Abstract
Background
Large tumor suppressor 2 (LATS2) gene is a putative tumor suppressor gene with potential roles in regulation of cell proliferation and apoptosis in lung cancer. The aim of this study is to explore the association of aberrant LATS2 expression with EGFR mutation and survival in lung adenocarcinoma (AD), and the effects of LATS2 silencing in both lung AD cell lines.
Methods
LATS2 mRNA and protein expression in resected lung AD were correlated with demographic characteristics, EGFR mutation and survival. LATS2-specific siRNA was transfected into four EGFR wild-type (WT) and three EGFR mutant AD cell lines and the changes in LATS2 expression and relevant signaling molecules before and after LATS2 knockdown were assayed.
Results
Fifty resected lung AD were included (M:F = 23:27, smokers:non-smokers = 19:31, EGFR mutant:wild-type = 21:29) with LATS2 mRNA levels showed no significant difference between gender, age, smoking and pathological stages while LATS2 immunohistochemical staining on an independent set of 79 lung AD showed similar trend. LATS2 mRNA level was found to be a significant independent predictor for survival status (disease-free survival RR = 0.217; p = 0.003; Overall survival RR = 0.238; p = 0.036). siRNA-mediated suppression of LATS2 expression resulted in augmentation of ERK phosphorylation in EGFR wild-type AD cell lines with high basal LATS2 expression, discriminatory modulation of Akt signaling between EGFR wild-type and mutant cells, and induction of p53 accumulation in AD cell lines with low baseline p53 levels.
Conclusions
LATS2 expression level is predictive of survival in patients with resected lung AD. LATS2 may modulate and contribute to tumor growth via different signaling pathways in EGFR mutant and wild-type tumors.
Keywords: Lung cancer, Large tumor suppressor 2 gene, Gene expression, Gene silencing, EGFR signaling
1. Introduction
LATS2 (large tumor suppressor 2), one of the two human homologues of Drosophila warts, is a putative tumor suppressor gene which encodes for a serine/threonine kinase [1]. As a component in the Hippo signaling pathway, LATS2 kinase plays a critical role in controlling organ size development and in coordinating cell proliferation and apoptosis [2,3]. As a putative tumor suppressor gene, LATS2 displays multiple mechanisms of actions in different cancer cell types, including cell cycle regulation by controlling G1/S and G2/M transition [4,5], induction of apoptosis by down-regulating anti-apoptotic proteins Bcl-2 and Bcl-XL [6], as well as maintenance of mitotic fidelity and genetic stability by interacting with other regulators of cell division, such as p53 [7] and Aurora kinases [8–10].
Dysregulation of LATS2 functions has been found in different types of tumors. The chromosomal location of LATS2 is mapped to 13q11–q12 where there is frequent loss of heterozygosity [11]. In breast cancer, LATS2 mRNA expression was down-regulated by promoter hypermethylation and this alteration was associated with large tumor size, high rate of metastasis and estrogen receptor and progesterone receptor negativity [12]. LATS2 may also play a role in the development of prostate cancer based on findings that reduced LATS2 expression occurred in prostate tumors and LATS2 negatively modulated androgen receptor-regulated gene transcription [13]. In malignant mesothelioma (MM), LATS2 was found to be inactivated in MM cells and this inactivation will lead to deregulated cell growth by allowing constitutive activation of the downstream transcription factor of LATS2 in the Hippo pathway, YAP [14,15]. The expression of LATS2 exhibits suppressive effects on mesothelioma cells.
In non-small cell lung cancer (NSCLC), occasional information has been reported on the effects of aberrant expression of LATS2 except its being one potential targets for microRNA-135b action to promote lung cancer metastasis [16]. LATS2 mutations are rare [17,18] but other mechanisms have been reported to cause down-regulation of its expression such as promoter hypermethylation [19] and micro-RNA regulation [20,21]. The relation between reduced LATS2 expression and lung cancer progression, and the underlying mechanisms remain unknown. In addition, we have previously found from expression profiling experiments that LATS2 showed differential expression between pulmonary adenocarcinomas with wild-type EGFR and ones bearing EGFR mutations at exons 18–21 [22]. In this study, we further validated the differential expression of LATS2 in lung adenocarcinoma (AD) tissues in relation to EGFR mutation status, as well as other clinicopathological factors including as smoking history and survival status. Furthermore, we silenced LATS2 expression by siRNA inference in several lung adenocarcinoma cell lines and examined LATS2 knockdown effects on EGFR downstream signaling pathways, Ras/Erk, PI3K/Akt, as well asp53 network.
2. Materials and methods
2.1. Human lung adenocarcinoma tissue
Newly diagnosed of lung adenocarcinoma patients were recruited prospectively before planned surgical resection. The protocol for lung cancer surgical specimen collection was approved by the Institutional Review Board Research Ethics Committee of the University of Hong Kong and Hong Kong Hospital Authority Hong Kong West Cluster. Resected lung adenocarcinoma (AD) tissues from Chinese patients were included in this study. Upon resection, these tissue samples were immediately submerged in RNAlater RNA Stabilization Reagent (Qiagen), frozen in liquid nitrogen, and stored at −80 °C until RNA extraction. An independent set of 79 archival paraffin blocks of lung adenocarcinomas with known patient demographics including age, gender and EGFR mutation status (but not smoking habits) collected successively at the Histopathology Laboratory of the Hong Kong and Sanatorium Hospital was used for immunohistochemical studies. All the tumor tissues used were collected from patients who underwent resection of lung tumors without prior treatment with EGFR tyrosine kinase inhibitors or any other form of anti-cancer treatment.
2.2. Human adenocarcinoma cell lines
Seven AD cell lines were cultured in RPMI 1640 (Gibco, USA) supplemented with 1% Penicillin–Streptomycin (Gibco, USA) and 2.5% or 10% fetal bovine serum (Gibco, USA). AD cell lines used in this study were HKULC-2, HKULC-4 [23], and H1648, H1650, H1975 and H2023 were from John D Minna M.D., University of Texas Southwestern Medical Center at Dallas; while PC9 was from PC Yang M.D., National Taiwan University. Three AD cell lines with EGFR mutations, which were H1975 with double mutations L858R and T790M, PC9 andH1650 bearing EGFR deletions at exon 19.
2.3. Direct sequencing for EGFR mutations
We utilized the historical standard for EGFR mutation testing, directing sequencing [24,25]. In order to enrich tumor cell content, we performed micro-dissection on formalin-fixed paraffin-embedded sections before DNA extraction. Then, exons 18–21 of EGFR were PCR-amplified by applying respective primers. Purified PCR product was analyzed by ABI 3730xl DNA analyzer and sequence data was reviewed by Sequence Scanner Software.
2.4. Real time PCR
Total RNA was extracted from frozen resected lung adenocarcinoma tissues by using the RNeasy kit (Qiagen, UK). After reverse transcription of total RNA via QuantiTect® kit (Qiagen, UK), 150 ng cDNA templates were used to detect LATS2 mRNA expression by using quantitative real time PCR (RT-qPCR) with the SYBR Green I method (Qiagen, UK). The primer sequences for LATS2 mRNA were: Forward, 5′-TGGCACCTACTCCCACAG-3′, and Reverse, 5′-CCAAGGGCTTTCTTCATCT-3′ [26]. The ribosomal 18S gene was chosen as an internal control, and the primer sequences were: Forward, 5′-AGGAATTGACGGAAGGGCAC-3′, and Reverse, 5′-GGACATCTAAGGGCATCACA-3′. Thermal cycle conditions were: 95 °C for 5 min followed by 40 cycles of amplification at 95 °C for 15 s per cycle, 58 °C (LATS2) or 60 °C (18S) for 45 s and 72 °C for 45 s. The dissociation curve analysis was carried out to exclude non-specific primer dimers. The relative expression level was determined as 2−ΔΔCt with relative to a reference sample (BS65.2N-KT, an immortalized normal bronchial epithelial cell line). The logarithmic values to base of two of these relative expression levels were used in the following statistical analysis.
2.5. Immunohistochemistry
Immunohistochemistry (IHC) of the LATS2 protein was performed on formalin-fixed, paraffin-embedded sections of an independent set of 79 paraffin blocks of lung AD different from those used for real-time PCR assay above.
Antigen retrieval was conducted in Tris–EDTA buffer (10 mM Tris, 1 mM EDTA, pH = 9) at 95 °C for 30 min. The goat polyclonal antibody against human LATS2 (dilution 1:200; Santa Cruz, USA) or negative control mouse IgG1 (dilution: 1:200; Dako, Denmark) was incubated with the sections overnight at 4 °C. After treating with the rabbit anti-goat/mouse secondary antibodies (dilution 1:400; Dako, Denmark) respectively at 37 °C for 30 min, the specimens were stained with DAB substrate chromogen system (Dako, Denmark) for 5 min. Scoring of immunohistochemical staining was performed by independent pathologists (Ximing Tang and Ignacio I Wistuba) without knowing the clinical annotations of the specimens. LATS2 expression H-scores based on the extension (0–100) and intensity (0, none; 1+, weak; 2+, moderate; and 3+, strong) of IHC staining was performed by two pathologists (X.T. and I.W.). If there is discrepancy between the two pathologists concerned, an independent third pathologist in the same laboratory who were not aware of the clinical details and previous H-scores by the first two pathologists. A final H score (0–300) was obtained by multiplying the intensity (1–3) and reactivity extension (0–100) for each case. Mean score was taken for the whole group of samples and samples with H scores above mean would be classified as of higher expression levels whereas those with H scores below mean would be classified as of lower expression levels.
2.6. Western blotting
Whole-cell lysates were prepared in 1× RIPA lysis buffer (Rock-land) with addition of 1% protease inhibitor cocktail (Sigma). Total protein (80 μg for Akt and 50 μg for other proteins) was fractionated using SDS-PAGE and thentransferred to nitrocellulose membranes (ExPro). After blocking with 5% non-fat milk in Tris buffered saline, membranes were incubated overnight at 4 °C with various primary antibodies and then probed with HRP-conjugated secondary antibodies (anti-rabbit or anti-mouse, Abcam) for 2 h at 4 °C. Primary antibodies to LATS2, ERK 1/2, p-ERK1/2, Akt, p-Akt (T308), p-Akt (S473) and p53 were from Cell Signaling. β-actin (Sigma) was used as a loading control. Immune complexes were visualized by using ECL detection kit (GE Healthcare, Japan), and the band intensity was quantified by Image J software.
2.7. LATS2 siRNA transfection
Cells were transfected using the Hiperfect reagent according to the manufacturer’s instructions (Qiagen, UK). The target sequence of LATS2 specific siRNA (siLATS2) is 5′-CTCCGCAAAGGGTACACTCAA-3′, and 5 nM siRNA was added to silence LATS2. One negative control siRNA (Qiagen, UK) was also included. Cultured cells were harvested for RNA or protein extraction at baseline and at 48 h.
2.8. Statistical analysis
Data were analyzed with SPSS 18.0 software. Differences between groups were estimated using the Chi-squared test, the Student’s t-test, or the log-rank test. Disease-free survival (DFS) and overall survival (OS) curves were calculated by the Kaplan–Meier method. Stepwise multiple regression models were built to determine the clinical parameters that independently predict either PFS or OS. Log rank tests were used to compare cumulative survival between different groups. All p values were two-sided and p < 0.05 was considered statistically significant. Cox proportional regression model was applied for multivariate analysis. A probability level of 0.05 was used to determine statistical significance.
3. Results
3.1. Demographics of lung cancer patients
Fifty patients were recruited prospectively before they underwent surgical resection, with 23 (46.9%) male and 26 (53.1%) female patients and with age range of 38–88 years (mean ± S.D., 64.1 ± 9.2 years). The ratio of non-smokers (61%) to ex- or current smokers (39%) was around 2 to 1. 28 tumors were EGFR wild-type (57%) with no mutation at exons 18–21 while 21 were EGFR mutants that showed at least one EGFR mutation in exons 18–21 (43%). The details can be found in Table 1.
Table 1.
Patient and tumor characteristics according to LATS2 mRNA expression (A) and LATS2 protein expression (B).
| (A) LATS2 mRNA expression
| |||
|---|---|---|---|
| High LATS2 expression | Low LATS2 expression | P-value | |
| N (%) | 23 (46.9) | 26 (53.1) | |
| Follow-up duration (day), mean ± S.D. | 1276 ± 404 | ||
| Gender, n (%), M:F | 23:26 | 0.907 | |
| Male | 11 (22.4) | 12 (24.5) | |
| Female | 12 (24.5) | 14 (28.6) | |
| Age, mean ± S.D. | 64.1 ± 9.2 | 0.336 | |
| 66.3 ± 7.6 | 62.2 ± 10.2 | ||
| Smoking, n (%), SM:NS | 19:30 | 0.590 | |
| SM | 8 (16.3) | 11 (22.4) | |
| NS | 15 (30.6) | 15 (30.6) | |
| Stage, n (%), I:II:III | 29:12:08 | 0.709 | |
| Stage I | 15 (30.6) | 14 (28.6) | |
| Stage II | 5 (10.2) | 7 (14.3) | |
| Stage III | 3 (6.1) | 5 (10.2) | |
| In groups | 0.706 | ||
| Earlier stage (I and II) | 20 (40.8) | 21 (42.9) | |
| Advanced stage (III) | 3 (6.1) | 5 (10.2) | |
| Relapse, n (%) | 0.015* | ||
| Yes | 7 (14.3) | 17 (34.7) | |
| No | 16 (32.7) | 9 (18.4) | |
| DFS (day), median | 919 | ||
| 1303 | 746 | 0.006* | |
| Death, n (%) | 0.044* | ||
| Yes | 3 (6.1) | 10 (20.4) | |
| No | 20 (40.8) | 16 (32.7) | |
| OS (day), median | 1095 | ||
| 1364 | 794 | 0.059 | |
| EGFR, n (%), Mut:WT | 21:28 | 0.283 | |
| Mutation (Mut) | 8 (16.3) | 13 (26.5) | |
| Wild type (WT) | 15 (30.6) | 13 (26.5) | |
| (B) LATS2 IHC staining
| |||
|---|---|---|---|
| Above mean | Below mean | P-value | |
| N (%) | 32 (50.8) | 31 (49.2) | |
| Gender, n (%), M:F | 36:27 | 0.021* | |
| Male | 18 (36) | 18 (36) | |
| Female | 13 (26) | 14 (28) | |
| EGFR, n (%), Mut:WT | 37:22 | 0.01* | |
| Mutation (Mut) | 18 (36) | 19 (38) | |
| Wild type (WT) | 11 (22) | 11 (22) | |
| (C) EGFR mutation status of samples included in this study
| |||
|---|---|---|---|
| Samples for mRNA measurement | Samples for IHC | ||
| EGFR wild-type | 28 | 22 | |
| Exon 18 | 4 | 3 | |
| Exon 19 | 8 | 10 | |
| Exon 20 | 1 | 1 | |
| EGFR mutation | Exon 21 | 7 | 6 |
| L858R + T790M | 1 | 1 | |
| Unknown | 0 | 27 | |
Statistically significant (p < 0.05).
3.2. LATS2 mRNA expression in lung AD patients
AD tissue samples expressing LATS2 mRNA at levels above the mean expression level (6.3; range 0–13.1) were assigned to the high expression group (mean expression value 9.4, n = 23), and samples with expression less than the mean value were considered as the low expression group (mean expression value 3.6, n = 26).
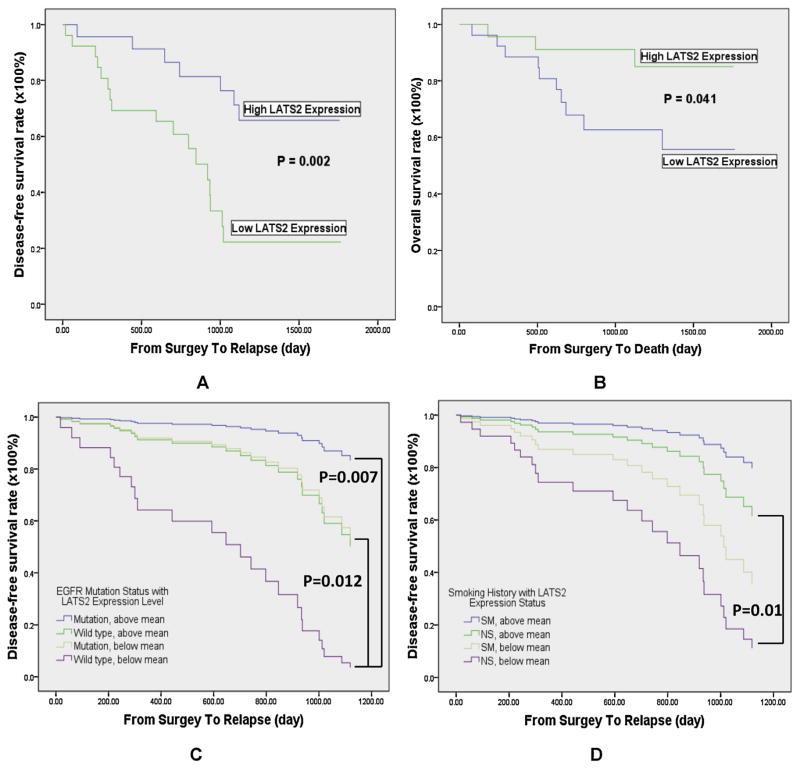
LATS2 mRNA levels showed no significant differences between different clinical parameters, including gender, age, smoking history, pathological stage and EGFR mutation status (Table 1). Survival analysis indicated that high LATS2 expression group had significantly longer disease-free survival (DFS) (p = 0.002, Fig. 1A) and overall survival (OS) (p = 0.041, Fig. 1B) than the low LATS2 expression group. Furthermore, LATS2 mRNA levels correlated with DFS (partial correlation ratio, 0.37; p = 0.012). Multivariate analysis further confirmed that LATS2 mRNA level was a significant prognostic factor for survival status (DFS: hazard ratio, 0.221; p = 0.003; OS: hazard ratio, 0.238; p = 0.036) independent of age, gender, smoking history and staging (Table 2). Meanwhile, pathological stage (hazard ratio, 5.102; p = 0.009), as well as the presence of EGFR mutations (hazard ratio, 0.207; p = 0.006) were also significant prognostic factor of DFS (Table 2). When dividing samples based on both EGFR mutation status and LATS2 level, patients with wild-type EGFR as well as expressing low LATS2 expression displayed
Fig. 1.
Survival analysis of disease-free survival (DFS) and overall survival (OS). (A) Significant difference in disease-free survival between high and low LATS2 expression groups was present. The p-value was estimated by the log-rank test. (B) AD patients with high LATS2 expression had a significantly better overall survival than those expressing low LATS2. (C) Cox regression analysis demonstrated that patients in the wild type EGFR together with low LATS2 expression group exhibited the worst DFS. (D) In non-smoker patients, high LATS2 expression is a significant predictor of better DFS. poor DFS (Fig. 1C). Inferior DFS was also observed in non-smokers exhibiting low LATS2 levels (Fig. 1D).
Table 2.
Multivariate analysis of disease-free survival (DFS) and overall survival (OS) (Cox proportional regression model).
| DFS
|
OS
|
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| Univariate analysis | ||||||
| Gender (female/male) | 1.129 | 0.497–2.566 | 0.772 | 2.364 | 0.769–7.270 | 0.133 |
| Age | 0.951 | 0.905–1.000 | 0.051 | 1.020 | 0.956–1.088 | 0.546 |
| Smoking history (SM/NS) | 1.157 | 0.477–2.807 | 0.747 | 0.561 | 0.187–1.678 | 0.301 |
| Stage (I – II/III) | 3.975 | 1.569–10.071 | 0.004* | 1.040 | 0.229–4.719 | 0.960 |
| EGFR mutation Status (WT/Mut) | 0.577 | 0.247–1.351 | 0.205 | 0.348 | 0.095–1.268 | 0.110 |
| LATS2 expression (low/high) | 0.269 | 0.110–0.662 | 0.004* | 0.281 | 0.077–1.029 | 0.055 |
| Multivariate analysis | ||||||
| Gender (female/male) | 2.431 | 0.654–9.039 | 0.185 | 2.246 | 0.448–11.251 | 0.325 |
| Age | 0.997 | 0.939–1.058 | 0.913 | 1.015 | 0.933–1.103 | 0.729 |
| Smoking history (SM/NS) | 2.119 | 0.564–7.961 | 0.266 | 1.482 | 0.327–6.724 | 0.610 |
| Stage (I – II/III) | 5.102 | 1.512–17.208 | 0.009* | 1.832 | 0.320–10.504 | 0.497 |
| EGFR mutation Status (WT/Mut) | 0.207 | 0.068–0.631 | 0.006* | 0.304 | 0.071–1.306 | 0.109 |
| LATS2 expression (low/high) | 0.221 | 0.081–0.605 | 0.003* | 0.238 | 0.062–0.908 | 0.036* |
HR, hazard ratio; CI, confidence interval; SM, smoker; NS, non-smoker; WT, wild type; Mut, mutant.
Statistically significant.
3.3. LATS2 protein expression in lung AD tumors
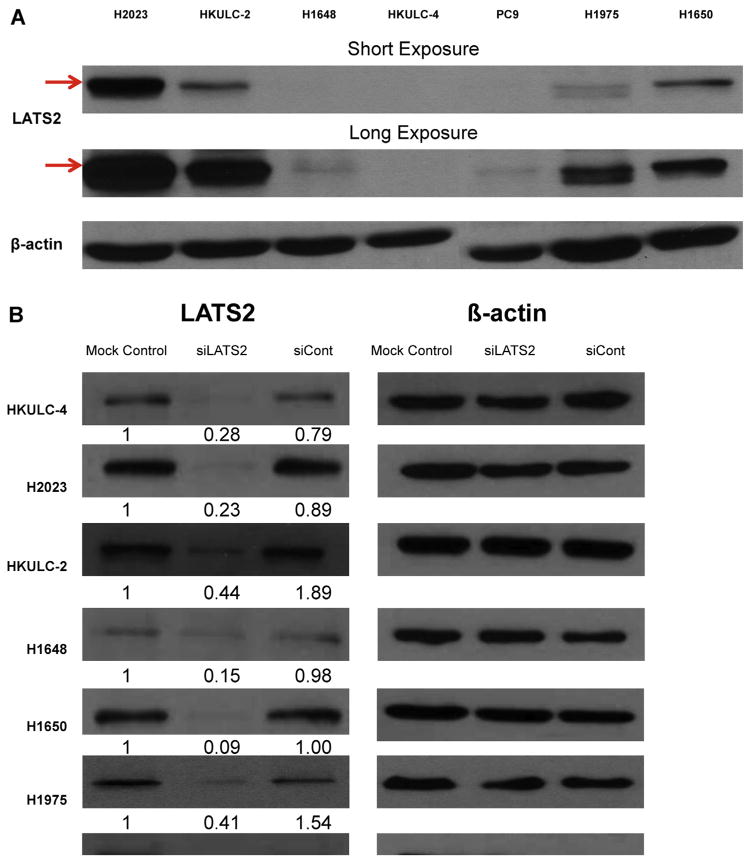
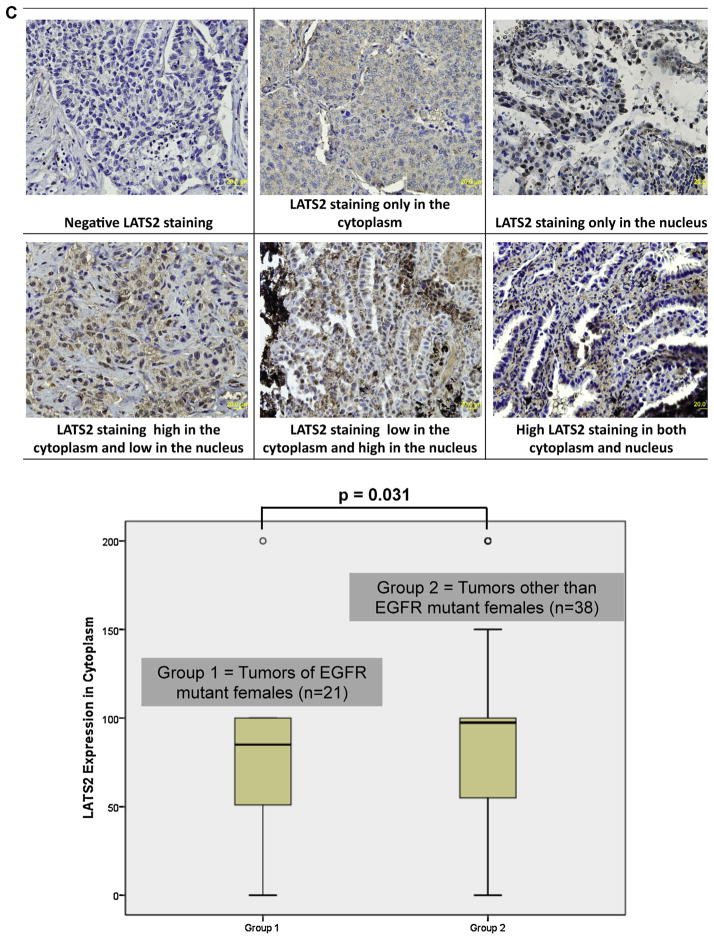
Immunohistochemistry (IHC) staining on the independent set of 79 paraffin blocks of AD indicated that LATS2 protein was expressed at relatively low levels in both the cytoplasm and the nucleus (Fig. 2C). After excluding 16 cases with zero H-score in both compartments, paired test revealed that LATS2 protein expression was significantly higher in the cytoplasm than in the nucleus (p < 0.001). There was an association between LATS2 IHC scores in the cytoplasm and gender with EGFR mutation status, in which female patients with EGFR mutations displayed modestly low levels of LATS2 cytoplasm expression (p = 0.031). In males, EGFR wild-type cohort expressed slightly higher cytoplasmic staining of LATS2. These observations with LATS2 IHC were consistent with the findings of LATS2 expression at mRNA levels among gender with EGFR mutation status groups in a different set of tumor specimens with real-time PCR assay done as described in the previous section. Nuclear LATS2 expression levels were generally low and no remarkable difference was found between all the clinical parameters examined.
Fig. 2.
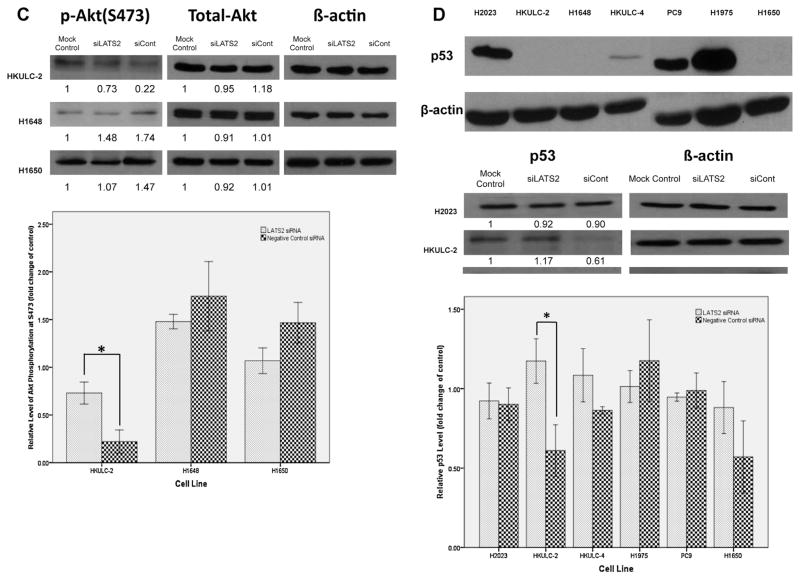
LATS2 protein expression in AD tumors and cell lines. (A) Before transfection, LATS2 protein expression was measured in each cell lines. (B) Cells were transfected with siRNA for LATS2 (siLATS2), Negative Control siRNA (siCont) or none (Mock Control) for 48hr. Number below each blot indicates fold change to mock control (n = 3). (C) Immunohistochemistry staining of LATS2 expression in clinical samples of AD. Female cases bearing EGFR mutations had moderately lower expression of LATS2 in the cytoplasm (mean expression value, 77.7 vs. 92.5, p = 0.031).
3.4. LATS2 mRNA expression in AD cell lines
Before transfection, LATS2 protein levels were quantified in each cell line (Fig. 2A). Basal LATS2 expression in H2023, HKULC-2 and H1650 were much higher, while H1648, HKULC-4, H1975 and PC9 exhibited relatively lower expression levels of LATS2. After being transfected with LATS2-specific siRNA (siLATS2) or Negative Control siRNA (siCont), LATS2 expression was successfully silenced in all cell lines confirmed with reduced levels of LATS2 protein (Fig. 2B).
3.5. LATS2 modulates ERK pathway in the EGFR wild-type AD cell lines with high LATS2 expression
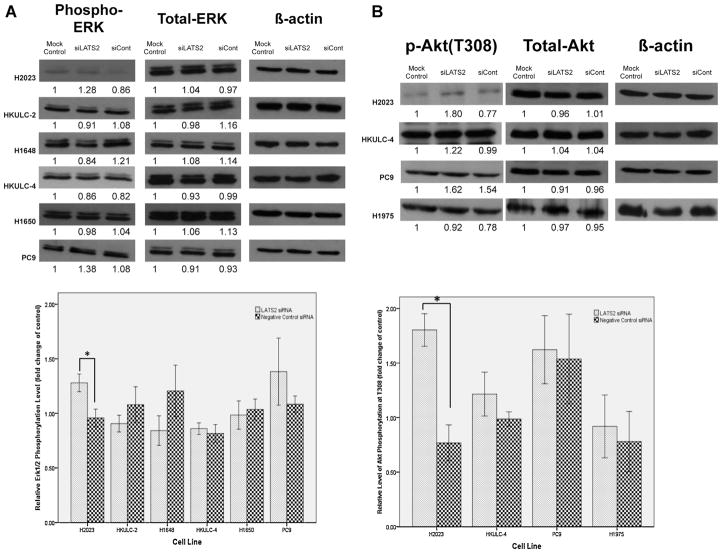
After successfully silencing LATS2 expression in AD cell lines, we first examined for changes in phosphorylation of mitogen-activated protein kinase, ERK1/2 (Fig. 3A). In most of the AD cell lines studied except for H1975 in which we failed to detect the activation of ERK1/2, the levels of phosphorylated ERK1/2 did not alter significantly with LATS2 silencing. Nevertheless, a modest increase in the phosphorylation status of ERK1/2 was observed in H2023.
Fig. 3.
The effects of LATS2 silencing on ERK activation, Aktphosphorylation and p53 level. Cells were transfected with siRNA for LATS2 (siLATS2), Negative Control siRNA (siCont) or none (Mock Control) for 48 h. Cellular lysates were analyzed by Western blotting with the indicated antibodies. Number below each blot indicates fold change to mock control (n = 3). One asterisk indicates p < 0.05 determined by Student’s t-test. (A) Changes in phosphorylated ERK levels in cell lines after 48-h transfection with siRNAs described above. (B) Analysis of Thr308-phosphorylated Akt levels in cells after transfected with siRNAs described above. (C) Levels of Ser473-phosphorylated Akt in HKULC-2, H1648 and H1650 cells with or without LATS2 knockdown. D) The top left picture showed the basal levels of p53 in different cell lines. Modulation of p53 levels in these cells after transfected with siRNAs described above was demonstrated in the lower left picture and in the graph.
3.6. LATS2 knockdown differentially influences Akt activation in different lung AD cell lines
As another EGFR-activated signaling cascade, Akt pathway could also be modulated by LATS2 kinase. siRNA silencing of LATS2 increased levels of Thr308-phosphorylated Akt in H2023 (Fig. 3B). Since phosphorylation of Akt at Thr308 could enhance Akt activity, relatively high basal LATS2 expression in this cell lines would inhibit Akt activation. Similar to the situation of ERK activity, LATS2 knockdown did not affect Akt phosphorylation in neither HKULC-4 which expressed very low level of LATS2 nor cell lines, PC9 and H1975, with EGFR mutations.
Phosphorylation at Ser473, which contributes to maximal Akt Activity, was also improved by LATS2 knockdown in HKULC-2 cells (Fig. 3C). Together with above results, LATS2 might diminish Akt activation in H2023 and HKULC-2cells. Both cell lines showed high LATS2 expression at baseline. In H1650, the addition of LATS2 siRNA slightly reduced the level of phosphorylated Aktat Ser473. Although the difference was not significant, it was still plausible that LATS2 may differentially influence Akt activation and the regulation might be independent of EGFR mutation status.
3.7. Silencing of LATS2 induces p53 accumulation in lung AD cell lines with low basal p53 levels
Previous reports have indicated that LATS2 is able to stabilize p53 thus facilitating p53-dependent checkpoint response to mitotic stress in breast cancer and osteosarcoma cells. However, in HKULC-2 cells, LATS2 knockdown markedly increased total p53 levels (Fig. 3D), suggesting that, instead of inducing p53 accumulation, LATS2 may actually downregulatep53 protein expression in these cells. Noticeably, HKULC-2 cells expressed very low amount of p53 at baseline (Fig. 3D). Another two cell lines, HKULC-4 and H1650, which also exhibited low basal p53 expression, LATS2 silencing likewise enhanced p53 levels although the differences were not statistically significant.
4. Discussion
In this study, we demonstrated that low LATS2 expression was an independent and significant predictor of poor overall survival. Furthermore, expression of LATS2 correlated with DFS of these lung adenocarcinoma patients (partial correlation ratio, 0.37; p = 0.012). These findings suggested that LATS2 may express tumor suppressive effects in lung adenocarcinoma.
In this study, patients with tumors harboring EGFR mutations exhibited longer median DFS than those with wild-type EGFR (median DFS: 33.4 vs. 27.2 months, respectively; p = 0.006). None of the recruited patients received TKI treatment. Other than complete resection for most of these early stage tumors, no treatment intervention was apparent. The influence of EGFR gene mutation on prognosis as shown in this study deserves further investigation. This unusual observation may stem from differences in ethnicity and pathological stage of recruited patients, as well as different detection methods of EGFR mutations, compared to reported data [27–29,30–32]. Since wild type EGFR also significantly predicted inferior DFS, it is not surprising that tumors with wild type EGFR and low LATS2 expression exhibited the shortest DFS (overall p = 0.004, Fig. 1C). In non-smokers, in which EGFR mutations are more common, significant difference in DFS was still observed between two LATS2 expression groups (p = 0.010, Fig. 1D). Similar observation has been made before, that knockdown of YAP (Yes kinase-associated protein), whose functions can be inhibited by LATS2, sensitizes cancer cells to EGFR-TKI erlotinib [33]. Further studies are warranted to investigate the relationships between LATS2 expression levels and therapeutic responses of these patients.
Immunohistochemistry suggested that AD tumors expressed LATS2 in both cytoplasm and nucleus. During interphase, LATS2 mainly remained in the cytoplasm, especially localized to the centrosome [8]. After phosphorylated by Aurora A kinase during mitosis, LATS2 could translocate to the chromosome and the central spindle, thus modulating chromosome segregation and cytokinesis [9,10,34]. To date, the best characterized function of LATS2 is the regulation of YAP/TAZ proto-proteins via canonical Hippo signaling mainly in breast, colon and hepatic cancers as well as in malignant pleural mesothelioma [15,35,36] but not lung adenocarcinomas. LATS2 might still plays critical roles in non-canonical Hippo signaling and even in Hippo independent pathways, such as the G-protein-coupled receptor (GPCR) signaling [37,38] and the K-ras/Raf-1 axis [39,40]. The exact roles, as well as the relationship between the subcellular localization and the biological functions of LATS2, in lung adenocarcinomas need further exploration.
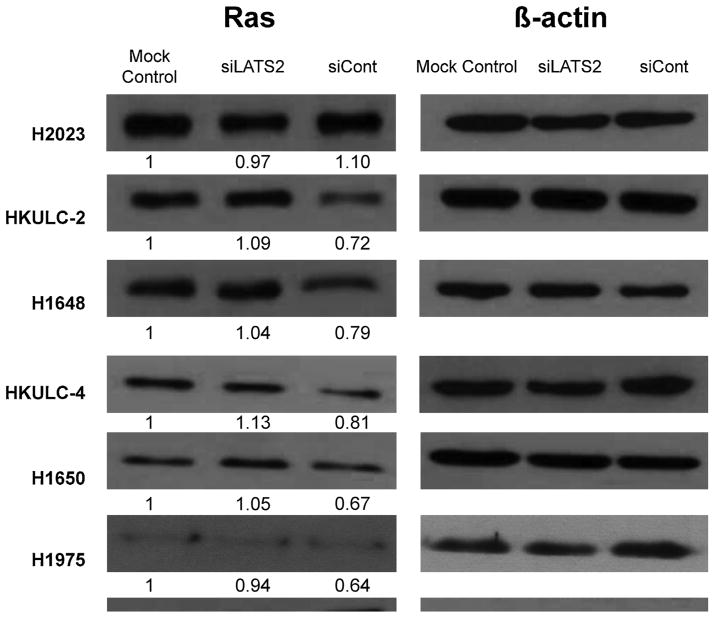
Our data suggested that LATS2 can suppress ERK signaling in EGFR wild-type lung adenocarcinoma cell lines with high basal LATS2 expression. Another study conducted in HeLa cells identified three down-regulated genes in siRNA-LATS1/2-treated cells, namely SPRED1, SPRY2 and SPRY4 [41]. These three proteins act as negative regulators of the pathway [42–45], indicating that LATS2 might inhibit ERK activation through regulating some members of the Sprouty proteins. Additionally, as an upstream activator of ERK1/2, any change in Ras protein levels after silencing of LATS2 may contribute to observed alterations in ERK1/2 activities. However, in siLATS2-treated H2023, no detectable change in Ras protein expression was observed (Fig. 4). This implies that LATS2 regulate ERK phosphorylation via other mechanisms in H2023. It is mentionable that, in most of lung AD cell lines studied LATS2 knockdown induced Ras protein expression (Fig. 4). The consequence of this modulated Ras expression by LATS2 needs further investigation.
Fig. 4.
Modulation of Ras expression by LATS2. (A) Cells were transfected with siRNA for LATS2 (siLATS2), Negative Control siRNA (siCont) or none (Mock Control) for 48 h. Cellular lysates were analyzed by Western blotting with the indicated antibodies. Number below each blot indicates fold change to mock control (n = 3).
The regulation of LATS2 on Akt signaling was more complicated and independent of EGFR mutation status. In two EGFR wild-type cell lines (H2023 and HKULC-2), LATS2 could reduce Akt activation by inhibiting Akt phosphorylation at either Thr308 or Ser473. This alternation may probably result from the ability of LATS2 to maintain the protein phosphatase-2A catalytic subunit (PP2A-C) level [39] which in turn leads to dephosphorylation of Akt Thr308 [46]. On the other hand, the interaction between LATS2 and Akt may be reciprocal and linked by mammalian sterile 20-like kinase-1 (Mst1). Apart from an upstream activator of LATS2, Mst1 has been reported to function as an inhibitor of Akt [47], and, meanwhile, Akt signaling leads to phosphorylation of Mst1 at Thr120 and limits Mst1-mediated tumor suppressive functions [48,49]. The role of LATS2 in this network deserves further study. However, LATS2 may also positively modulate Akt activity since silencing of LATS2 in H1650 triggered a mild decrease in the levels of phosphorylated Akt. As H1650 exhibits loss of PTEN gene [50] and constitutive activation of Akt, downregulation of LATS2in this cell line, which may in turn inhibit Akt activity, is possible to compensate this abnormal signaling transduction.
In this study, LATS2 may suppress p53 expression, which is contradictory to previous studies that LATS2 can cause p53 induction in response to mitotic apparatus damage [7] as well as oncogenic activation [51]. That may be partially explained by the absence of mitotic or oncogenic stress introduced to cells in this study. On the other hand, in NSCLC tumors, downregulated LATS2 mRNA levels have been found to be highly correlated with lower expression of Mdm2 which leads to p53 ubiquitination and degradation [17,18], implying that diminished LATS2 expression might positively regulate p53 protein level, as seen in HKULC-2, HKULC-4 and H1650. All of three cell lines express very low amount of p53 at baseline, while two of them, HKULC-2 and H1650, show relatively high basal LATS2 levels. So it is reasonable to deduce that the low basal p53 expression may partially result from the corresponding high basal expression of LATS2. Additionally, based on two studies that Snail has been found to suppress p53 in A549 lung carcinoma cell line [52] and LATS2 can act as a positive regulator of Snail1 protein [53], we could speculate that LATS2 may negatively mediate p53 activity through Snail. Further research can include these two candidates, MDM2 and Snail, to disclose the comprehensive interaction between LATS2 and p53 in lung adenocarcinoma cells. Moreover, since silencing of LATS2 would induce p53 accumulation, we might postulate that LATS2 is linked to drug sensitivity through regulation of p53-mediated processes [54].
In conclusion, relatively higher level of LATS2 gene correlates with the better survival in patients with stage resected lung AD. In vitroLATS2 knockdown suggested that this differential expression could impact EGFR-activated signaling network, particularly in high LATS2 expression group, and p53 pathway. LATS2 also appeared to modulate different signaling pathways via phosphorylated ERK or Akt in EGFR wild-type and EGFR mutant lung AD cell lines respectively. Thus, LATS2 gene expression in lung AD patients warrant further investigations for being a potential biomarker for survival status in lung cancer patients of both early and advanced stages as well as the role of LATS2 expression in modulating different signaling pathways in either EGFR mutant or EGFR wild-type lung cancer.
Acknowledgments
Funding
This study was supported in part by the Hong Kong SK Yee Medical Grant 2012 (No. 210210) and the University of Hong Kong Seed Funding 2011.
We would like to thank Professor PC Yang, National Taiwan University, for providing the lung cancer cell line (PC9) used in this study.
Footnotes
Conflict of interest statement
All the authors in this manuscript declare no conflict of interest that could inappropriately influence or bias this piece of work.
References
- 1.Hori T, Takaori-Kondo A, Kamikubo Y, Uchiyama T. Molecular cloning of a novel human protein kinase, kpm, that is homologous to warts/lats, a Drosophila tumor suppressor. Oncogene. 2000;19:3101–9. doi: 10.1038/sj.onc.1203659. [DOI] [PubMed] [Google Scholar]
- 2.Matsui Y, Nakano N, Shao D, Gao S, Luo W, Hong C, et al. Lats2 is a negative regulator of myocyte size in the heart. Circ Res. 2008;103:1309–18. doi: 10.1161/CIRCRESAHA.108.180042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–97. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 4.Kamikubo Y, Takaori-Kondo A, Uchiyama T, Hori T. Inhibition of cell growth by conditional expression of kpm, a human homologue of Drosophila warts/lats tumor suppressor. J Biol Chem. 2003;278:17609–14. doi: 10.1074/jbc.M211974200. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Pei J, Xia H, Ke H, Wang H, Tao W, et al. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene. 2003;22:4398–405. doi: 10.1038/sj.onc.1206603. [DOI] [PubMed] [Google Scholar]
- 6.Ke H, Pei J, Ni Z, Xia H, Qi H, Woods T, et al. Putative tumor suppressor Lats2 induces apoptosis through downregulation of Bcl-2 and Bcl-x(L) Exp Cell Res. 2004;298:329–38. doi: 10.1016/j.yexcr.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 7.Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M, et al. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toji S, Yabuta N, Hosomi T, Nishihara S, Kobayashi T, Suzuki S, et al. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells. 2004;9:383–97. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- 9.Yabuta N, Mukai S, Okada N, Aylon Y, Nojima H. The tumor suppressor Lats2 is pivotal in Aurora A and Aurora B signaling during mitosis. Cell Cycle. 2011;10:2724–36. doi: 10.4161/cc.10.16.16873. [DOI] [PubMed] [Google Scholar]
- 10.Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem. 2007;282:19259–71. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- 11.Yabuta N, Fujii T, Copeland NG, Gilbert DJ, Jenkins NA, Nishiguchi H, et al. Structure, expression, and chromosome mapping of LATS2, a mammalian homologue of the Drosophila tumor suppressor gene lats/warts. Genomics. 2000;63:263–70. doi: 10.1006/geno.1999.6065. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, et al. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–5. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- 13.Powzaniuk M, McElwee-Witmer S, Vogel RL, Hayami T, Rutledge SJ, Chen F, et al. The LATS2/KPM tumor suppressor is a negative regulator of the androgen receptor. Mol Endocrinol. 2004;18:2011–23. doi: 10.1210/me.2004-0065. [DOI] [PubMed] [Google Scholar]
- 14.Mizuno T, Murakami H, Fujii M, Ishiguro F, Tanaka I, Kondo Y, et al. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31:5117–22. doi: 10.1038/onc.2012.5. [DOI] [PubMed] [Google Scholar]
- 15.Murakami H, Mizuno T, Taniguchi T, Fujii M, Ishiguro F, Fukhi T, et al. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 2011;71:873–83. doi: 10.1158/0008-5472.CAN-10-2164. [DOI] [PubMed] [Google Scholar]
- 16.Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun. 2013;4:1877. doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 17.Strazisar M, Mlakar V, Glavac D. LATS2 tumour specific mutations and down-regulation of the gene in non-small cell carcinoma. Lung Cancer. 2009;64:257–62. doi: 10.1016/j.lungcan.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Strazisar M, Mlakar V, Glavac D. The expression of COX-2, hTERT, MDM2, LATS2 and S100A2 in different types of non-small cell lung cancer (NSCLC) Cell Mol Biol Lett. 2009;14:442–56. doi: 10.2478/s11658-009-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki H, Hikosaka Y, Kawano O, Yano M, Fujii Y. Hypermethylation of the large tumor suppressor genes in Japanese lung cancer. Oncol Lett. 2010;1:303–7. doi: 10.3892/ol_00000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai JH, She TF, Juang YM, Tsay YG, Huang AH, Yu SL, et al. Comparative proteomic profiling of human lung adenocarcinoma cells (CL 1–0) expressing miR-372. Electrophoresis. 2012;33:675–88. doi: 10.1002/elps.201100329. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 2010;120:1298–309. doi: 10.1172/JCI39566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam DC, Girard L, Beer DG, Gerald WL, Kris M, Sihoe AD, et al. Expression of LATS2 and STK17B distinguishes epidermal growth factor receptor (EGFR) gene mutations in exons 19 and 21 from those in 18 and 20 in lung adenocarcinomas. J Thorac Oncol. 2011;6:S734. [Google Scholar]
- 23.Lam DC, Girard L, Suen WS, Chung LP, Tin VP, Lam WK, et al. Establishment and expression profiling of new lung cancer cell lines from Chinese smokers and lifetime never-smokers. J Thorac Oncol. 2006;1:932–42. [PubMed] [Google Scholar]
- 24.Ellison G, Zhu G, Moulis A, Dearden S, Speake G, McCormak R, et al. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol. 2013;66:79–89. doi: 10.1136/jclinpath-2012-201194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma ES, Ng WK, Wong CL. EGFR gene mutation study in cytology specimens. Acta Cytol. 2012;56:661–8. doi: 10.1159/000343606. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Hu CF, Chen J, Yan LX, Zeng YX, Shao JY, et al. LATS2 is demethylated and overexpressed in nasopharyngeal carcinoma and predicts poor prognosis. BMC Cancer. 2010;10:538. doi: 10.1186/1471-2407-10-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YT, Seong YW, Jung YJ, Jeon YK, Park IK, Kang CH, et al. The presence of mutations in epidermal growth factor receptor gene is not a prognostic factor for long-term outcome after surgical resection of non-small-cell lung cancer. J Thorac Oncol. 2013;8:171–8. doi: 10.1097/JTO.0b013e318277a3bb. [DOI] [PubMed] [Google Scholar]
- 28.Liu WS, Zhao LJ, Pang QS, Yuan ZY, Li B, Wang P, et al. Prognostic value of epidermal growth factor receptor mutations in resected lung adenocarcinomas. Med Oncol. 2014;31:771. doi: 10.1007/s12032-013-0771-9. [DOI] [PubMed] [Google Scholar]
- 29.Kosaka T, Yatabe Y, Onozato R, Kuwano H, Mitsudomi T. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol. 2009;4:22–9. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- 30.D’Angelo SP, Janjigian YY, Ahye N, Riely GJ, Chaft JE, Sima CS, et al. Distinct clinical course of EGFR-mutant resected lung cancers: results of testing of 1118 surgical specimens and effects of adjuvant gefitinib and erlotinib. J Thorac Oncol. 2012;7:1815–22. doi: 10.1097/JTO.0b013e31826bb7b2. [DOI] [PubMed] [Google Scholar]
- 31.Izar B, Sequist L, Lee M, Muzikansky A, Heist R, Iafrate J, et al. The impact of EGFR mutation status on outcomes in patients with resected stage I non-small cell lung cancers. Ann Thorac Surg. 2013;96:962–8. doi: 10.1016/j.athoracsur.2013.05.091. [DOI] [PubMed] [Google Scholar]
- 32.Lee YJ, Park IK, Park MS, Choi HJ, Cho BC, Chung KY, et al. Activating mutations within the EGFR kinase domain: a molecular predictor of disease-free survival in resected pulmonary adenocarcinoma. J Cancer Res Clin Oncol. 2009;135:1647–54. doi: 10.1007/s00432-009-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang JM, Nagatomo I, Suzuki E, Mizuno T, Kumagai T, Berezov A, et al. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene. 2012;32:2220–9. doi: 10.1038/onc.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Iyer J, Chowdhury A, Ji M, Xiao L, Yang S, et al. KIBRA regulates aurora kinase activity and is required for precise chromosome alignment during mitosis. J Biol Chem. 2012;287:34069–77. doi: 10.1074/jbc.M112.385518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–57. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 36.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–71. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–43. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–91. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilili GK, Kyriakis JM. Mammalian Ste20-like kinase (Mst2) indirectly supports Raf-1/ERK pathway activity via maintenance of protein phosphatase-2A catalytic subunit levels and consequent suppression of inhibitory Raf-1 phosphorylation. J Biol Chem. 2010;285:15076–87. doi: 10.1074/jbc.M109.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matallanas D, Romano D, Al-Mulla F, O’Neill E, Al-Ali W, Crespo P, et al. Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Mol Cell. 2011;44:893–906. doi: 10.1016/j.molcel.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Visser S, Yang X. Identification of LATS transcriptional targets in HeLa cells using whole human genome oligonucleotide microarray. Gene. 2010;449:22–9. doi: 10.1016/j.gene.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Bundschu K, Walter U, Schuh K. Getting a first clue about SPRED functions. Bioessays. 2007;29:897–907. doi: 10.1002/bies.20632. [DOI] [PubMed] [Google Scholar]
- 43.Lo TL, Fong CW, Yusoff P, McKie AB, Chua MS, Leung HY, et al. Sprouty and cancer: the first terms report. Cancer Lett. 2006;242:141–50. doi: 10.1016/j.canlet.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 44.Sutterluty H, Mayer CE, Setinek U, Attems J, Ovtcharov S, Mikula M, et al. Down-regulation of Sprouty2 in non-small cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and -independent mechanisms. Mol Cancer Res. 2007;5:509–20. doi: 10.1158/1541-7786.MCR-06-0273. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Dominguez CA, Martinez N, Gragera T, Perez-Rodriguez A, Retana D, Leon G, et al. Sprouty2 and Spred1-2 proteins inhibit the activation of the ERK pathway elicited by cyclopentenone prostanoids. PLoS ONE. 2011;6:e16787. doi: 10.1371/journal.pone.0016787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andjelkovic M, Jakubowicz T, Cron P, Ming XF, Han JW, Hemmings BA, et al. Activation and phosphorylation of a pleckstrin homology domain containing protein kinase (RAC-PK/PKB) promoted by serum and protein phosphatase inhibitors. Proc Natl Acad Sci USA. 1996;93:5699–704. doi: 10.1073/pnas.93.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cinar B, Fang PK, Lutchman M, Di Vizio D, Adam RM, Pavlova N, et al. The pro-apoptotic kinase Mst1 and its caspase cleavage products are direct inhibitors of Akt1. EMBO J. 2007;26:4523–34. doi: 10.1038/sj.emboj.7601872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collak FK, Yagiz K, Luthringer DJ, Erkaya B, Cinar B. Threonine-120 phosphorylation regulated by phosphoinositide-3-kinase/Akt and mammalian target of rapamycin pathway signaling limits the antitumor activity of mammalian sterile 20-like kinase 1. J Biol Chem. 2012;287:23698–709. doi: 10.1074/jbc.M112.358713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan Z, Kim D, Shu S, Wu J, Guo J, Xiao L, et al. Phosphoinositide 3-kinase/Akt inhibits MST1-mediated proapoptotic signaling through phosphorylation of threonine 120. J Biol Chem. 2010;285:3815–24. doi: 10.1074/jbc.M109.059675. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Chen G, Kronenberger P, Teugels E, Umelo IA, De Greve J. Targeting the epidermal growth factor receptor in non-small cell lung cancer cells: the effect of combining RNA interference with tyrosine kinase inhibitors or cetuximab. BMC Med. 2012;10:28. doi: 10.1186/1741-7015-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aylon Y, Yabuta N, Besserglick H, Buganim Y, Rotter V, Nojima H, et al. Silencing of the Lats2 tumor suppressor overrides a p53-dependent oncogenic stress checkpoint and enables mutant H-Ras-driven cell transformation. Oncogene. 2009;28:4469–79. doi: 10.1038/onc.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SH, Lee SJ, Jung YS, Xu Y, Kang HS, Ha NC, et al. Blocking of p53-Snail binding, promoted by oncogenic K-Ras, recovers p53 expression and function. Neoplasia. 2009;11:22–31. doi: 10.1593/neo.81006. 6p following 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K, Rodriguez-Aznar E, Yabuta N, Owen RJ, Mingot JM, Nojima H, et al. Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. EMBO J. 2012;31:29–43. doi: 10.1038/emboj.2011.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawahara M, Hori T, Chonabayashi K, Oka T, Sudol M, Uchiyama T, et al. Kpm/Lats2 is linked to chemosensitivity of leukemic cells through the stabilization of p73. Blood. 2008;112:3856–66. doi: 10.1182/blood-2007-09-111773. [DOI] [PubMed] [Google Scholar]