Abstract
Malaria, HIV, and tuberculosis (TB) collectively account for several million deaths each year, with all three ranking among the top ten killers in low-income countries. Despite being caused by very different organisms, malaria, HIV, and TB present a suite of challenges for mathematical modellers that are particularly pronounced in these infections, but represent general problems in infectious disease modelling, and highlight many of the challenges described throughout this issue. Here, we describe some of the unifying challenges that arise in modelling malaria, HIV, and TB, including variation in dynamics within the host, diversity in the pathogen, and heterogeneity in human contact networks and behaviour. Through the lens of these three pathogens, we provide specific examples of the other challenges in this issue and discuss their implications for informing public health efforts.
Introduction
Taken together, malaria, HIV, and tuberculosis (TB) infect more than a third of the global population and are responsible for almost three million deaths each year (1–3). Substantial investments in the prevention and treatment of these three pathogens have led to significant reductions in morbidity and mortality worldwide over the past century (1–3). Throughout this advancement, mathematical modelling has been a key tool in helping to understand transmission dynamics and in predicting the impact of control programs (4–10). For example, vector control as a way to effectively reduce malaria was recognized through population-level compartmental modelling more than 100 years ago (11) and the importance of CD4+ lymphocytes as sites for HIV proliferation was predicted using simple models nearly two decades ago (12). Although these and other models have provided valuable insights, incomplete understanding of the biology and transmission of these three pathogens remains a significant hurdle to the development of useful mathematical frameworks; new theoretical approaches and improved integration of a variety of different kinds of data are needed. Here, we use malaria, HIV, and TB to examine unifying mathematical challenges across the field of infectious disease modelling, despite their biological differences, to provide concrete examples reflecting general problems in the field, and to consider the role that modelling can play to inform public health efforts. We focus our attention on relatively simple models, exposing the data gaps and uncertainties that create fundamental challenges in designing basic model structures and parameterization, rather than on large-scale simulations, which often suffer from the same knowledge gaps as simpler frameworks but are less transparent and can be difficult to interpret. Indeed, we propose that, in general, while simple models often do not capture the biological complexities of these infections, more complex models may lack the data for parameterization and validation, presenting a paradox for modellers. We identify challenges in the following main areas: variation in dynamics within the host, pathogen genetic diversity, and heterogeneity in human contact networks and behaviour. Throughout, we reference specific modelling challenges addressed in depth elsewhere in this issue (referred to by article and challenge number).
1. Understanding infection dynamics in the host
The majority of models designed to inform policy on malaria, HIV, and TB are based upon population-level compartmental models, which generally assume single categories of infected and immune people, a fixed rate of recovery, and simple estimates for the duration of infectiousness (13–15). The most commonly used Susceptible-Infected-Recovered (SIR) compartmental models were developed to study outbreaks of acute immunizing infections among immunologically naïve populations, often describing the potential for an epidemic through summary statistics such as the reproductive number, R0. These assumptions must be modified for endemic pathogens exhibiting variable infection dynamics in the host (Article 15 (16) Challenge #2), heterogeneous immunological states between hosts (Article 11 (17) Challenge #4), and significant spatial and temporal variation in risk (Article 20 (18) Challenge #1). At the level of an individual, all three of these pathogens cause a wide range of clinical and infection outcomes related to host genetic heterogeneity, coinfection, or previous exposure (Article 11 (17) Challenge #5), and this creates extensive variability in both the infection length and the dynamics of infectiousness of an individual (Figure 1). Therefore, assumptions of a constant rate of loss of infectiousness and a uniform recovery rate may not adequately represent the infectious population underlying transmission.
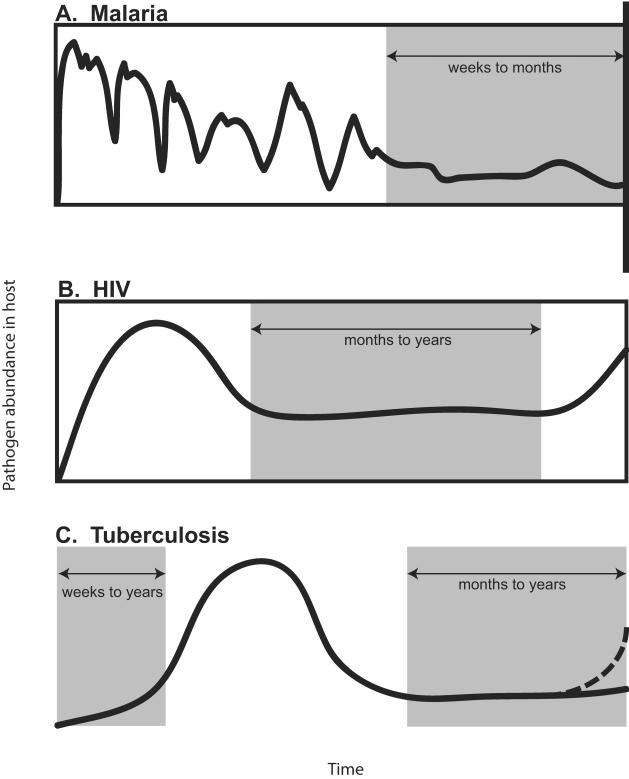
Figure 1. Pathogen dynamics within individual infections.
Grey shaded areas denote periods where the infection is asymptomatic, either due to chronicity in malaria (A) and HIV (B) or latency as in TB (C). Despite a lack of symptoms, particularly during chronic infections, individuals may still be infectious and contribute to spread of the pathogen.
In malaria, sustained parasite proliferation in the blood by the most virulent species Plasmodium falciparum may lead to a chronic phase of highly variable intensity and duration (19, 20) (Figure 1A). Repeated and simultaneous infections with different antigenically diverse parasites lead to a “semi-immune” status in older children and adults in endemic regions that is protective against severe disease (21); however, little is known about how patterns of exposure alter the distribution of chronic periods and the extent of the infectious population (22–26) (Article 8 (27) Challenge #6). Most infected individuals in endemic regions harbour multiple clones and have a complex history of exposure (Article 15 (16) Challenge #5), and although compartments can be added to SIR frameworks to account for the accumulation of multiple exposures, basic parameters are lacking to describe these individuals including relative infectiousness to mosquitoes, susceptibility to new infections, and rates of clearance of infection (Article 11 (17) Challenge #5; Article 18 (28) Challenge #1). Ultimately, this means that the probability of a mosquito bite leading to infection is poorly defined, so feedback between human and mosquito components of the model may be mis-specified, complicating estimates of the impact of different interventions (22).
Infection with HIV is incurable, which removes the need to estimate heterogeneous parameters of recovery and immunity, but complex within-host dynamics lead to variable infectious periods (Figure 1B). An HIV infection is characterized by an initial acute phase with rapid pathogen growth, followed by stabilization at a set point viral load that plays a critical role in determining the duration of the chronic period (29) but is highly heterogeneous among individuals, likely the result of complex and poorly understood interplay between the host immune system and the virus (30) (Article 13 (31) Challenge #3). As a result, the progression from chronic, low-density infections to full-blown AIDS and the variable length of chronic infections remain difficult to incorporate within epidemiological models (Figure 1B). In particular, including individual heterogeneity in the rate of transition through these different states of disease and infectiousness will require a better understanding of the complex mechanisms determining the dynamics of HIV within an individual.
The dominant source of variability in the within-host dynamics of tuberculosis infections results from heterogeneity in the timing of active disease. Most new infections result in an asymptomatic “latent” infection, with innate immunity controlling bacillary growth in the lung (32) (see (33) for a novel view on latent infections). Although latent infection confers immunity to active disease arising from reinfection (34), a small proportion of those who are latently infected progress to active disease (32). The lack of data on the distribution of times between becoming infectious to self-cure, disease or diagnosis, with indirect estimates suggesting that this period can range from a few months to several years (35), makes realistic incorporation of the initiation of active disease into models difficult (Article 22 (36) Challenge #3) (Figure 1C). It is clear that TB progresses much more rapidly in HIV-positive than in HIV-negative individuals (37), emphasizing the important and understudied dynamics that may result from co-infection (Article 13 (31) Challenge #2).
Heterogeneous within-host dynamics for all three infections make it difficult to establish the timing and duration of latent or chronic infection periods, where transmission potential may differ significantly from acute infection. As a result, modelling the impact of individual-level interventions on population-level transmission is challenging. Given the paucity of data on the dynamics of natural infections in endemic regions (Article 22 (36) Challenge #3), standard model frameworks frequently assume that varying previous exposure does not alter distributions of infectious periods in malaria, that infectiousness is not related to within-host infection dynamics for HIV, and that TB progression rates are randomly distributed. Note that we have focused here on simple smodels, but the same uncertain parameters governing within-host dynamics must also be estimated in more complex frameworks (Article 15 (16) Challenge #7; Article 23 (38)). A more thorough understanding of within host infection dynamics of malaria, HIV, and TB will lead to a more accurate assessment of the infectious populations responsible for transmission, a crucial quantity when evaluating interventions to inform policy decisions.
2. Incorporating pathogen genetic variation
Although very different organisms cause malaria, HIV, and TB, pathogen genetic diversity is a major obstacle to the design and use of vaccines and therapeutics for all three, hindering control and elimination prospects. Any model incorporating multiple pathogen strains faces the challenge of keeping track of people infected with, and immune to, different strains (Article 11 (17)), as well as defining the antigenic relationships between strains, their rate of genetic change (Articles 13 (31) and 14 (39)), genetic bottlenecks resulting from transmission (Article 15 (16) Challenge #1), and the consequences and frequency of superinfection (Article 15 (16) Challenge #5; Article 16 (40) Challenge #6).
The malaria parasite exhibits remarkable genetic diversity, especially among gene families such as the var genes, which are involved in phenotypic variation within the host and associated with disease outcome (41). Diversity among these gene families is continually generated through frequent recombination, a mode of diversification that is difficult to accommodate within modelling frameworks (Article 14 (39) Challenge #5). Indeed, malaria parasite genomic diversity is so great in endemic regions that we still lack adequate genotype markers, making it nearly impossible to track genotypes in a population or to sensitively establish the multiplicity of infection (42). Model complexities arising from frequent superinfection include a rapid increase in the number of categories necessary to account for individuals harbouring varying numbers of genotypes and insufficient data to accurately assign immunological relationships against parasite genotypes. Most models incorporating superinfection assume independence of strains with a fixed time since infection for each genotype, neglecting the impact of superinfection on duration of infection and relative infectiousness to mosquitoes (Article 15 (16) Challenge #5; Article 16 (40) Challenge #6). In particular, parameterizing the outcome of competition between strains within a single host plays an important role for understanding how variation created in a single infection carries over to the next human host, as the mosquito vector acts as a critical genetic bottleneck (43) (Article 15 (16)).
Rapid diversification within and between hosts poses significant challenges for HIV, where every possible point mutation may arise in a single day during HIV proliferation (44), and this complex within-host diversity is subsequently shaped through pressure from the host's immune system and drug treatment (Article 15 (16) Challenge #2). On a population level, the antigenic Env glycoprotein exhibits extensive diversity, which increases by 1–2% every year (45). Modellers must decide whether to include both within-host and population-level models of viral diversification. In the absence of an explicit within-host component, assumptions must be made about how diversity generated during an individual infection relates to the subset of viral genomes that are successfully transmitted (Article 14 (39) Challenge #7). Despite a recent rise in interest, there remains insufficient data about the relationship between within host diversification and transmission for HIV, so current theoretical frameworks make strong assumptions about the bottleneck imposed by transmission (Article 15 (16) Challenge #1) and the impact of superinfection (Article 15 (16) Challenges #5 and #6).
Although Mycobacterium tuberculosis is one of the least diverse bacterial pathogens, among the few of its loci that vary are genes that harbour mutations coding for drug resistance (46). Evidence that different drug resistant strains and lineages have different mutation rates, different fitness costs and elicit different levels of immunity suggests the epidemiological dynamics of drug resistant TB will require a better understanding of the biology of strain differences and the inclusion of multiple strains in models (47) (Article 14 (39) Challenge #6). Selection within the host and resulting emergence of drug resistance at a population level is not well understood although the relationship between pathogen diversity and the immune system is known to be critical. Models must be adapted to include evolution and dynamics of pathogen genotypes across scales in order to understand the emergence and maintenance of diverse populations of Mycobactrium tuberculosis (Article 13 (31) Challenge #4). Consequences for TB epidemiology depend on measuring and understanding fitness across pathogen generations, for which little conclusive data exists (48). Characterizing the impact of genetic diversity on the fitness of drug resistant TB strains will allow for incorporation into strain models.
Pathogen genetic diversity is often ignored within prevailing models of malaria, HIV, and TB despite the variety of strains and genotypes known to circulate at the population level. As antigenic variation among, and competition between, strains will alter their frequency in the population and transmission overall, the inclusion of multiple genetic variants will be important for many policy questions, particularly in relation to drug resistance.
3. Accounting for heterogeneous contact rates and changes in behaviour
The inclusion of realistic human behaviours relating to the spread of disease and the efficacy of interventions remains a challenge for many infectious disease models (Articles 18 (28) and 20 (18)), but represents a particular problem for malaria, HIV, and TB. Heterogeneities from dynamic contact networks, such as sexual contacts in HIV and indirect contacts in vector-borne infections like malaria, as well as a wide range of behaviours that influence exposure and effectiveness of treatment, are challenging to accurately parameterize (49, 50) (Article 18 (28); Article 19 (51) Challenges #3 and #4). Travel of infected individuals, which is often difficult to measure, spreads infections within and between countries and regions (Article 4 (52) Challenge #6; Article 20 (18) Challenge #3), jeopardizing the sustainability of control programs and prospects for elimination (53) (Article 3 (54)). In all three cases, vulnerable groups may exhibit fundamentally different epidemiological dynamics than the general population, and must be modelled and parameterized separately.
Malaria requires a vector for transmission, in contrast to many pathogens including HIV and TB, so contacts between people are by definition indirect (Figure 2A). The variability of vector habitats and hosts means that malaria prevalence shows strong geographical heterogeneity (Article 6 (55) Challenge #2), with transmission being focused in so-called “hotspots” or in mobile populations that are exposed to infection through forestry or agricultural work (56–58). These heterogeneities are obscured by prevalence estimates that are generally derived on large spatial scales, and decrease the predictive power of mathematical models of transmission (59) (Article 20 (18) Challenges #1 and #5). Complex dynamics may emerge from the interaction of seasonal travel for agricultural work and seasonal changes in the environmental drivers of mosquito populations, and are difficult to accommodate in current model frameworks. Additional variation arises as many individuals do not seek treatment or use preventive measures like bed nets despite widespread availability, which makes it difficult to predict the impact of interventions and the timelines required for control and elimination.
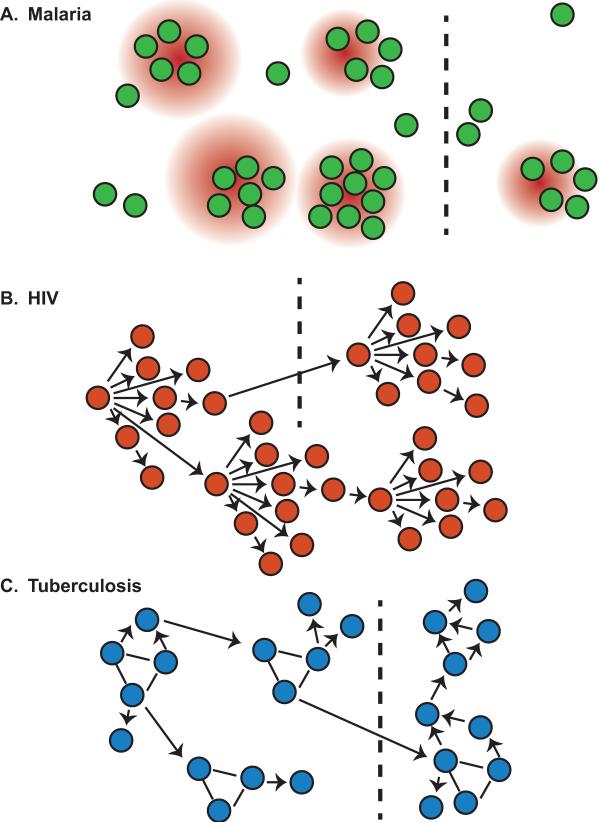
Figure 2. Networks for spread of infection.
Malaria (A) transmission between individuals (circles) requires the mosquito vector, resulting in “hotspots” for transmission driven by heightened exposure to malaria-infected mosquitoes in certain regions (red circles). For HIV (B) and TB (C) transmission occurs between individuals in a directed fashion indicated by arrows. Dotted black lines indicate spatially separated populations.
HIV spreads unevenly through communities defined by distinct behaviours (Figure 2B), such as sex workers, men who have sex with men, and injecting drug users (60–64). Measuring and quantifying the heterogeneities in these networks remains challenging (Article 21 (65)); for example, partnership acquisition rates are needed to generate realistic contact networks in models, but generally only cross-sectional data is available on the total number of partners each individual has over a specified time period (66). These different social groups may also exhibit systematic variations in behaviours affecting treatment-seeking, adherence to treatment, and exposure, such as the use of condoms or the dropout of people over the so-called treatment cascade (67), which can further alter the risk and course of infection. Even when detailed data is available, the incorporation of temporally changing interactions is challenging to include and nearly impossible to accurately parameterize. Static and highly simplified contact patterns are often used in the absence of data on the complex and dynamic network structures that actually underlie HIV transmission.
For the spread of TB (Figure 2C), many occupational and behavioural risk factors that enhance transmission have been clearly identified, focusing attention on the homeless and substance users in low burden settings in the US and Europe (68), and on health care workers, drivers of crowded vehicles, prisoners, migrants, those working in mines and residents of overcrowded slums in high burden countries (69). Collecting reliable data on patterns of movement and migration is difficult, particularly across borders among refugee and migrant communities, who are at high risk of infection (Article 22 (36)). Among those infected, behaviour strongly influences the duration of infectiousness because treatment seeking depends on access to care and recognition of symptoms, which overlap with other chronic lung conditions including smoking (70). All of these factors lead to difficulty identifying the infectious population, while the lack of granularity in incidence and prevalence data and the inability to directly capture values for behavioural parameters make valid parameterization of models challenging (Article 23 (38) Challenge #4).
Despite the known importance of behaviour on disease prevalence, the extent of the infectious population, and the emergence of drug resistance, the social factors that drive individual behaviours are poorly understood. What level of behaviour change is necessary to alter an epidemic (Article 4 (52) Challenge #1) and the level of detailed data required to measure this change (Article 4 Challenges #2 and #4) remain open questions. Social and spatial aspects are often not accounted for in traditional model frameworks, which generalize behaviour on a population level, and this not only distorts epidemic dynamics but also underestimates re-exposure and reinfection among groups most at risk (71) (Articles 16 (40) and 19 (51)). Furthermore, most models treat behaviour as static, although modifications of behaviour are likely to change as perception of risk shifts along with transmission. Incorporating variation in behaviour over the course of an intervention or in the case of multiple interventions is essential if models are to reliably predict the impact of the interventions or calculate their cost-effectiveness (72, 73) (Article 2 (54) Challenge #5).
Conclusion
As we move towards control and potentially elimination for malaria, HIV, and TB, mathematical models provide a powerful framework to consider the possible impact of interventions, identify areas where further empirical work is needed, and focus on the important policy and research questions. Critical knowledge gaps for the effective application of models for policy include spatial and temporal variations in disease prevalence and transmission intensity, host-pathogen interactions and infection outcome, and human behaviour. Most importantly, better communication between modellers and experimentalists or field workers will be needed to refine the questions, determine which data are most important and must urgently needed, and to ensure that the analytical work leads to better policy and better control of all three infections (Article 2 (27, 74) Challenges #4 and #6).
Malaria, HIV, and TB prompt mathematical challenges in infectious disease modelling.
There exist many critical knowledge gaps to accurately parameterize models.
Complex within-host dynamics leads to variation in infectiousness among individuals.
Pathogen genetic diversity alters prevalence and transmission of pathogens.
Heterogeneous behaviour influences exposure and treatment effectiveness.
Acknowledgments
The authors acknowledge funding from the following sources: Award U54GM088558 from the National Institute Of General Medical Sciences (LMC, COB), Award RO1MH087328-03 (http://grants.nih.gov/grants/about_grants.htm) (NNA) and Award NIAID AI 007433 (http://www.niaid.nih.gov/researchfunding/pages/default.aspx) (NNA). CD is a staff member of the World Health Organization; the authors alone are responsible for the views expressed in this publication, which do not necessarily represent the decisions, policy or views of WHO. SG is a royal Society Wolfson Research Fellow and an ERC Advanced Investigator (ERC Advanced Grant - DIVERSITY).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.WHO . Global update on HIV treatment 2013: Results, impacts and opportunities. World Health Organization; Geneva, Switzerland: 2013. [Google Scholar]
- 2.WHO . Global Tuberculosis Report 2013. World Health Organization; Geneva, Switzerland: 2013. [Google Scholar]
- 3.WHO . World Malaria Reprot 2013. World Health Organization; Geneva, Switzerland: 2013. [Google Scholar]
- 4.Anderson RM, Gupta S, May RM. Potential of community-wide chemotherapy or immunotherapy to control the spread of HIV-1. Nature. 1991;350(6316):356–9. doi: 10.1038/350356a0. doi: 10.1038/350356a0. PubMed PMID: 2008214. [DOI] [PubMed] [Google Scholar]
- 5.Dye C, Glaziou P, Floyd K, Raviglione M. Prospects for tuberculosis elimination. Annual review of public health. 2013;34:271–86. doi: 10.1146/annurev-publhealth-031912-114431. doi: 10.1146/annurev-publhealth-031912-114431. PubMed PMID: 23244049. [DOI] [PubMed] [Google Scholar]
- 6.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328(5980):856–61. doi: 10.1126/science.1185449. doi: 10.1126/science.1185449. PubMed PMID: 20466923. [DOI] [PubMed] [Google Scholar]
- 7.Garnett GP, Cousens S, Hallett TB, Steketee R, Walker N. Mathematical models in the evaluation of health programmes. Lancet. 2011;378(9790):515–25. doi: 10.1016/S0140-6736(10)61505-X. doi: 10.1016/S0140-6736(10)61505-X. PubMed PMID: 21481448. [DOI] [PubMed] [Google Scholar]
- 8.Macdonald G. Theory of the eradication of malaria. Bulletin of the World Health Organization. 1956;15(3–5):369–87. PubMed PMID: 13404426; PubMed Central PMCID: PMC2538288. [PMC free article] [PubMed] [Google Scholar]
- 9.mal ERACGoM A research agenda for malaria eradication: modeling. PLoS medicine. 2011;8(1):e1000403.. doi: 10.1371/journal.pmed.1000403. doi: 10.1371/journal.pmed.1000403. PubMed PMID: 21283605; PubMed Central PMCID: PMC3026697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stover J. Influence of mathematical modeling of HIV and AIDS on policies and programs in the developing world. Sexually transmitted diseases. 2000;27(10):572–8. doi: 10.1097/00007435-200011000-00005. PubMed PMID:11099072. [DOI] [PubMed] [Google Scholar]
- 11.Ross R. The Prevention of Malaria. E.P. Dutton and Company; New York: 1910. [Google Scholar]
- 12.Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373(6510):123–6. doi: 10.1038/373123a0. doi: 10.1038/373123a0. PubMed PMID:7816094. [DOI] [PubMed] [Google Scholar]
- 13.Dowdy DW, Dye C, Cohen T. Data needs for evidence-based decisions: a tuberculosis modeler's 'wish list'. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2013;17(7):866–77. doi: 10.5588/ijtld.12.0573. doi: 10.5588/ijtld.12.0573. PubMed PMID: 23743307; PubMed Central PMCID: PMC4041555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson LF, White PJ. A review of mathematical models of HIV/AIDS interventions and their implications for policy. Sexually transmitted infections. 2011;87(7):629–34. doi: 10.1136/sti.2010.045500. doi: 10.1136/sti.2010.045500. PubMed PMID 21685191. [DOI] [PubMed] [Google Scholar]
- 15.Reiner RC, Jr., Perkins TA, Barker CM, Niu T, Chaves LF, Ellis AM, George DB, Le Menach A, Pulliam JR, Bisanzio D, Buckee C, Chiyaka C, Cummings DA, Garcia AJ, Gatton ML, Gething PW, Hartley DM, Johnston G, Klein EY, Michael E, Lindsay SW, Lloyd AL, Pigott DM, Reisen WK, Ruktanonchai N, Singh BK, Tatem AJ, Kitron U, Hay SI, Scott TW, Smith DL. A systematic review of mathematical models of mosquito-borne pathogen transmission: 1970–2010. Journal of the Royal Society, Interface / the Royal Society. 2013;10(81):20120921. doi: 10.1098/rsif.2012.0921. doi: 10.1098/rsif.2012.0921. PubMed PMID: 23407571; PubMed Central PMCID: PMC3627099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gog JR, Pellis L, Wood JL, McLean AR, Arinaminpathy N, Lloyd-Smith JO. Epidemics. Seven challenges in modelling pathogen dynamics within-host and across scales. [DOI] [PubMed] [Google Scholar]
- 17.Wikramaratna PS, Kucharski A, Gupta S, Anderson RM, McLean AR, Gog JR. Epidemics. Five challenges in modelling interacting strain dynamics. [DOI] [PubMed] [Google Scholar]
- 18.Riley S, Eames K, Isham V, Mollison D, Trapman P. Epidemics. Five challenges for Spatial Epidemic Models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bull PC, Lowe BS, Kortok M, Molyneux CS, Newbold CI, Marsh K. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nature medicine. 1998;4(3):358–60. doi: 10.1038/nm0398-358. PubMed PMID: 9500614; PubMed Central PMCID: PMC3836255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newbold CI, Craig AG, Kyes S, Berendt AR, Snow RW, Peshu N, Marsh K. PfEMP1, polymorphism and pathogenesis. Annals of tropical medicine and parasitology. 1997;91(5):551–7. doi: 10.1080/00034989760923. PubMed PMID:9329992. [DOI] [PubMed] [Google Scholar]
- 21.Bull PC, Berriman M, Kyes S, Quail MA, Hall N, Kortok MM, Marsh K, Newbold CI. Plasmodium falciparum variant surface antigen expression patterns during malaria. PLoS pathogens. 2005;1(3):e26. doi: 10.1371/journal.ppat.0010026. doi: 10.1371/journal.ppat.0010026. PubMed PMID: 16304608; PubMed Central PMCID: PMC1287908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen E, Buckee CO. Modeling the human infectious reservoir for malaria control: does heterogeneity matter? Trends in parasitology. 2013;29(6):270–5. doi: 10.1016/j.pt.2013.03.009. doi: 10.1016/j.pt.2013.03.009. PubMed PMID: 23597499; PubMed Central PMCID: PMC3665612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Churcher TS, Bousema T, Walker M, Drakeley C, Schneider P, Ouedraogo AL, Basanez MG. Predicting mosquito infection from Plasmodium falciparum gametocyte density and estimating the reservoir of infection. eLife. 2013;2:e00626. doi: 10.7554/eLife.00626. doi: 10.7554/eLife.00626. PubMed PMID: 23705071; PubMed Central PMCID: PMC3660740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Killeen GF, Ross A, Smith T. Infectiousness of malaria-endemic human populations to vectors. The American journal of tropical medicine and hygiene. 2006;75(2 Suppl):38–45. doi: 10.4269/ajtmh.2006.75.2_suppl.0750038. PubMed PMID:16931814. [DOI] [PubMed] [Google Scholar]
- 25.Lin JT, Saunders DL, Meshnick SR. The role of submicroscopic parasitemia in malaria transmission: what is the evidence? Trends in parasitology. 2014;30(4):183–90. doi: 10.1016/j.pt.2014.02.004. doi: 10.1016/j.pt.2014.02.004. PubMed PMID: 24642035; PubMed Central PMCID: PMC4049069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindblade KA, Steinhardt L, Samuels A, Kachur SP, Slutsker L. The silent threat: asymptomatic parasitemia and malaria transmission. Expert review of anti-infective therapy. 2013;11(6):623–39. doi: 10.1586/eri.13.45. doi: 10.1586/eri.13.45. PubMed PMID:23750733. [DOI] [PubMed] [Google Scholar]
- 27.Cunniffe NJ, Koskella B, Metcalf J, Parnell S, Gottwald T, Gilligan C. Epidemics. Challenges in modelling plant disease. [DOI] [PubMed] [Google Scholar]
- 28.Pellis L, Ball F, Bansal S, Eames K, House T, Isham V, Trapman P. Epidemics. Challenges for network epidemic models. [DOI] [PubMed] [Google Scholar]
- 29.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(44):17441–6. doi: 10.1073/pnas.0708559104. doi: 10.1073/pnas.0708559104. PubMed PMID: 17954909; PubMed Central PMCID: PMC2077275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perreau M, Levy Y, Pantaleo G. Immune response to HIV. Current opinion in HIV and AIDS. 2013;8(4):333–40. doi: 10.1097/COH.0b013e328361faf4. doi: 10.1097/COH.0b013e328361faf4. PubMed PMID:23743723. [DOI] [PubMed] [Google Scholar]
- 31.Metcalf J, Birger R, Funk S, Kouyos RD, Lloyd-Smith JO, Jansen VAA. Epidemics. Five challenges in evolution and infectious diseases. [DOI] [PubMed] [Google Scholar]
- 32.Ernst JD. The immunological life cycle of tuberculosis. Nature reviews Immunology. 2012;12(8):581–91. doi: 10.1038/nri3259. doi: 10.1038/nri3259. PubMed PMID:22790178. [DOI] [PubMed] [Google Scholar]
- 33.Orme IM. A new unifying theory of the pathogenesis of tuberculosis. Tuberculosis. 2014;94(1):8–14. doi: 10.1016/j.tube.2013.07.004. doi: 10.1016/j.tube.2013.07.004. PubMed PMID: 24157189; PubMed Central PMCID: PMC3877201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrews JR, Noubary F, Walensky RP, Cerda R, Losina E, Horsburgh CR. Risk of progression to active tuberculosis following reinfection with Mycobacterium tuberculosis. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2012;54(6):784–91. doi: 10.1093/cid/cir951. doi: 10.1093/cid/cir951. PubMed PMID: 22267721; PubMed Central PMCID: PMC3284215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiemersma EW, van der Werf MJ, Borgdorff MW, Williams BG, Nagelkerke NJ. Natural history of tuberculosis: duration and fatality of untreated pulmonary tuberculosis in HIV negative patients: a systematic review. PloS one. 2011;6(4):e17601. doi: 10.1371/journal.pone.0017601. doi: 10.1371/journal.pone.0017601. PubMed PMID: 21483732; PubMed Central PMCID: PMC3070694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lessler J, Edmunds J, Halloran ME, Hollingsworth TD, Lloyd AL. Epidemics. Six Challenges for Model-Driven Data Collection in Experimental and Observational Studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawn SD, Churchyard G. Epidemiology of HIV-associated tuberculosis. Current opinion in HIV and AIDS. 2009;4(4):325–33. doi: 10.1097/COH.0b013e32832c7d61. doi: 10.1097/COH.0b013e32832c7d61. PubMed PMID:19532072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Angelis D, Presanis AM, Birrell PJ, Scalia-Tomba G, House T. Epidemics. Five challenges in infectious disease modelling using data from multiple sources. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frost SDW, Pybus OG, Gog JR, Viboud C, Bonhoeffer S, Bedford T. Epidemics. Challenges in Phylodynamics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts MG, Andreasen V, Lloyd AL, Pellis L. Epidemics. Eight challenges for deterministic epidemic models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston GL, Smith DL, Fidock DA. Malaria's missing number: calculating the human component of R0 by a within-host mechanistic model of Plasmodium falciparum infection and transmission. PLoS computational biology. 2013;9(4):e1003025. doi: 10.1371/journal.pcbi.1003025. doi: 10.1371/journal.pcbi.1003025. PubMed PMID: 23637586; PubMed Central PMCID: PMC3630126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, O'Brien J, Djimde A, Doumbo O, Zongo I, Ouedraogo JB, Michon P, Mueller I, Siba P, Nzila A, Borrmann S, Kiara SM, Marsh K, Jiang H, Su XZ, Amaratunga C, Fairhurst R, Socheat D, Nosten F, Imwong M, White NJ, Sanders M, Anastasi E, Alcock D, Drury E, Oyola S, Quail MA, Turner DJ, Ruano-Rubio V, Jyothi D, Amenga-Etego L, Hubbart C, Jeffreys A, Rowlands K, Sutherland C, Roper C, Mangano V, Modiano D, Tan JC, Ferdig MT, Amambua-Ngwa A, Conway DJ, Takala-Harrison S, Plowe CV, Rayner JC, Rockett KA, Clark TG, Newbold CI, Berriman M, MacInnis B, Kwiatkowski DP. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487(7407):375–9. doi: 10.1038/nature11174. doi: 10.1038/nature11174. PubMed PMID: 22722859; PubMed Central PMCID: PMC3738909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conway DJ, Roper C, Oduola AM, Arnot DE, Kremsner PG, Grobusch MP, Curtis CF, Greenwood BM. High recombination rate in natural populations of Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(8):4506–11. doi: 10.1073/pnas.96.8.4506. PubMed PMID: 10200292; PubMed Central PMCID: PMC16362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perelson AS, Essunger P, Ho DD. Dynamics of HIV-1 and CD4+ lymphocytes in vivo. Aids. 1997;11(Suppl A):S17–24. PubMed PMID:9451962. [PubMed] [Google Scholar]
- 45.Novitsky V, Lagakos S, Herzig M, Bonney C, Kebaabetswe L, Rossenkhan R, Nkwe D, Margolin L, Musonda R, Moyo S, Woldegabriel E, van Widenfelt E, Makhema J, Essex M. Evolution of proviral gp120 over the first year of HIV-1 subtype C infection. Virology. 2009;383(1):47–59. doi: 10.1016/j.virol.2008.09.017. doi: 10.1016/j.virol.2008.09.017. PubMed PMID: 18973914; PubMed Central PMCID: PMC2642736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGrath M, Gey van Pittius NC, van Helden PD, Warren RM, Warner DF. Mutation rate and the emergence of drug resistance in Mycobacterium tuberculosis. The Journal of antimicrobial chemotherapy. 2014;69(2):292–302. doi: 10.1093/jac/dkt364. doi: 10.1093/jac/dkt364. PubMed PMID:24072169. [DOI] [PubMed] [Google Scholar]
- 47.Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nature genetics. 2013;45(7):784–90. doi: 10.1038/ng.2656. doi: 10.1038/ng.2656. PubMed PMID: 23749189; PubMed Central PMCID: PMC3777616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dye C, Williams BG. Slow elimination of multidrug-resistant tuberculosis. Science translational medicine. 2009;1(3):3ra8. doi: 10.1126/scitranslmed.3000346. doi: 10.1126/scitranslmed.3000346. PubMed PMID:20368167. [DOI] [PubMed] [Google Scholar]
- 49.Kato M, Granich R, Bui DD, Tran HV, Nadol P, Jacka D, Sabin K, Suthar AB, Mesquita F, Lo YR, Williams B. The potential impact of expanding antiretroviral therapy and combination prevention in Vietnam: towards elimination of HIV transmission. Journal of acquired immune deficiency syndromes. 2013;63(5):e142–9. doi: 10.1097/QAI.0b013e31829b535b. doi: 10.1097/QAI.0b013e31829b535b. PubMed PMID: 23714739; PubMed Central PMCID: PMC3814627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller JC. Spread of infectious disease through clustered populations. Journal of the Royal Society, Interface / the Royal Society. 2009;6(41):1121–34. doi: 10.1098/rsif.2008.0524. doi: 10.1098/rsif.2008.0524. PubMed PMID: 19324673; PubMed Central PMCID: PMC2817154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ball F, Britton T, House T, Isham V, Mollison D, Pellis L, Scalia-Tomba G. Epidemics. Challenges for Metapopulation Models of Epidemics. [DOI] [PubMed] [Google Scholar]
- 52.Funk S, Bansal S, Bauch CT, Eames K, Edmunds J, Galvani AP, Klepac P. Epidemics. Challenges in incorporating the dynamics of behaviour in infectious disease models. [DOI] [PubMed] [Google Scholar]
- 53.Buckee CO, Wesolowski A, Eagle NN, Hansen E, Snow RW. Mobile phones and malaria: modeling human and parasite travel. Travel medicine and infectious disease. 2013;11(1):15–22. doi: 10.1016/j.tmaid.2012.12.003. doi: 10.1016/j.tmaid.2012.12.003. PubMed PMID: 23478045; PubMed Central PMCID: PMC3697114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klepac P, Funk S, Hollingsworth TD, Metcalf J. Epidemics. Six challenges in the eradication of infectious diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hollingsworth TD, Pulliam J, Funk S, Truscott JE, Isham V, Lloyd AL. Epidemics. Seven challenges for modelling indirect transmission: vector-borne diseases, macroparasites and neglected tropical diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bousema T, Drakeley C, Gesase S, Hashim R, Magesa S, Mosha F, Otieno S, Carneiro I, Cox J, Msuya E, Kleinschmidt I, Maxwell C, Greenwood B, Riley E, Sauerwein R, Chandramohan D, Gosling R. Identification of hot spots of malaria transmission for targeted malaria control. The Journal of infectious diseases. 2010;201(11):1764–74. doi: 10.1086/652456. doi: 10.1086/652456. PubMed PMID: 20415536. [DOI] [PubMed] [Google Scholar]
- 57.Ahmed S, Galagan S, Scobie H, Khyang J, Prue CS, Khan WA, Ram M, Alam MS, Haq MZ, Akter J, Glass G, Norris DE, Nyunt MM, Shields T, Sullivan DJ, Sack DA. Malaria hotspots drive hypoendemic transmission in the Chittagong Hill Districts of Bangladesh. PloS one. 2013;8(8):e69713. doi: 10.1371/journal.pone.0069713. doi: 10.1371/journal.pone.0069713. PubMed PMID: 23936345; PubMed Central PMCID: PMC3735545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carrara VI, Lwin KM, Phyo AP, Ashley E, Wiladphaingern J, Sriprawat K, Rijken M, Boel M, McGready R, Proux S, Chu C, Singhasivanon P, White N, Nosten F. Malaria burden and artemisinin resistance in the mobile and migrant population on the Thai-Myanmar border, 1999–2011: an observational study. PLoS medicine. 2013;10(3):e1001398. doi: 10.1371/journal.pmed.1001398. doi: 10.1371/journal.pmed.1001398. PubMed PMID: 23472056; PubMed Central PMCID: PMC3589269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hasibeder G, Dye C. Population dynamics of mosquito-borne disease: persistence in a completely heterogeneous environment. Theoretical population biology. 1988;33(1):31–53. doi: 10.1016/0040-5809(88)90003-2. PubMed PMID:2897726. [DOI] [PubMed] [Google Scholar]
- 60.Gupta S, Anderson RM, May RM. Networks of sexual contacts: implications for the pattern of spread of HIV. Aids. 1989;3(12):807–17. PubMed PMID:2517202. [PubMed] [Google Scholar]
- 61.Eaton JW, Hallett TB, Garnett GP. Concurrent sexual partnerships and primary HIV infection: a critical interaction. AIDS and behavior. 2011;15(4):687–92. doi: 10.1007/s10461-010-9787-8. doi: 10.1007/s10461-010-9787-8. PubMed PMID: 20890654; PubMed Central PMCID: PMC3520057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garnett GP, Anderson RM. Balancing sexual partnerships in an age and activity stratified model of HIV transmission in heterosexual populations. IMA journal of mathematics applied in medicine and biology. 1994;11(3):161–92. doi: 10.1093/imammb/11.3.161. PubMed PMID:7822888. [DOI] [PubMed] [Google Scholar]
- 63.Tanser F, Barnighausen T, Hund L, Garnett GP, McGrath N, Newell ML. Effect of concurrent sexual partnerships on rate of new HIV infections in a high-prevalence, rural South African population: a cohort study. Lancet. 2011;378(9787):247–55. doi: 10.1016/S0140-6736(11)60779-4. doi: 10.1016/S0140-6736(11)60779-4. PubMed PMID: 21763937; PubMed Central PMCID: PMC3141142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zuma K, Lurie MN, Williams BG, Mkaya-Mwamburi D, Garnett GP, Sturm AW. Risk factors of sexually transmitted infections among migrant and non-migrant sexual partnerships from rural South Africa. Epidemiology and infection. 2005;133(3):421–8. doi: 10.1017/s0950268804003607. PubMed PMID: 15962548; PubMed Central PMCID: PMC2870265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eames K, Bansal S, Frost SDW, Riley S. Epidemics. Challenges in measuring networks for use in modelling. [DOI] [PubMed] [Google Scholar]
- 66.Abuelezam NN, Rough K, Seage GR., 3rd. Individual-based simulation models of HIV transmission: reporting quality and recommendations. PloS one. 2013;8(9):e75624. doi: 10.1371/journal.pone.0075624. doi: 10.1371/journal.pone.0075624. PubMed PMID: 24098707; PubMed Central PMCID: PMC3787035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Losina E, Bassett IV, Giddy J, Chetty S, Regan S, Walensky RP, Ross D, Scott CA, Uhler LM, Katz JN, Holst H, Freedberg KA. The “ART” of linkage: pre-treatment loss to care after HIV diagnosis at two PEPFAR sites in Durban, South Africa. PloS one. 2010;5(3):e9538. doi: 10.1371/journal.pone.0009538. doi: 10.1371/journal.pone.0009538. PubMed PMID: 20209059; PubMed Central PMCID: PMC2832018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lonnroth K, Jaramillo E, Williams BG, Dye C, Raviglione M. Drivers of tuberculosis epidemics: the role of risk factors and social determinants. Social science & medicine. 2009;68(12):2240–6. doi: 10.1016/j.socscimed.2009.03.041. doi: 10.1016/j.socscimed.2009.03.041. PubMed PMID:19394122. [DOI] [PubMed] [Google Scholar]
- 69.Bates I, Fenton C, Gruber J, Lalloo D, Lara AM, Squire SB, Theobald S, Thomson R, Tolhurst R. Vulnerability to malaria, tuberculosis, and HIV/AIDS infection and disease. Part II: Determinants operating at environmental and institutional level. The Lancet infectious diseases. 2004;4(6):368–75. doi: 10.1016/S1473-3099(04)01047-3. doi: 10.1016/S1473-3099(04)01047-3. PubMed PMID:15172345. [DOI] [PubMed] [Google Scholar]
- 70.Storla DG, Yimer S, Bjune GA. A systematic review of delay in the diagnosis and treatment of tuberculosis. BMC public health. 2008;8:15. doi: 10.1186/1471-2458-8-15. doi: 10.1186/1471-2458-8-15. PubMed PMID: 18194573; PubMed Central PMCID: PMC2265684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perkins TA, Scott TW, Le Menach A, Smith DL. Heterogeneity, mixing, and the spatial scales of mosquito-borne pathogen transmission. PLoS computational biology. 2013;9(12):e1003327. doi: 10.1371/journal.pcbi.1003327. doi: 10.1371/journal.pcbi.1003327. PubMed PMID: 24348223; PubMed Central PMCID: PMC3861021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dye C, Williams BG. Eliminating human tuberculosis in the twenty-first century. Journal of the Royal Society, Interface / the Royal Society. 2008;5(23):653–62. doi: 10.1098/rsif.2007.1138. doi: 10.1098/rsif.2007.1138. PubMed PMID: 17690054; PubMed Central PMCID: PMC3226985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Long EF, Stavert RR. Portfolios of biomedical HIV interventions in South Africa: a cost-effectiveness analysis. Journal of general internal medicine. 2013;28(10):1294–301. doi: 10.1007/s11606-013-2417-1. doi: 10.1007/s11606-013-2417-1. PubMed PMID: 23588668; PubMed Central PMCID: PMC3785647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Metcalf J, Edmunds J, Lessler J. Epidemics. Six challenges in public health and modelling. [DOI] [PubMed] [Google Scholar]