Abstract
Filamentous growth is a foraging response that occurs in fungal species. It allows fungal pathogens to invade cells and tissues of a host organism. Budding yeast undergoes filamentous growth and can invade semisolid agar plates, penetrating the agar surface. These cells cannot be removed by rinsing with water, and form an invasive scar. The plate-washing assay is an easy first test for filamentous growth and is performed at low cost with minimal reagents. The assay is versatile: it can be used as a teaching tool, is amenable to high-throughput genetic analysis, and is used to evaluate filamentous growth in different fungal species including pathogens like Candida albicans.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
RECIPES: Please see the end of this protocol for recipes indicated by <R>. Additional recipes can be found online at http://cshprotocols.cshlp.org/site/recipes.
Reagents
Distilled water, sterile
SD agar plates <R>
SD liquid medium <R>
-
Yeast strains of interest
The Σ1278b background undergoes filamentous growth (Gimeno et al. 1992). Commonly used laboratory strains have lost the ability to undergo filamentous growth (Liu et al. 1996). A mutant defective for agar invasion (e.g., flo11Δ) should be included in the assay as a negative control. Maintain stocks on SD plates (not YEPD). YEPD agar plates <R>
Equipment
Digital camera
Flat toothpicks, sterile
Glass tubes with metal tops, sterile
ImageJ software (http://imagej.nih.gov; Schneider et al. 2012)
Incubator set at 30°C
Innoculation loop or long wooden toothpick, sterile
Light box
Light microscope with DIC optics and 100× objective
Microcentrifuge
Microcentrifuge tubes, sterile
Microscope oil
Microscope slides and coverslips
Pipette tips, sterile
Shaking incubator or a shaker in a 30°C room
METHOD
-
Using a long wooden toothpick or inoculation loop, inoculate yeast from freshly patched SD plates (from freezer stocks or a plate stored at 4°C) into 5 mL of SD medium in a glass tube with a metal top and grow until saturation is reached.
We typically grow cultures for 16 h (overnight) at 30°C with shaking at 225 rpm. Synthetic (SD) medium reduces clumpiness by minimizing expression of FLO11. Cell aggregation complicates quantitating cell density and culture manipulation. Remove 1 mL of cells from the overnight culture. Pellet the cells by centrifugation at 16,000g for 1 min at 20°C. Resuspend the cells in 1 mL of sterile distilled H2O.
-
Spot 10 μL of cells directly onto YEPD agar plates. If testing multiple strains (and/or serial dilutions from tubes) on a single plate, spot cells equidistant from each other and the edges of the plate.
Colonies that have more access to nutrients (e.g., along an edge) will invade better than colonies in the center of the plate. Invasive growth occurs more robustly when fewer colonies are spotted.A quick and commonly used alternative to spotting is to patch cells in quadrants on YEPD plates using a toothpick. Patching is quicker, whereas spotting is more quantitative. -
Incubate plates at 30°C for 2–10 d until invasive growth occurs.
See Discussion. -
Photograph the colonies.
See Figure 1. -
Wash the plates in a stream of water. Use the same flow rate and water temperature for all plates. Wash each colony for the same period of time; do not wash more invasive colonies longer.
This step can be separated into a “soft” and “hard” wash. For the soft wash, use a stream of water. For the hard wash, rub your finger over the colonies to remove cells from the plate. Perform the soft wash first. If informative, photograph the plate after the soft wash. If working with a pathogen, wear gloves for the hard wash. -
Photograph the plates by transmitted light, using a light box, to reveal details of the invasive pattern.
The invasive scar will be at a different focal plane than the colony. Change the focus of the camera accordingly.See Figure 1. -
Examine the invaded cells by microscopy. Gently scrape the invasive scar with a toothpick to excise invaded cells. Place the cells onto a slide spotted with 7 μL of H2O and add a coverslip. Examine the cells by DIC at 100× magnification under oil immersion microscopy.
Cells undergoing filamentous growth form chains of elongated and connected cells. The localization of proteins in filamentous cells can be determined with GFP fusion proteins and fluorescent microscopy. Additionally, excised cells can be evaluated by molecular techniques such as RNA-seq, DNA microarray analysis, or RT-PCR. -
Quantitate agar invasion.
Analyze photographs of colonies and washed plates by densitometry using ImageJ software. Use the invert function and subtract background values.
-
Examine multiple replicates and perform statistical analysis to assess whether differences in agar invasion are statistically significant.
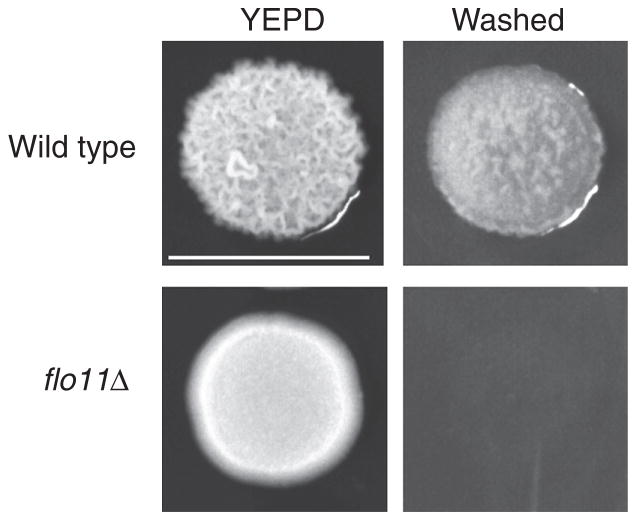
FIGURE 1.
Sample results from a plate-washing assay. Colonies of the Σ1278b background grown on YEPD have a ruffled appearance (top left). Washing the plate in a stream of water reveals an invasive scar (top right). A mutant lacking the FLO11 gene (flo11Δ) that is defective for filamentous growth has a smooth pattern (bottom left) and is defective for agar invasion (bottom right). Bar, 1 cm.
DISCUSSION
Agar invasion occurs after 2–10 d depending on the strain (different Σ1278b derivatives show different degrees of agar invasion) and the number of colonies spotted per plate. Washing multiple plates over a time series will establish the optimal incubation time. Some mutants/strains show hyperinvasive growth, which can be measured by washing plates at early time points (1–2 d), before the invasive growth seen in wild-type cells. Even cells that are defective for agar invasion (e.g., cells lacking a MAPK pathway) show agar invasion over an extended time period.
Colonies become ruffled over time because of cell adhesion mediated by FLO11 (Granek and Magwene 2010). Colony ruffling typically correlates with invasive growth (Fig. 1). Longer incubation (1–2 wk) results in exaggerated colony ruffling, robust invasive growth, and an increase in cell length, which can facilitate quantitation of these phenotypes.
RELATED INFORMATION
This assay has been adapted for high-throughput genetic analysis (Ryan et al. 2012) and for measuring filamentous growth in Candida albicans (Warenda et al. 2003).
RECIPES
SD Agar Plates
2% Glucose
6.7 g/L yeast nitrogen base without amino acids
-
Amino acids
20 mg/L histidine
120 mg/L leucine
60 mg/L lysine
20 mg/L arginine
20 mg/L tryptophan
20 mg/L tyrosine
40 mg/L threonine
20 mg/L methionine
50 mg/L phenylalanine
20 mg/L uracil
20 mg/L adenine
20 g/L agar
Include any additional amino acids that may be required for the growth of an auxotrophic strain. Prepare in H2O and sterilize by autoclaving. Fill sterile Petri dishes with ~25 mL of autoclaved medium.
SD Liquid Medium
2% glucose
6.7 g/L yeast nitrogen base without amino acids
-
Amino acids:
20 mg/L histidine
120 mg/L leucine
60 mg/L lysine
20 mg/L arginine
20 mg/L tryptophan
20 mg/L tyrosine
40 mg/L threonine
20 mg/L methionine
50 mg/L phenylalanine
20 mg/L uracil
20 mg/L adenine
Include any additional amino acids that may be required for the growth of an auxotrophic strain. Prepare in H2O and sterilize by autoclaving.
YEPD Agar Plates
| Reagent | Quantity |
|---|---|
| Bacto-agar (2%) | 20 g |
| YEPD liquid medium | 1 L |
Add Bacto-agar to YEPD liquid medium in a 2-L flask and autoclave. Fill sterile Petri dishes with 30–40 mL of autoclaved medium.
Yeast Extract-Peptone-Dextrose Growth Medium (YEPD)
| Reagent | Quantity (for 1 L) | Final concentration (w/v) |
|---|---|---|
| Bacto peptone | 20 g | 2% |
| Yeast extract | 10 g | 1% |
| Dextrose | 20 g | 2% |
| H2O | to 1 L |
Sterilize by autoclaving.
Acknowledgments
P.J.C. is supported by a U.S. Public Health Service grant (GM098629).
References
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Granek JA, Magwene PM. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 2010;6:e1000823. doi: 10.1371/journal.pgen.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin ZY, Cox MJ, Vizeacoumar F, Cheung D, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337:1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warenda AJ, Kauffman S, Sherrill TP, Becker JM, Konopka JB. Candida albicans septin mutants are defective for invasive growth and virulence. Infect Immun. 2003;71:4045–4051. doi: 10.1128/IAI.71.7.4045-4051.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan J, Raspor P. Quantitative agar-invasion assay. J Microbiol Methods. 2008;73:100–104. doi: 10.1016/j.mimet.2008.02.009. [DOI] [PubMed] [Google Scholar]