template RNA, and 10 units/μL SuperScript III reverse transcriptase (Invitrogen) in RNAse-free water. Primers were annealed to RNA targets by heating to 65°C for 5 minutes in the absence of buffer, DTT, and polymerase, which were added upon incubation at 55°C for 1 hour. Reaction tubes were kept on the thermal cycler at 55°C while adding reagents in order to avoid cooling of the sample, which could lead to non-specific annealing of RT primers. Reactions were pooled after inactivating the polymerase by heating to 75°C for 20 minutes, 95°C for 1 minute, and adding EDTA (10 mM final).
The absence of a biotin-labeled oligonucleotide during RT allowed us to capture cDNAs in a single step using biotinylated oligonucleotides complementary to primer extended sequences (Supplementary Tables 7 and 9). Pooled and purified cDNA templates were distributed into separate tubes for singleplex end-point PCR of each target using primers listed in Supplementary Tables 7 and 9. Thermal cycling parameters were identical to those described for miRNAs above, except for use of an annealing temperature of 63°C instead of 60°C for the first cycle.
Next-generation sequencing
Templates were prepared for Ion Torrent sequencing using the automated Ion OneTouch System (Life Technologies). Gel-purified amplicons were diluted to the concentration recommended by the manufacturer prior to loading on the instrument. Automated emulsion PCR enabled massively parallel clonal amplification onto Ion Sphere Particles (ISPs). To minimize polyclonal ISPs, template dilution was adjusted to achieve between 10% and 30% template-positive ISPs. The OneTouch Enrichment System was used to isolate template-positive ISPs, which were then loaded onto a semiconductor chip for sequencing. Depending on the desired sequence depth, either a 314 low-capacity chip or a 318 high-capacity chip was used. Sequencing was carried out on an Ion Torrent PGM (Life Technologies) using a 200 bp reagent kit.
Binning and counting of sequences
To determine the number of reads belonging to each target-barcode bin, we used the Torrent Mapping Alignment Program (TMAP) provided as part of the TorrentSuite Software (version 4.0). Uploading of three files was necessary for analysis of a given data set: a text file containing user-defined barcodes and adapter sequences, a FASTA format file listing miRNA or mRNA reference sequences, and a BED file defining target regions. After performing alignment of reads to target reference sequences, the coverage analysis plug-in module was run, and the resulting barcode-amplicon coverage matrix was downloaded. This matrix contained read counts for each bin, and could be opened and further manipulated in Microsoft Excel.
Since down-sampling of sequence data was not possible within the TorrentSuite software, we used an alternative approach to obtain binned counts from defined subsets of reads for Supplementary Figure 1e. The “countifs” function in Microsoft Excel was exploited for this purpose. An important difference with this approach compared to the TMAP analysis was that only perfect sequence matches were counted. Thus, to minimize the probability of an imperfect match due to sequencer error, we used short reference sequences of ~10–12 nucleotides. Reference sequences were chosen to extend beyond the sequence contained in any single primer to avoid counting of spurious PCR products (e.g. primer dimers). Care was also taken to ensure that each reference sequence matched only a single target. Supplementary Table 10 provides an illustration of how the “countifs” function was used.
Normalization and standardization of binned sequence counts
To generate heat maps displaying the rose image in Supplementary Figures 1a and 1b, counts from two replicate experiments were averaged for each of the 9,216 data bins. Counts were then normalized across rows and columns relative to the known total amounts of dispensed synthetic RNAs. First, counts in a given row were multiplied by the ratio of the sum of counts to the total amount of RNA dispensed in that row. Second, the resulting values in a given column were multiplied by the ratio of the sum of values to the total amount of RNA dispensed in that column. Finally, the binary logarithms of these normalized values were calculated and plotted on a heat map.
The normalization and standardization of miRNA and mRNA measurements from human tissues, cell lines, and blood samples (Figs. 2a,b and 3; Supplementary Figs. 4–8, and 11) was performed as previously described12–14 with some modifications. First, replicate values were averaged for each data bin. Second, to equalize the total counts produced by different singleplex PCRs for each target, the values across a given row were multiplied by a common factor to make the sum of values in that row equal to 1000. Third, flooring of the data was achieved by adding 0.01 to all bins (thus eliminating 0 values). This was analogous to the common practice in qRT-PCR experiments of transforming Cq values greater than 35 to 35. Fourth, to normalize miRNA levels we used the mean expression value for all miRNAs in a given sample as the normalization factor13, 14. mRNAs from irradiated blood samples were normalized relative to the mean expression values of two housekeeping genes, ACTB and GAPDH. Fifth, log10(fold-change) values were calculated for all data bins. Sixth, mean centering was performed by subtracting the row average from each value. Finally, values were autoscaled by dividing each value by the standard deviation across the row.
To determine the absolute quantity of miRNAs in normal human tissues (Supplementary Fig. 10), a quantitative reference standard sample containing ~15,000 copies of each synthetic miRNA was reverse-transcribed and competitively amplified with 50 ng tissue-derived total RNA samples. Since tagged cDNAs derived from the reference and test samples were pooled and amplified in the same reaction volume, the ratio of sequence counts reflected the relative abundance of the reference and test RNAs. Because the reference standard contained a known quantity of synthetic RNA, the absolute quantity could be estimated for the test samples. All samples were analyzed in 3 technical replicates. Read counts were averaged for the replicates. The average count for a target in a given tissue sample was divided by the average count for the same target in the reference sample. The resulting value was then multiplied by 15,000, yielding an estimate of the number of miRNA copies per 50 ng total RNA in that tissue sample. Log10-transformed values were plotted on a heat map.
Within the NCI-60 cell lines, we found several miRNAs that showed poor expression, consistent with prior studies9, 15–17. Such miRNAs were excluded from consideration if in more than 85% of cell lines, they had published Cq values > 33 or our measurements produced raw read counts < 10 (Supplementary Fig. 5). The same set of miRNAs was then used for comparisons with other quantitative platforms (Supplementary Fig. 6–8).
Plotting of heat maps
All heat maps were generated without clustering, using TreeView software (downloaded for free from the website of Dr. Michael Eisen’s lab: http://rana.lbl.gov/EisenSoftware.htm). Raw Cq values from published qRT-PCR studies9, 10 were obtained from the miRNA body map website (www.mirnabodymap.org). The values were floored at 35 and were subjected to the same normalization and standardization steps as outlined above, beginning at the fourth step. Standardized values of published and measured data were plotted on separate heat maps using identical color scale and contrast parameters. Split-pixel maps were created by erasing half of each pixel on one map, and then overlaying it on the second map using Adobe Illustrator and Photoshop.
Analysis of mRNAs in MAQC samples
Target genes for mRNA analysis were chosen from among the 48 genes that were commonly tested across all 3 quantitative (non-microarray) platforms reported in the MAQC data sets18. Among these 48 genes, we chose 30 whose expression was measured at consistent levels (having a low coefficient of variance) across the 3 platforms. The targeted genes are listed in Supplementary Table 5.
Binned sequence counts from quadruplicate experiments were averaged for each of the four MAQC samples (A, B, C, and D). The mean counts for a given gene were multiplied by a common factor to make the sum of values for that gene equal to 1000. No flooring was applied. Since only 30 targets were analyzed, normalization relative to the global mean expression level across a sample would not be recommended. Expression values for a given sample were thus normalized relative to average measurements of POLR2 and ACTB reference genes for that sample.
Normalized expression values were used to calculate the fold-change for all 30 genes between the Human Universal Reference RNA (sample A) and the Human Brain Reference RNA (sample B). Relative accuracy was calculated as described in the main text, based on measurements of samples C and D.
Supplementary Material
Acknowledgments
We thank J. Deluca and A. Blanchard for help with oligonucleotide synthesis; S. Mane and N. DaSilva for assistance with next-generation sequencing; P. Gareiss and M. Salcius for help with robotic liquid handling; S. Holbeck (US National Cancer Institute) for providing RNA from the NCI-60 cancer cell lines; M. Saba for assistance with creating figures; and J. Steitz, D. Brash and P. Glazer for valuable comments on the manuscript. Research was supported by the Yale Cancer Center, The Honorable Tina Brozman Foundation, a Rudolph Anderson fellowship, a Leslie Warner fellowship, and Clinical and Translational Science Award grants UL1 TR000142 and KL2 TR000140 from the National Center for Advancing Translational Sciences, a component of the US National Institutes of Health.
Footnotes
Competing Financial Interests
A provisional patent application has been filed covering the described method, with A.A.P. listed as an inventor. A.N. and A.B. have no competing financial interests.
Author Contributions
All authors contributed to planning of experiments, analysis of data, and writing of the paper. A.A.P. conceived the design and supervised the study. A.N. performed the majority of experimental work, and A.B. performed experiments related to radiation biodosimetry.
References
- 1.Nagalakshmi U, et al. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilhelm BT, et al. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 3.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 4.Cloonan N, et al. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Gerstein M, Snyder M. Nature reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 7.Schena M, Shalon D, Davis RW, Brown PO. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Nucleic acids research. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaur A, et al. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 10.Mestdagh P, et al. Nucleic Acids Res. 2011;39:e136. doi: 10.1093/nar/gkr646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Y, Ridzon D, Wong L, Chen C. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willems E, Leyns L, Vandesompele J. Anal Biochem. 2008;379:127–129. doi: 10.1016/j.ab.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 13.Mestdagh P, et al. Genome biology. 2009;10:R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Haene B, Mestdagh P, Hellemans J, Vandesompele J. Methods Mol Biol. 2012;822:261–272. doi: 10.1007/978-1-61779-427-8_18. [DOI] [PubMed] [Google Scholar]
- 15.Blower PE, et al. Mol Cancer Ther. 2007;6:1483–1491. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, et al. Mol Cancer Ther. 2010;9:1080–1091. doi: 10.1158/1535-7163.MCT-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokilde R, et al. Mol Cancer Ther. 2011;10:375–384. doi: 10.1158/1535-7163.MCT-10-0605. [DOI] [PubMed] [Google Scholar]
- 18.Canales RD, et al. Nat Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 19.Shippy R, et al. Nat Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul S, Amundson SA. Int J Radiat Oncol Biol Phys. 2008;71:1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paul S, et al. Radiat Res. 2011;175:257–265. doi: 10.1667/RR2420.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen C, et al. Nucleic acids research. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.