Summary
We describe here the existence of a heregulin-HER3 autocrine loop, and the contribution of heregulin-dependent, HER2-mediated HER3 activation to gefitinib insensitivity in non-small cell lung cancer (NSCLC). ADAM17 protein, a major ErbB ligand sheddase, is upregulated in NSCLC and is required not only for heregulin-dependent HER3 signaling, but also for EGFR ligand-dependent signaling in NSCLC cell lines. A selective ADAM inhibitor, INCB3619, prevents the processing and activation of multiple ErbB ligands, including heregulin. In addition, INCB3619 inhibits gefitinib-resistant HER3 signaling and enhances gefitinib inhibition of EGFR signaling in NSCLC. These results show that ADAM inhibition affects multiple ErbB pathways in NSCLC and thus offers an excellent opportunity for pharmacological intervention, either alone or in combination with other drugs.
Introduction
The ErbB family of receptor tyrosine kinases and their ligands are important regulators of tumor cell proliferation, angiogenesis, and metastasis (Gschwind et al., 2004; Yarden and Sliwkowski, 2001). There are four receptors in the ErbB family: EGFR (HER1 or ErbB1), HER2 (neu or ErbB2), HER3 (ErbB3), and HER4 (Erb4). Among them, EGFR, HER2, and HER4 have tyrosine kinase activities; HER3 has a truncated kinase domain, which is not functional, and thus can signal only in the context of receptor heterodimerization (Guy et al., 1994). Eleven ligands have been reported to bind to the ErbB receptor family, including epidermal growth factor (EGF), transforming growth factor α (TGFα), heparin binding EGF-like ligand (HB-EGF), amphiregulin (AR), betacellulin (BTC), epiregulin (EPR), epigen (EPG), and heregulin (HRG)/neuregulin (NRG) family members (Harris et al., 2003). These ligands bind directly to EGFR, HER3, or HER4, leading to the formation of hetero- or homodimers that trigger multiple downstream signaling cascades, including Ras-ERK and PI3K-Akt pathways (Yarden and Sliwkowski, 2001). Although HER2 lacks a functional ligand binding domain, it is the preferred partner for heterodimerization upon ligand binding (Graus-Porta et al., 1997; Mosesson and Yarden, 2004).
While multiple ligands, including EGF, TGFα, HB-EGF, AR, BTC, EPR, and EPG, can bind EGFR, heregulin is the only known ligand for HER3. Heterodimerization of HER3 with HER2 results in the formation of the most oncogenically active ErbB receptor complex upon heregulin stimulation; coexpression of HER3 and HER2, but not HER3 and EGFR, synergizes to transform NIH3T3 cells (Alimandi et al., 1995; Holbro et al., 2003). In addition, treatment of transformed cell lines with anti-HER3 antibodies has been shown to reduce their proliferation and migration in vitro, in part through the alteration of HER2-HER3 dimerization (van der Horst et al., 2005), suggesting that this pathway is important for promoting tumorigenesis. However, the properties and clinical significance of a putative autocrine mechanism involving heregulin, HER3, and HER3’s heterodimerization partners have not been well characterized.
ErbB ligands are structurally and functionally related membrane proteins that can be proteolytically cleaved and released from cells for signaling through autocrine and paracrine mechanisms (Normanno et al., 2001; Borrell-Pages et al., 2003). This cleavage event is critical for the activation of the ligands under a variety of circumstances and is thought to be mediated by ADAMs (a disintegrin and metalloproteases) (Blobel, 2005; Gee and Knowlden, 2003; Sahin et al., 2004; Zhou et al., 2005), which are zinc-dependent membrane-associated metalloproteases. More specifically, ADAM17-deficient cells have been shown to be defective in the shedding of TGFα, HB-EGF, EPR, AR, and heregulin (Merlos-Suarez et al., 2001; Montero et al., 2000; Peschon et al., 1998; Sahin et al., 2004; Sunnarborg et al., 2002; Horiuchi et al., 2005). Although ADAM17 has been suggested to be the major ErbB ligand sheddase, ADAM10 is potentially the main sheddase of EGF and BTC in mouse embryonic fibroblasts (Sahin et al., 2004) and possibly other ligands under specific circumstances. It is apparent that these ADAMs regulate the availability of ErbB ligands by liberating the soluble functional forms and, as a result, activate ErbB signaling.
Several anti-EGFR agents, including antibodies and small molecule kinase inhibitors, have been approved recently to treat various human cancers (Baselga and Arteaga, 2005; Herbst et al., 2004). The chimeric monoclonal antibody cetuximab (Erbitux), which blocks EGFR ligand binding, has been shown to improve the survival of patients with metastatic colon cancer and has shown efficacy in patients with non-small cell lung cancer (NSCLC) (Baselga and Arteaga, 2005), clinically validating the approach of targeting EGFR ligands to treat these cancers. The EGFR tyrosine kinase inhibitors gefitinib (Iressa) and erlotinib (Tarceva) have also shown efficacy in patients with NSCLC (Herbst et al., 2004), particularly in a subgroup of patients that have mutations in the EGFR kinase domain that are more susceptible to inhibition by these molecules (Lynch et al., 2004; Paez et al., 2004; Pao et al., 2004). Unlike other growth factor receptors, many of these mutant EGFR proteins are not constitutively active and are still sensitive to ligand stimulation (Lynch et al., 2004; Riemenschneider et al., 2005).
Despite the efficacy of the antibodies and kinase inhibitors targeting the EGFR pathway, the majority of patients do not experience long-term benefit from these therapies. Data from preclinical models suggest that activation of alternative ErbB pathways can bypass specific blockade and that combination therapies are more effective than any single agent against the EGFR pathway (Huang et al., 2004; Matar et al., 2004; Normanno et al., 2002). Alternative strategies targeting the ErbB pathway are still needed. A potent ADAM inhibitor would fulfill this role by controlling the availability of multiple ErbB ligands and could potentially be used in combination with various anti-ErbB agents. It should be noted that many of the existing broad-spectrum metalloprotease inhibitors, such as marimastat, inhibit not only MMPs (matrix metalloproteinases) but also ADAMs in vitro (Roghani et al., 1999) and thereby prevent the processing of multiple ErbB ligands (Dong et al., 1999; O-Charoenrat et al., 2002). However, their clinical development has been hampered by side effects, many of which are likely due to MMP inhibition (Coussens et al., 2002; Zhou et al., 2005). In addition, it remains to be tested whether a selective ADAM inhibitor can also inhibit multiple ErbB ligand processing and signaling.
Since the importance of the ErbB pathway and the molecular understanding of its sensitivity to EGFR antagonists are best documented in NSCLC, and NSCLC is the leading cause of death from cancer in both men and women in the United States (Jemal et al., 2005), we focused our current study in this disease setting and tried to address the following questions. Does the activation of alternative ErbB signaling pathways, specifically those involving heregulin and HER3, lead to resistance to anti-EGFR agents such as gefitinib in NSCLC? Which ADAM should we target to inhibit the HER3 pathway and overcome heregulin-dependent gefitinib resistance? Can we find an ADAM inhibitor that is more selective than marimastat but still capable of inhibiting cleavage of multiple ErbB ligands? Can the selective ADAM inhibitor be used in combination with gefitinib and chemotherapeutic agents in NSCLC?
Results
A HER3 autocrine stimulatory loop in NSCLC
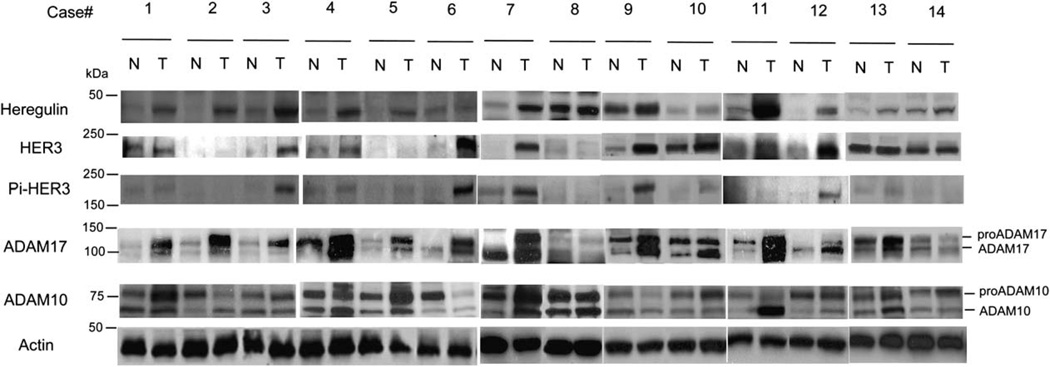
To test our hypothesis that the heregulin-HER3 autocrine loop may be involved in human lung tumorigenesis, we examined the expression of heregulin and HER3 in NSCLC tumors using antibodies against heregulin and HER3. Fourteen fresh NSCLC tumors, of which seven were squamous cell carcinomas (cases 1, 2, 4, 5, 7, 9, and 10) and seven were adenocarcinomas (cases 3, 6, 8, 11, 12, 13, and 14), and their autologous-matched nonmalignant lung tissues were obtained from patients undergoing tumor resection as part of their treatment. As shown in Figure 1, nine out of fourteen freshly resected NSCLC tissues presented with increased expression of heregulin compared with nonmalignant lung tissue control, while seven out of fourteen had increased expression of HER3. To show directly that the HER3 autocrine stimulatory loop was active in NSCLC, we used a phosphospecific antibody against Tyr-1289 of HER3, which is phosphorylated upon heregulin binding and is in a YXXM motif that participates in direct signaling to PI3 kinase (Kim et al., 1994). As shown in Figure 1, at least five out of fourteen freshly resected NSCLC tissues had activated HER3, and all of them coexpressed both HER3 and heregulin, suggesting the existence of a HER3 autocrine stimulatory loop in NSCLC.
Figure 1.
Protein expression of heregulin, HER3, ADAM17, and ADAM10, and activation phosphorylation of HER3 in NSCLC tumor samples
Whole-cell extracts were prepared using tumor tissue (T) and adjacent nonmalignant lung tissue (N) from NSCLC patients (cases 1 to 14) and subjected to SDS-PAGE and Western blot with various antibodies. β-Actin expression was used as a loading control.
Heregulin and HER2-mediated HER3 activation correlate with insensitivity to gefitinib among NSCLC cell lines
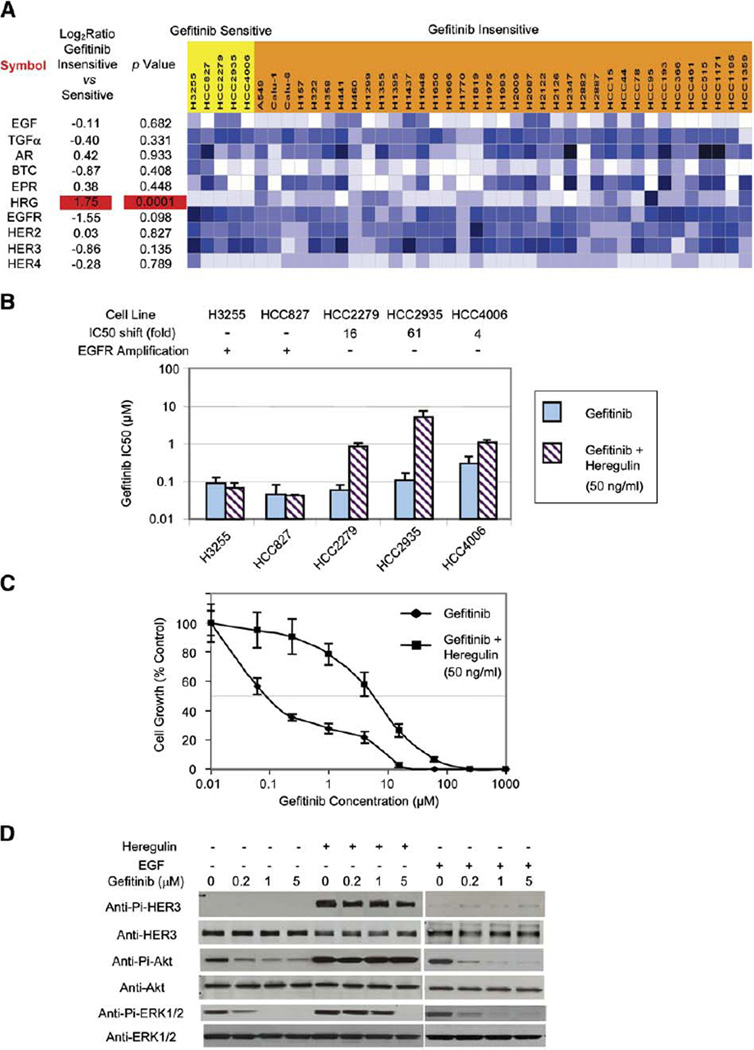
Because the clinical significance of HER3 activation in NSCLC is currently unknown, we used NSCLC cell lines to ask whether HER3 activation could contribute to drug resistance. We recently evaluated gene expression in 44 different NSCLC cell lines using microarrays and determined their in vitro sensitivity to gefitinib (L.G., M.P., I. Sekine, S. Zachariah, C. Lam, M. Wong, D. Beer, W. Gerald, A.F.G., and J.D.M., unpublished data; M.P., L.G., and J.D.M., unpublished data). To examine the effects of ErbB ligands and receptors on gefitinib sensitivity, we compared their mRNA levels against gefitinib sensitivity across the 44 NSCLC cell lines. Among various genes examined (Figure 2A), only the expression of heregulin (HRG) significantly correlated with insensitivity to gefitinib (p<0.001). Interestingly, HER3 expression very weakly correlated with gefitinib sensitivity (p = 0.135), suggesting that it is the heregulin-induced HER3 activation rather than the level of HER3 expression that leads to gefitinib insensitivity.
Figure 2.
Heregulin contributes to gefitinib resistance in NSCLC cell lines
A: Heregulin expression correlates with gefitinib insensitivity in NSCLC cell lines. Microarray expression data are presented on 42 NSCLC cell lines, including five gefitinib-sensitive cell lines (IC50 < 0.4 µM, highlighted in yellow) and 37 gefitinib-insensitive cell lines (IC50 > 4.0 µM, highlighted in orange). Calu-3 and H820 cell lines have IC50s of 1.0 and 3.0 µM, respectively, and their microarray data are not shown. Log ratios and p values are shown on the left. Expression levels are color coded such that darker colors correspond to higher levels.
B: Effects of soluble heregulin on the gefitinib sensitivities of five gefitinib-sensitive cell lines. Shown are gefitinib IC50s with error bars (SD; n = 8) of various cell lines with and without heregulin (50 ng/ml), and the IC50 shift caused by adding heregulin. While H3255 and HCC827 have genomic amplification of EGFR, HCC2279, HCC2935, and HCC4006 do not.
C: Addition of soluble heregulin makes the gefitinib-sensitive cell line HCC2935 more than 60-fold less sensitive to gefitinib. Cells with or without heregulin (50 ng/ml) added were treated for 96 hr in the presence of increasing concentrations of gefitinib, and their growth was then measured using the MTS assay and plotted as a percentage of the growth of untreated cells (control) with error bars (SD; n = 3).
D: Heregulin-induced HER3 and Akt phosphorylation can bypass the gefitinib blockade in HCC2935. HCC2935 cells were serum starved for 6 hr and then grown in the presence of various concentration of gefitinib for 12 hr followed by heregulin (50 ng/ml) or EGF (50 ng/ml) stimulation for 30 min. Western blots are shown for phospho- and total HER3, ERK1/2, and Akt.
Upon ligand binding, HER3 can form heterodimers with both EGFR and HER2. To further examine our hypothesis, we added soluble heregulin to five gefitinib-sensitive cell lines: all of them have EGFR mutations, but none of them expresses significant levels of heregulin (Figure 2A). Among them, three cell lines without EGFR amplification (HCC2935, HCC2279, and HCC4006) showed increased resistance to gefitinib after heregulin was added (Figure 2B). The change is particularly dramatic for HCC2935, the only gefitinib-sensitive line in our study that expresses more HER2 than EGFR (Figure 2A; M.P., L.G., and J.D.M., unpublished data). As shown in Figure 2C, the addition of soluble heregulin shifts the IC50 of gefitinib from 110 nM to 6.71 µM. The addition of soluble EGF at similar concentrations did not have much impact on gefitinib sensitivity in the HCC2935 cell line (data not shown). Furthermore, while the constitutive Akt phosphorylation and EGF-induced Akt phosphorylation in HCC2935 are sensitive to gefitinib, heregulin-induced HER3 phosphorylation and Akt phosphorylation are not inhibitable by gefitinib at concentrations up to 5 µM (Figure 2D). On the contrary, H3255 and HCC827, both of which have genomic amplification of the EGFR locus (Tracy et al., 2004; Amann et al., 2005) and presumably predominant EGFR homodimers or heterodimers, do not change their sensitivity to gefitinib after the addition of either heregulin (Figure 2B) or EGF (data not shown). Since gefitinib is selective toward EGFR, heregulin-dependent HER3 activation may bypass gefitinib inhibition when HER2-HER3 is one of the major heterodimers present.
A549 is an example of a HER3 autocrine line that is insensitive to gefitinib and dependent on EGFR and HER2 for HER3 signaling
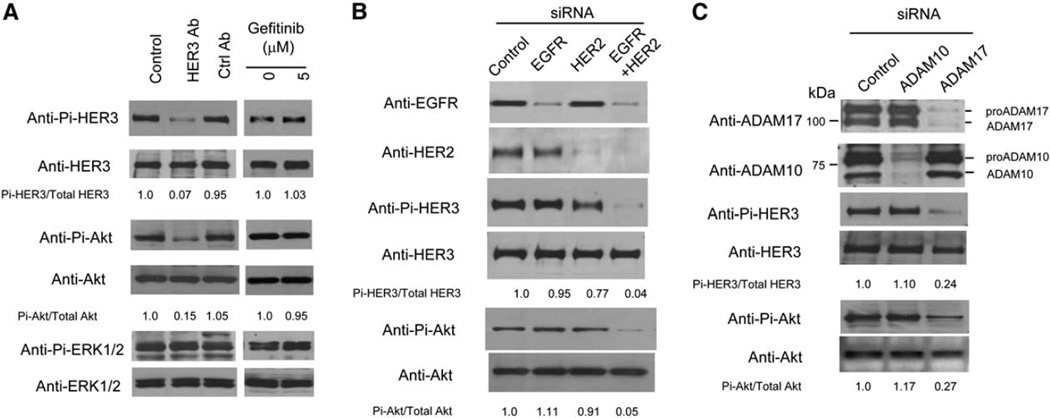
Among the few NSCLC cell lines expressing high levels of heregulin, HER3, and HER2 (Figure 2A), A549 has been previously suggested to be a heregulin-HER3 autocrine line (Gollamudi et al., 2004). We confirmed that A549 has constitutively active HER3 under serum-free conditions (Figure 3A). To further characterize A549 as a heregulin-HER3 autocrine line, we exogenously added anti-HER3 antibody that can block heregulin binding to HER3. As shown in Figure 3A, blocking of heregulin binding inhibits the constitutive phosphorylation of HER3 as well as Akt downstream in the pathway, suggesting that heregulin functions as an autocrine growth factor in A549. On the other hand, the constitutive ERK activity in A549 was not affected by blocking heregulin binding and may be mediated by mutant K-ras (Valenzuela and Groffen, 1986). In contrast to the anti-HER3 antibody, gefitinib does not have a significant impact on this autocrine pathway at the concentrations tested (Figure 3A), implicating the role of HER2 in this autocrine loop as well. This hypothesis is supported by RNA interference experiments: knockdown of either EGFR or HER2 is not sufficient to significantly inhibit the HER3 autocrine loop in A549, while knocking down both leads to a dramatic reduction in both HER3 and Akt phosphorylation (Figure 3B).
Figure 3.
Characterization of A549 as a HER3 autocrine line and its dependence on ADAM17
A: Effects of anti-HER3 antibody and gefitinib on phosphorylation of HER3 and downstream Akt and ERK1/2 in A549. A549 cells were serum starved for 6 hr and then grown with various treatments for 48 hr. Western blots are shown for phospho- and total HER3, Akt, and ERK1/2. Phospho- and total Akt and HER3 signals were quantitated, and the ratios are presented relative to control.
B: Both EGFR and HER2 are required for heregulin-dependent HER3 signaling in A549. A549 cells were transfected with appropriate siRNAs twice, serum starved for 48 hr, and then harvested for Western blot.
C: ADAM17 is important for heregulin-dependent HER3 signaling in A549. siRNA experiments were performed as in B.
ADAM17 protein correlates with HER3 activation in NSCLC and is required for HER3 signaling in A549 cells
ADAM17 was suggested to be a major sheddase for heregulin (Horiuchi et al., 2005) and, therefore, could contribute to the autocrine activation of HER3 in NSCLC. To study the role of ADAM17 in NSCLC, we analyzed the expression of ADAM17 in NSCLC tumors using antibodies against ADAM17. As shown in Figure 1, ten out of fourteen freshly resected NSCLC tissues had increased expression of ADAM17 compared with autologous-matched nonmalignant lung tissue control. In addition, there was a high rate of activation of HER3 in tumors with increased ADAM17 levels compared with those tumors with low ADAM17 expression (p ≤ 0.05). Although many patients have similar ADAM10 protein expression in tumors versus control, several of them (including cases 2, 4, and 11) have a higher ratio of the processed versus the proform of ADAM10 in the tumor samples, indicative of ADAM10 activation in these patients. However, ADAM10 activation does not correlate with HER3 activation (Figure 1), and neither do the other ADAMs previously implicated in ErbB ligand processing (Figure S1 in the Supplemental Data available with this article online).
To test if either ADAM17 or ADAM10 is involved in HER3 autocrine signaling in A549, we used RNA interference to selectively knock down the expression level of either protein. As shown in Figure 3C, ADAM10 knockdown had no significant effect on HER3 or Akt activity in A549, whereas knockdown of ADAM17 mirrored the effect of the anti-HER3 antibody and affected both HER3 and Akt phosphorylation. On the other hand, knocking down ADAM9 or ADAM15, two ADAMs with significant expression in A549, had no apparent effect on HER3 autocrine signaling in A549 cells (Figure S2). Together, these results support ADAM17 as a potential target for modulating the HER3 autocrine loop in NSCLC.
Inhibition of ADAM protease activity prevents the cleavage of heregulin and EGFR ligands
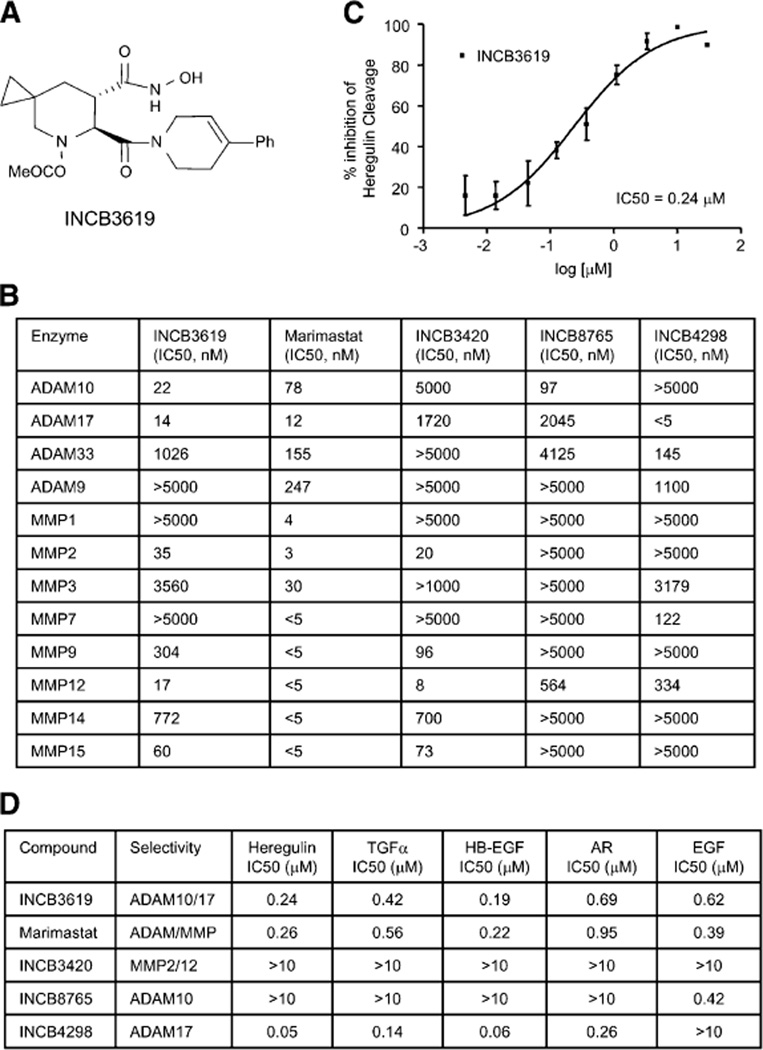
To study the impact of ADAM inhibition on shedding of heregulin as well as EGFR ligands and its potential to inhibit multiple ErbB pathways in the clinic, we identified a selective, orally bioavailable small molecule inhibitor, INCB3619 (Figure 4A; Liu et al., 2006), which has potent inhibitory activity against both ADAM10 and ADAM17 and a better selectivity profile than marimastat across a panel of metalloproteases (Figure 4B). The selectivity and inhibitory activity of the structurally related molecules INCB8765, INCB4298, and INCB3420 are also presented in Figure 4B. Since INCB3619 has some MMP2 and MMP12 inhibitory activities, INCB3420, which lacks ADAM10 and ADAM17 activity, was identified as a negative control. As shown in Figure 4C, the dual ADAM inhibitor INCB3619 inhibits heregulin cleavage with an IC50 value of 0.24 µM. In addition to heregulin, INCB3619 can prevent the cleavage of TGFα, HB-EGF, AR, EGF (Figure 4D), and HER2 (Liu et al., 2006) similar to marimastat. In agreement with data obtained from studies using knockout cells (Sahin et al., 2004; Horiuchi et al., 2005), we found that the ADAM17-selective inhibitor INCB4298 inhibits the shedding of heregulin, TGFα, HB-EGF, and AR, whereas the ADAM10-selective inhibitor INCB8765 blocks EGF ligand processing. These results suggest that pharmacological intervention of ADAM activity has a similar effect to knocking out the protein. In contrast, the MMP2/12-active, ADAM10/17-inactive INCB3420 did not block the shedding of any tested ErbB ligands (Figure 4D), further strengthening the argument that it is ADAMs, particularly ADAM17 and ADAM10, not MMPs, that are involved in ErbB ligand shedding. To maximize the effect on ErbB ligand processing and signaling, the dual inhibitor INCB3619 was chosen for further characterization.
Figure 4.
Selective ADAM inhibitor INCB3619 inhibits the cleavage of heregulin and EGFR ligands
A: Chemical structure of INCB3619.
B: Selectivity and inhibition profile of INCB3619 and related ADAM inhibitors against a panel of metalloproteases.
C: INCB3619 inhibits the heregulin cleavage measured by ELISA assay. Data are presented as mean percentage inhibition values with error bars (SD; n = 4).
D: Effects of several ADAM inhibitors on the cleavage of various ErbB ligands.
IC50s are presented here.
INCB3619 abrogates heregulin activation and heregulin-dependent HER3 signaling and increases gefitinib sensitivity in A549 cells
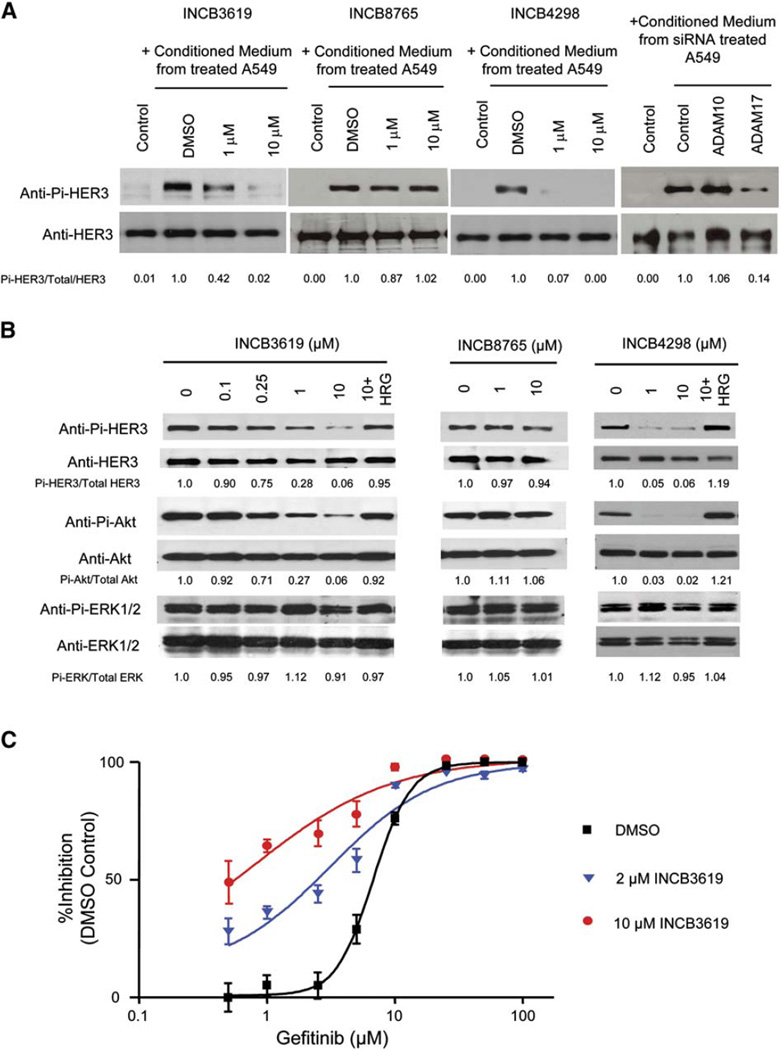
To examine the effect of ADAMs and their inhibitors on the functional release and activation of heregulin, conditioned medium from A549 cells treated with inhibitors or transfected with siRNAs was added to the medium of a reporter cell, MCF-7, which expresses HER3 but insignificant amount of heregulin. As shown in Figure 5A, INCB3619 inhibits the heregulin release and activation, measured by HER3 activation in MCF-7, similar to the effect of ADAM17 siRNA. The ADAM17-selective inhibitor INCB4298 showed a similar effect, whereas the ADAM10-selective inhibitor INCB8765 or ADAM10 siRNA had no effect. As a control, the heregulin neutralizing antibody and the anti-HER3 antibody can block the activity in A549 conditioned medium (Figure S3), confirming that it is the heregulin activity that is measured in the MCF-7 reporter cell assay.
Figure 5.
ADAM inhibitors block the activation of heregulin and HER3 autocrine signaling in A549
A: Effect of ADAM inhibition or knockdown on the heregulin activation from A549 measured by the MCF-7 reporter assay. From left: control, medium alone; DMSO, conditioned medium from A549 treated with DMSO; 1 µM or 10 µM, conditioned medium from A549 treated with different compounds at indicated concentrations. Far right panel: control, medium alone; control, ADAM10, or ADAM17, conditioned medium from A549 transfected with control siRNA, ADAM10 siRNA, or ADAM17 siRNA. Phosphoand total HER3 signals were quantitated, and the ratios are presented relative to control.
B: Effect of ADAM inhibition on heregulin-dependent HER3 signaling in A549 cells. A549 cells were serum starved for 4 hr before being treated with indicated concentrations of INCB3619, INCB8765, and INCB4298 for 72 hr. For INCB3619 and INCB4298, 50 ng/ml of heregulin was also added to reverse the signaling effect.
C: INCB3619 sensitizes A549 cells to gefitinib. A549 cells in serum-free medium were treated with various concentrations of gefitinib and INCB3619 for 96 hr, and their viability was measured and plotted as a percentage of viable cells relative to DMSO mock treatment. Data are presented as mean values with error bars (SD; n = 3).
Similar to antibody blockage of heregulin binding to HER3, INCB3619 can inhibit the heregulin-dependent HER3-Akt pathway but not ERK activity in A549 cells; the ADAM17-selective inhibitor INCB4298 has a similar effect (Figure 5B). Addition of soluble heregulin can bypass the inhibition, consistent with the hypothesis that heregulin cleavage is the target of inhibition. As controls, the ADAM10-selective inhibitor INCB8765 (Figure 5B) and the ADAM10/17-inactive inhibitor INCB3420 (data not shown) did not have a significant impact on this pathway.
Since a HER3 autocrine mechanism is believed to contribute to the gefitinib insensitivity in A549 cells, we tested whether INCB3619 can sensitize A549 cells to gefitinib. As shown in Figure 5C, INCB3619 shifts the IC50 of gefitinib in A549 cells quite significantly, further supporting the role of a heregulin-HER3 autocrine mechanism in gefitinib insensitivity; however, we cannot rule out the possibility that the K-ras mutation in A549 also contributes to the gefitinib resistance.
INCB3619 induces apoptosis in A549 cells and has antitumor activity in the A549 xenograft model
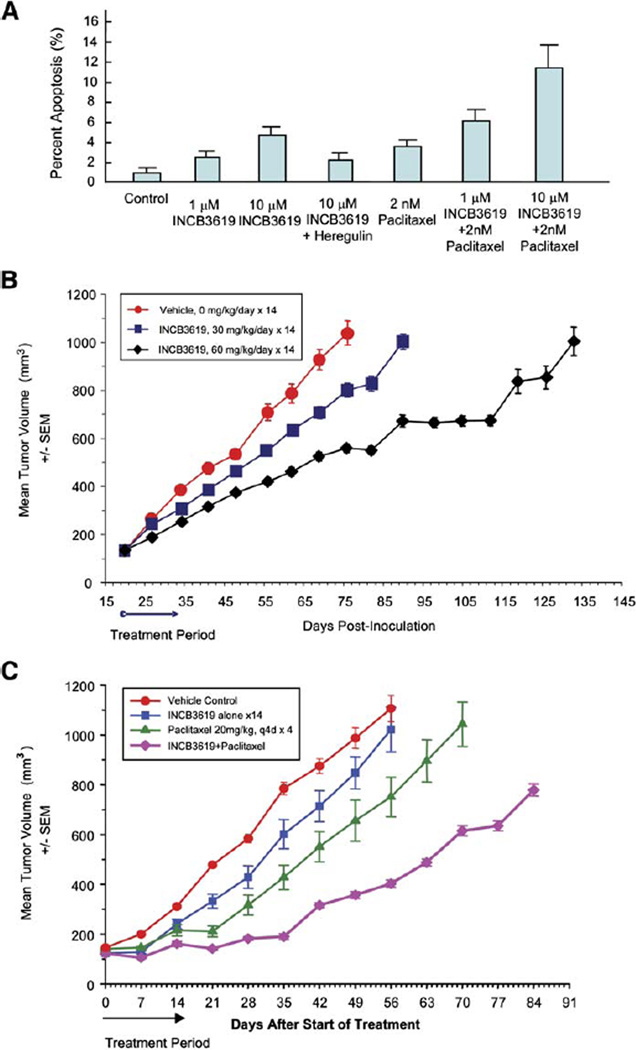
Consistent with its impact on the Akt pathway, INCB3619 can induce apoptosis in A549 cells (Figure 6A). The addition of heregulin can reverse the effect, suggesting that its effects are most likely pathway specific. Since INCB3619 impacts the survival branch of the ErbB pathway in A549, we reasoned that our inhibitor should synergize with a cytotoxic agent in this cell line. As shown in Figure 6A, INCB3619 can reduce the apoptotic threshold for paclitaxel in A549 cells.
Figure 6.
INCB3619 induces apoptosis in A549 cells and has antitumor activity in the A549 xenograft model
A: INCB3619 can increase apoptosis and reduce the apoptotic threshold for paclitaxel in A549. Shown are the mean percentages of apoptosis with error bars (SD; n = 3).
B: The effect of INCB3619 on the growth of A549 human NSCLC xenograft in BALB/c nu/nu mice. INCB3619 was administered subcutaneously using ALZET osmotic pumps at 30 or 60 mg/kg/day for 14 days. The experiment has been repeated twice, and one set of data is shown here. Shown are mean tumor volumes with error bars (SEM; n = 7).
C: Combination effect of INCB3619 and paclitaxel in treatment of A549 human NSCLC xenografts in CD-1 nu/nu mice. INCB3619 was administered subcutaneously using ALZET osmotic pumps at a dose of 50 mg/kg/day for 14 days. Paclitaxel was administered intravenously at a dose of 20 mg/kg, every 4 days for a total of four injections. Shown are mean tumor volumes with error bars (SEM; n = 7).
To extend these observations, we determined whether INCB3619 can impact tumor growth as a single agent and when combined with paclitaxel in the A549 xenograft model. As shown in Figure 6B, there is a significant tumor growth inhibition and delay (p = 0.016, compared with control group) at a dose of 60 mg/kg/day, a dose that produces a steady-state plasma drug level of nearly 1.5 µM, based on historical pharmacokinetic data. When adjusted for serum protein binding, the effective drug concentration is approximately 0.5 µM, consistent with the in vitro IC50 for heregulin cleavage inhibition by INCB3619 (Figure 4C). In these studies, the compound itself was well tolerated, based on clinic observations and body weight changes in the drug-treated mice (data not shown). Furthermore, paclitaxel alone (20 mg/kg, every 4 days, four doses in total) had a moderate antitumor activity (p = 0.03), while INCB3619 (50 mg/kg/day) had a weak activity (p = 0.2) in the A549 xenograft model in CD-1 nu/nu mice (Figure 6C). The lower activity of the compound alone observed in this experimental setting may be due to less drug exposure in CD-1 nu/nu mice compared to BALB/c nu/nu mice used in the previous experiment (Figure 6B). Nevertheless, the combination of INCB3619 and paclitaxel significantly inhibited tumor growth (p = 0.0001), with no additional toxicity observed over that seen with paclitaxel alone (data not shown). Together, these results suggest that the ADAM inhibitor INCB3619 inhibits the gefitinib-resistant HER3 autocrine mechanism, possesses antitumor activity in vivo, and sensitizes tumors to chemotherapeutic agents.
INCB3619 inhibits the EGFR ligand signaling in the EGFR autocrine cell line NCI-H1666 and sensitizes it to gefitinib
The EGFR autocrine mechanism can be targeted by anti-EGFR agents such as gefitinib and cetuximab. However, many NSCLC lines expressing EGFR ligands, especially those with wild-type EGFR, are not particularly sensitive to gefitinib (Figure 2A). To study the effect of our selective ADAM inhibitor on EGFR signaling and gefitinib sensitivity, we searched for an EGFR ligand-dependent NSCLC autocrine cell line. NCI-H1666 is among those putative EGFR-driven autocrine cell lines with wild-type EGFR and limited sensitivity to gefitinib and cetuximab (Tracy et al., 2004; Amann et al., 2005; Mukohara et al., 2005). Using various siRNAs and C225 (the murine version of cetuximab), we indeed found that NCI-H1666 is an ADAM17-mediated, EGFR ligand-dependent autocrine NSCLC cell line with intermediate sensitivity to gefitinib (Figure S4).
We studied the impact of ADAM inhibitors on the functional release and activation of EGFR ligands from NCI-H1666 cells, which express TGFα, AR, and EPR (Figure 2A), using an A431 reporter cell assay. As shown in Figure 7A, INCB3619 inhibits EGFR ligand activation, as measured by EGFR phosphorylation on Tyr-1173 in A431. Similar to C225, INCB3619 inhibits ligand-dependent activation of the EGFR-ERK pathway in NCI-H1666 cells (Figure 7B); addition of soluble TGFα can bypass this inhibition. As a control, the ADAM10-selective inhibitor INCB8765 does not have a significant impact on either ligand activation or EGFR signaling, while the ADAM17-selective inhibitor INCB4298 has a similar effect to INCB3619. These results suggest that INCB3619 abrogates EGFR autocrine signaling in NCI-H1666, most likely through inhibiting ADAM17-mediated processing of several EGFR ligands.
Figure 7.
INCB3619 inhibits EGFR autocrine signaling in NCI-H1666 and sensitizes it to gefitinib
A: INCB3619 inhibits EGFR ligand activation from NCI-H1666 measured by the A431 reporter assay. Left panel: control, buffer; TGFα, 10 ng/ml TGFα; TGFα + C225, TGFα added to the A431 cells pretreated with C225 for 10 min. Right panel: control, medium alone; DMSO, conditioned medium from NCI-H1666 treated with DMSO for 48 hr; 2 µM 3619 or 10 µM 3619, conditioned medium from NCI-H1666 treated with INCB3619 at indicated concentrations for 48 hr. Phospho- and total EGFR signals were quantitated, and the ratios are presented relative to control.
B: INCB3619 inhibits EGFR ligand-dependent signaling in NCI-H1666 cells. A549 cells were serum starved for 4 hr before being treated with indicated concentrations of various compounds for 72 hr. For INCB3619, 50 ng/ml of TGFα was added to reverse the signaling effect.
C: INCB3619 sensitizes NCI-H1666 cells to gefitinib. NCI-H1666 cells in serum-free medium were treated with various concentrations of gefitinib and INCB3619 for 72 hr, and their viability was measured and plotted as a percentage of viable cells relative to DMSO mock treatment. Data are presented as mean values with error bars (SD; n = 3).
D: Combination effect of INCB3619 and gefitinib on the EGFR autocrine signaling. NCI-H1666 cells in serum-free medium were treated with DMSO alone, 2 µM INCB3619, 2 µM INCB3619 in the presence of 50 ng/ml TGFα, 0.5 µM gefitinib, or 0.5 µM gefitinib plus 2 µM INCB3619. Phospho- and total ERK1/2 signals were quantitated, and the ratios are presented relative to control.
INCB3619 inhibits the proliferation of NCI-H1666 cells under serum-free conditions (Figure S5). Since both INCB3619 and gefitinib inhibit NCI-H1666 proliferation, presumably by blocking targets at different levels in the same pathway, we tested whether the inhibition of EGFR ligand cleavage can sensitize NCI-H1666 to gefitinib. Figure 7C shows the combined effect of INCB3619 and gefitinib: under serum-free condition, 0.53 µM of gefitinib is needed to inhibit proliferation by 50%; with 2 µM INCB3619, 6-fold less (0.087 µM) gefitinib achieves an equivalent effect. Using the concentration near the IC50 for each compound, we tested the combined effect of the sheddase inhibitor and gefitinib on ErbB signaling. As shown in Figure 7D, both 2 µM INCB3619 and 0.5 µM gefitinib significantly reduced ERK phosphorylation, while the combined effect is even more dramatic. Our results with NCI-H1666 cells suggest that the ADAM inhibitor INCB3619 can inhibit EGFR signaling and be used in combination with other anti-EGFR agents.
Discussion
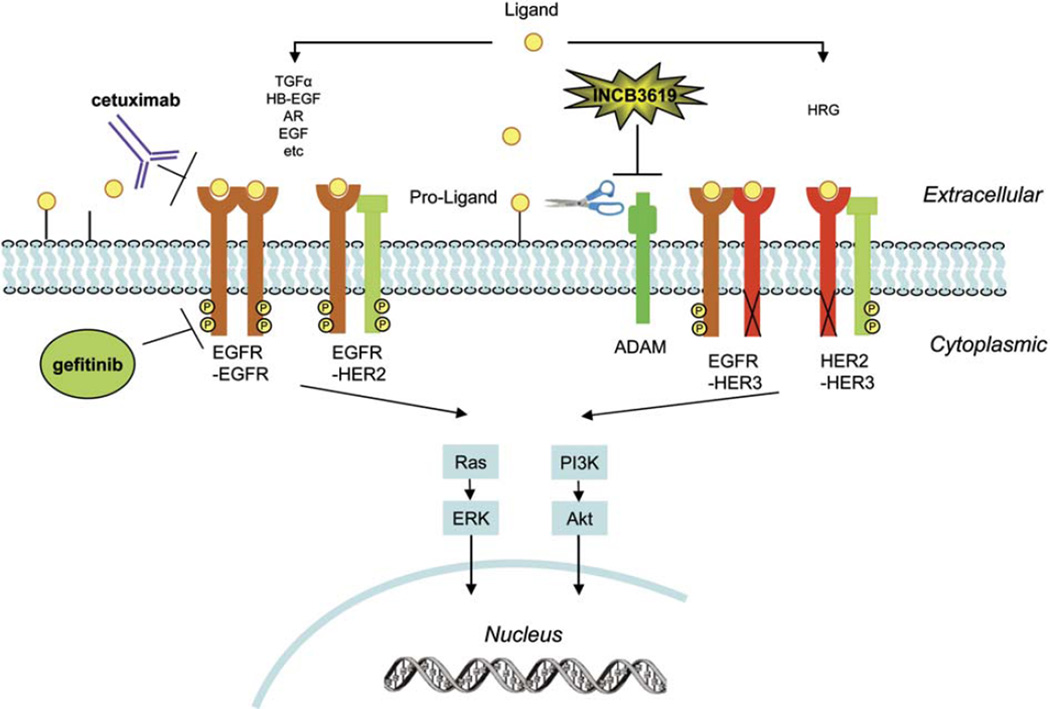
Cancer cells synthesize and secrete various growth factors and can also express their cognate receptors, allowing the secreted growth factors to stimulate tumor growth through autocrine mechanisms (Barnes and Sato, 1980; Carpenter and Cohen, 1990; Osborne and Arteaga, 1990; Sporn and Todaro, 1980). EGFR and its ligands form one of the best-defined autocrine growth loops in human tumors (Salomon et al., 1995), and their coexpression correlates with aggressive disease and poor prognosis in several types of tumors, including NSCLC (Salomon et al., 1995; Umekita et al., 2000; Tateishi et al., 1990; Gorgoulis et al., 1992). Based on these observations, the concept has developed that preventing ligand binding to EGFR, just like inhibiting EGFR kinase activity, should inhibit the proliferation of epithelial-derived tumor cells (Mendelsohn, 1997). With various anti-EGFR antibodies having demonstrated clinical activity in cancer, this hypothesis has gained considerable support (Baselga and Arteaga, 2005). In this paper, we discuss a complementary approach that prevents the release and activation of ligands for both EGFR and HER3 (Figure 8).
Figure 8.
The ErbB signaling network and sites of therapeutic intervention
ADAM-mediated proteolytic cleavage releases the ligands, which bind to the ErbB receptor through autocrine and/or paracrine mechanisms and lead to receptor dimerization, triggering multiple downstream signaling cascades, including Ras-ERK and PI3K-Akt pathways. While cetuximab blocks the binding of several EGFR ligands to EGFR, gefitinib inhibits EGFR kinase activity. On the other hand, small molecule inhibitors of ADAM proteases, such as INCB3619, inhibit the availability of multiple ErbB ligands, including EGFR ligands and heregulin.
One potential mechanism of resistance to anti-EGFR agents is the activation of ligands that target the alternate ErbB receptors. We demonstrate here the existence of a HER3 autocrine stimulatory loop in NSCLC using tissue samples. Although HER3 expression has been shown to correlate with gefitinib sensitivity among NSCLC cell lines (Engelman et al., 2005), genomic gain of HER3 is not associated with increased sensitivity to gefitinib in advanced NSCLC patients (Cappuzzo et al., 2005). The lack of a strong association with gefitinib sensitivity is not surprising because HER3 lacks kinase activity and only signals in the context of a receptor heterodimer upon ligand binding. Therefore, the best way to evaluate the impact of HER3is in combination with its ligand and partnering ErbB receptors, particularly EGFR and HER2. In our study, we found that HER3 expression very weakly correlates with gefitinib sensitivity, while the expression of heregulin shows a significant correlation with gefitinib insensitivity. Subsequent studies with NSCLC cell lines suggested that heregulin-dependent HER3 activation can bypass the inhibition of gefitinib, and presumably other EGFR specific agents as well, when HER2-HER3 is one of the major heterodimers present.
ADAMs might be attractive targets for the treatment of cancer due to their role in shedding ErbB ligands (Blobel, 2005; Zhou et al., 2005). We found that the ADAM17 protein level is elevated, while the activated form of ADAM10 is present at higher levels in NSCLC. Further, we found a correlation between ADAM17 protein level and HER3 activation in NSCLC, similar to that observed between ADAM17 overexpression and EGFR activation in human breast cancers (Borrell-Pages et al., 2003). Our studies with both siRNAs and ADAM inhibitors suggest that ADAM17 is important for both heregulin-dependent ErbB signaling in A549 cells and EGFR ligand-dependent ErbB signaling in NCI-H1666 cells, further validating ADAM17 as a drug target in NSCLC. TIMP3, a negative regulator of ADAMs, including ADAM17, was found to be downregulated at the mRNA level in NSCLC (data not shown), at least partially due to aberrant TIMP3 promoter methylation (Bachman et al., 1999; Zochbauer-Muller et al., 2001).Overexpression of TIMP-3, which would be expected to reduce ErbB ligand shedding, has been shown to suppress tumor growth in vivo (Bian et al., 1996); therefore, addition of an ADAM inhibitor would essentially restore the negative regulation normally exercised by TIMP-3.
Compared with many available metalloprotease inhibitors, such as marimastat, the ADAM inhibitor INCB3619 is more selective for the ADAM family, is not active against many members of the MMP family of metalloproteases, and has a far superior safety profile in preclinical models, including the lack of musculoskeletal side effects (J.S.F., E.C., M. Hansbury, X. Liu, X. Gu, M. Pan, B.-B.S.Z., G.Y., Q. Wang, Y.L., S. Thomas, J. Grandis, J. Zhuo, W.Y., R.C.N., S.M.F., P.A.S., and K.V., unpublished data). One consideration associated with utilizing an ADAM17 and ADAM10 dual inhibitor is that ADAM10 has also been suggested to be involved in ErbB ligand shedding under certain circumstances (Prenzel et al., 1999; Sahin et al., 2004; Yan et al., 2002). In addition, only dual inhibitors, not the more selective inhibitors, have inhibited the shedding and activation of all ligands tested in our study. Although both NSCLC ErbB autocrine lines that we studied depend on ADAM17 for ligand-dependent signaling, we cannot rule out the possibility that other NSCLC ErbB autocrine lines or autocrine mechanisms in certain NSCLC patients might rely on ADAM10, particularly since ADAM10 is also activated in NSCLC. In addition, the dual ADAM inhibitor can affect ErbB signaling pathways during tumor progression by inhibiting ErbB receptor cleavage (Liu et al., 2006). An additional benefit of targeting ADAM10 could come from its role in angiogenesis and/or metastasis (Zhou et al., 2005).
Although we focused our in vitro studies on autocrine activation of various ErbB pathways, paracrine mechanisms also exist in xenograft models and in patients, although their contribution is less clear in most cases. Since the same proteases are involved in these processes, we expect that an ADAM inhibitor will inhibit both potential sources of ligand. In some model systems, membrane bound ErbB ligands can also engage in juxtacrine signaling (Brachmann et al., 1989; Wong et al., 1989), which was recently shown to require ADAM activity as well (Borrell-Pages et al., 2003). Consistent with this hypothesis, INCB3619 inhibits heregulin and EGFR ligand-mediated ErbB signaling, even under high-cell density conditions, which favor juxtacrine signaling (data not shown).
Inhibition of the ErbB ligand processing step has different functional consequences in different systems. In A549 cells, INCB3619 can inhibit the HER3 and Akt activity, similar to knocking down both EGFR and HER2 in this cell line. Importantly, INCB3619 can sensitize A549 cells to gefitinib and paclitaxel and has antitumor activity alone and in combination with paclitaxel in an A549 xenograft model. NCI-H1666 cells, although possessing wild-type EGFR, have intermediate sensitivity to gefitinib. We demonstrate here that INCB3619 inhibits cell proliferation in the NCI-H1666 cell line and sensitizes NCI-H1666 cells to gefitinib, potentially translating to improved efficacy of gefitinib among those NSCLC patients with stable disease or partial response to gefitinib in the clinic. On the other hand, INCB3619 does not affect cell proliferation or ErbB signaling in nontransformed lung epithelial cells (Figure S6) and is inactive in a xenograft model (MDA-MB-231) that is not dependent on ErbB signaling for growth (J.S.F., E.C., M. Hansbury, X. Liu, X. Gu, M. Pan, B.-B.S.Z., G.Y., Q. Wang, Y.L., S. Thomas, J. Grandis, J. Zhuo, W.Y., R.C.N., S.M.F., P.A.S., and K.V., unpublished data), further demonstrating its selectivity.
In summary, the therapeutic use of ADAM inhibitors, such as INCB3619, has the potential to control the availability of numerous ErbB ligands and overcome gefitinib resistance caused by the HER3 autocrine mechanism, and thereby offer important advantages over existing anti-EGFR strategies by abrogating the ability to signal through multiple ErbB pathways (Figure 8). Clearly, strategies targeting ADAMs might be an important complement to existing anti-ErbB approaches and could potentially be used in combination with various anti-ErbB and chemotherapeutic agents. Current ongoing clinical evaluation of our selective ADAM inhibitor is only beginning to address the scope of its therapeutic potential in cancer treatment.
Experimental procedures
Cell lines
The cell lines described in this manuscript, except for A549, were established at the National Cancer Institute (NCI-H series) or at the Hamon Center for Therapeutic Oncology Research, University of Texas Southwestern Medical Center (HCC series). These cell lines have been deposited for distribution in the American Type Culture Collection (ATCC) (http://www.atcc.org/). A549 was obtained from ATCC.
Compounds
INCB3619, methyl (6S,7S)-7-[(hydroxyamino)carbonyl]-6-[(4-phenyl-3,6-dihy-dropyridin-1(2H)-yl)carbonyl]-5-azaspiro[2.5]octane-5-carboxylate, INCB8765, (1R,3S,4S)-3-[(hydroxyamino)carbonyl]-4-[(4-phenylpiperidin-1-yl)carbonyl]cyclohexyl pyrrolidine-1-carboxylate, INCB3420, (1S,2R)-N-hydroxy-2-({[4-(pyridin-4-yloxy)phenyl]sulfonyl}amino)cyclohexanecarboxamide, and related compound INCB4298 were synthesized by the Chemistry Department of Incyte Corporation (Wilmington, DE), according to the methods described in U.S. patent Ser. No. 60/543,501 and W.Y., J. Zhuo, D. Burns, M. Xu, F. Zhang, and B. Metcalf (unpublished data). Gefitinib (Iressa) and paclitaxel (Taxol) were purchased from Hanna Pharmaceuticals (Wilmington, DE). Marimastat was obtained from British Biotech (Oxford, UK).
Antibodies and proteins
All antibodies were purchased from commercial sources as described here: antibodies to ERK1/2 (p44/42 ERK kinase), phospho-ERK1/2 (Thr-202/Tyr-204), Akt, phospho-Akt, cleaved Caspase-3 (Asp-175), EGFR, phospho-EGFR (Tyr-1173), phospho-EGFR (Tyr-1068), HER2, phospho-HER3/ErbB3 (Tyr-1289), and monoclonal, HRP-conjugated goat anti-rabbit IgG and anti-mouse IgG were purchased from Cell Signaling (Beverly, MA). Anti-HER3/ErbB3 (Ab6), used for Western blot, and anti-HER3/ErbB3 (Ab5, CloneH3.105.5), used to block heregulin binding to HER3, were from Neo-Markers (Fremont, CA). Anti-heregulin was from R&D systems (Minneapolis, MN). Anti-ADAM17 was from Calbiochem (San Diego, CA). Anti-ADAM10 was from CHEMICON (Temecula, CA). Mouse monoclonal antibody C225 (EGFR Ab2) was from NeoMarkers (Fremont, CA).
Purified recombinant soluble heregulin, EGF, and TGFα were purchased from R&D systems (Minneapolis, MN).
NSCLC tissue specimen preparation and Western blot analysis
Fresh NSCLC tissue and adjacent nonmalignant lung tissue from patients undergoing curative primary resection of their tumors were collected at the time of surgery and immediately snap-frozen in liquid nitrogen (UCSF IRB approval #H8714-15319-07A). These tissue samples were kept at −170°C in a liquid nitrogen freezer before use. T-PER tissue protein extraction reagent was purchased from Pierce (Rockford, IL) and used for preparation of whole-cell extract.
Microarray analysis of NSCLC cell lines
RNA fluorescent labeling reaction and hybridization were performed using the Affymetrix Gene Chips HG-U133A and HG-U133B according to the manufacturer’s instructions (http://www.affymetrix.com/). The arrays consist of 22,283 (HG-U133A) and 22,645 (HG-U133B) probe sets, which together amount to 23,583 unique genes based on Unigene build 173. Microarray analysis was performed using Affymetrix Microarray Suite 5.0 and in-house Visual Basic software MATRIX 1.26. Array data were median normalized, and replicate genes were combined by averaging. Samples (or averages of samples) were then compared against each other by calculating log ratios for each gene, and statistical significance was presented as a p value calculated by Student’s t test. The microarray data have been uploaded to GEO (Gene Expression Omnibus), and the accession number is GSE-4824.
RNA interference studies
All siRNAs were obtained from Dharmacon (Lafayette, CO). Sequences for ADAM17 interfering RNAs (siRNA) were 5′-GAACAAGUGUAAAUUAUUGU U-3′ (ADAM17 RNAi #1), 5′-GAUCAUCGCUUCUACAGAUUU-3′ (ADAM17 RNAi #2), 5′-UAUGGGAACUCUUGGAUUAUU-3′ (ADAM17 RNAi #3), 5′-GA GGAAGCAUCUAAAGUUUUU-3′ (ADAM17 RNAi #4). To minimize nonspecific effects of interfering RNAs, non-pathway-related siRNAs were used as negative control. Transfection of siRNAs into cell lines was achieved using Lipofectamine 2000 (Invitrogen, CA).
In vitro metalloprotease assays and ErbB ligand cleavage assays
Human MMPs, ADAM9, ADAM10, and ADAM33 were purchased from R&D Systems (Minneapolis, MN), and partially purified porcine ADAM17 was obtained from spleen as described (Moss et al., 1997). Both MMP and ADAM assays were performed using substrates and conditions supplied by R&D systems (Minneapolis, MN).
All ErbB ligand ELISA kits are from R&D Systems (Minneapolis, MN). Heregulin release was measured by ELISA in supernatants collected from heregulin-transfected CHO cells following stimulation with PMA (phorbol myristate acetate) for 1 hr. HB-EGF release was measured in supernatants collected from HB-EGF transfected CHO cells following stimulation with PMA for 45 min. The HB-EGF protein was tagged with alkaline phosphatase so that levels could be measured using a chemiluminescent substrate. TGFα release and AR release were measured by ELISA in supernatants collected from HCT116 cells following stimulation with PMA for 30 min. EGF release was measured by ELISA in supernatants collected from H460 cells following stimulation with PMA for 72 hr.
Reporter cell assay measuring the activation of ErbB ligand
A549 or NCI-H1666 cells were grown in 75 cm2 culture flasks in RPMI medium containing 10% FCS until >50% confluency, washed, then treated in serumfree medium. The medium was collected after 48 or 72 hr, concentrated by a centrifugal filter device (Centriplus, Amicon, MA), then subjected to functional testing using MCF-7 or A431 cells. For testing of heregulin activity, MCF-7 cells (approximately 200,000 cells per well in 12-well plates) were allowed to adhere in DMEM with 10% FCS overnight and then were growth arrested for 8–16 hr in serum-free medium. Concentrated, conditioned medium (100 µl) was added to the growth-arrested cells, cells were lysed 15 min post-medium addition, and 20 µg of proteins were subjected to Western blotting for phosphorylated HER3. For testing of EGFR ligands, similar experiments were performed using NCI-H1666, and the activity was detected using phosphospecific antibody against Tyr-1173 of EGFR in A431 cells.
Ligand-dependent ErbB pathway analysis
A549 or NCI-H1666 cells were grown in 6-well plates in RPMI medium with 10% FCS until 30%–40% confluency and then treated in serum-free medium. The cells were harvested 48–72 hr later for Western blot. The intensities of Western blot signals were quantitated using QuantiScan software (Biosoft, Cambridge, UK).
Apoptosis assay by caspase immunostaining
Twenty thousand cells were seeded into each well of double-chamber slides (Nalge Nunc International, IL), then treated in serum-free medium next day. After 96 hr the cells were fixed with methanol at −20°C for 10 min and then blocked with PBS containing 1%BSA for 1 hr. Apoptotic cells were detected by the cleaved caspase-3 (Asp-175) antibody (fluorescein conjugate) (Cell Signaling Technology, MA); total cell numbers were measured by Hoechst staining.
Proliferation assay
Cell growth was evaluated by MTS assays (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega, WI) or cell viability assays (CellTiter-Glo Luminescent Cell Viability Assay, Promega, WI). Cells were plated 24 hr prior to addition of compounds, and assays were performed in 96-well plates.
Xenograft studies in nude mice
Female BALB/c nu/nu or CD-1 nu/nu mice were obtained from Charles River Laboratories (Wilmington, MA). When the mice were 7–8 weeks of age, each mouse was inoculated with 1 × 107 tumor cells in 0.2 ml of medium subcutaneously in the right flank. The treatments were started at 7–10 days after inoculation when the tumor size reached approximately 80–180 mm3, and each treatment group contained seven animals. Tumor sizes were measured weekly in two dimensions using a caliper, and the volume (mm3) was calculated using the formula V = 0.5a × b2, where a and b are the long and short diameters of the tumor, respectively. The statistical significance was analyzed using the unpaired two-tailed Student’s t test. The standard error of the mean (SEM) was calculated based on the standard deviation (SD) within each group. All procedures involving animals were approved by Incyte Animal Care and Use Committee (ACUC).
Supplementary Material
SIGNIFICANCE.
Agents that inhibit EGFR function, such as gefitinib and cetuximab, have shown modest therapeutic efficacy in patients with NSCLC. One mechanism of drug resistance is the activation of alternative ErbB pathways, such as signaling through HER3, as demonstrated herein. Recently, ADAMs, particularly ADAM17, have emerged as upstream activators of ErbB ligands. We demonstrate here that a selective ADAM inhibitor blocks the cleavage of multiple ErbB ligands and thereby inhibits the activation of multiple ErbB pathways, including HER3 in NSCLC. A strategy targeting ADAMs may be an important complement to existing anti-ErbB approaches and could be used in combination with various anti-ErbB and chemotherapeutic agents.
Acknowledgments
We gratefully acknowledge Dr. Xiangdong Liu and Mr. Mike Hansbury for helpful discussion and generous technical help during the course of this work, and Dr. Philip Liu for advice and technical help with siRNA experiments. We thank Drs. Kermit Carraway, Cary Lai, David Nethery, and Ruth Lupu for helpful advice on the heregulin experiments. The work in J.D.M.’s group was supported by Lung Cancer SPORE grant P50CA70907 and the Gilson Longenbaugh Foundation. The work in D.M.J.’s group was supported by the Larry Hall Memorial Trust and the Kazan, McClain, Edises, Abrams, Fernandez, Lyons & Farrise Foundation.
Footnotes
Supplemental data
The Supplemental Data include Supplemental Experimental Procedures and six supplemental figures and can be found with this article online at http://www.cancercell.org/cgi/content/full/10/1/39/DC1/.
Authors affiliated with Incyte declare competing financial interests.
References
- Alimandi M, Romano A, Curia MC, Muraro R, Fedi P, Aaronson SA, Di Fiore PP, Kraus MH. Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene. 1995;10:1813–1821. [PubMed] [Google Scholar]
- Amann J, Kalyankrishna S, Massion PP, Ohm JE, Girard L, Shigematsu H, Peyton M, Juroske D, Huang Y, Stuart Salmon J, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65:226–235. [PubMed] [Google Scholar]
- Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- Barnes D, Sato G. Serum-free cell culture: a unifying approach. Cell. 1980;22:649–655. doi: 10.1016/0092-8674(80)90540-1. [DOI] [PubMed] [Google Scholar]
- Baselga J, Arteaga CL. Critical update and emerging trends in epidermal growth factor receptor targeting in cancer. J. Clin. Oncol. 2005;23:2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- Bian J, Wang Y, Smith MR, Kim H, Jacobs C, Jackman J, Kung HF, Colburn NH, Sun Y. Suppression of in vivo tumor growth and induction of suspension cell death by tissue inhibitor of metalloproteinases (TIMP)-3. Carcinogenesis. 1996;17:1805–1811. doi: 10.1093/carcin/17.9.1805. [DOI] [PubMed] [Google Scholar]
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- Borrell-Pages M, Rojo F, Albanell J, Baselga J, Arribas J. TACE is required for the activation of the EGFR by TGF-α in tumors. EMBO J. 2003;22:1114–1124. doi: 10.1093/emboj/cdg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann R, Lindquist PB, Nagashima M, Kohr W, Lipari T, Napier M, Derynck R. Transmembrane TGF-α precursors activate EGF/TGF-α receptors. Cell. 1989;56:691–700. doi: 10.1016/0092-8674(89)90591-6. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Toschi L, Domenichini I, Bartolini S, Ceresoli GL, Rossi E, Ludovini V, Cancellieri A, Magrini E, Bemis L, et al. HER3 genomic gain and sensitivity to gefitinib in advanced non-small-cell lung cancer patients. Br. J. Cancer. 2005;93:1334–1340. doi: 10.1038/sj.bjc.6602865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G, Cohen S. Epidermal growth factor. J. Biol. Chem. 1990;265:7709–7712. [PubMed] [Google Scholar]
- Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- Dong J, Opresko LK, Dempsey PJ, Lauffenburger DA, Coffey RJ, Wiley HS. Metalloprotease-mediated ligand release regulates autocrine signaling through the epidermal growth factor receptor. Proc. Natl. Acad. Sci. USA. 1999;96:6235–6240. doi: 10.1073/pnas.96.11.6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Janne PA, Mermel C, Pearberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC. ErbB-3 mediates phosphoinositide 3 kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc. Natl. Acad. Sci. USA. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee JM, Knowlden JM. ADAM metalloproteases and EGFR signalling. Breast Cancer Res. 2003;5:223–224. doi: 10.1186/bcr637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollamudi M, Nethery D, Liu J, Kern JA. Autocrine activation of ErbB2/ErbB3 receptor complex by NRG-1 in non-small cell lung cancer cell lines. Lung Cancer. 2004;43:135–143. doi: 10.1016/j.lungcan.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Gorgoulis V, Aninos D, Mikou P, Kanavaros P, Karameris A, Joardanoglou J, Rasidakis A, Veslemes M, Ozanne B, Spandidos DA. Expression of EGF, TGF-α and EGFR in squamous cell lung carcinomas. Anticancer Res. 1992;12:1183–1187. [PubMed] [Google Scholar]
- Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat. Rev. Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., III Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc. Natl. Acad. Sci. USA. 1994;91:8132–8136. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RC, Chung E, Coffey RJ. EGF receptor ligands. Exp. Cell Res. 2003;284:2–13. doi: 10.1016/s0014-4827(02)00105-2. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Fukuoka M, Baselga J. Gefitinib—a novel targeted approach to treating cancer. Nat. Rev. Cancer. 2004;4:956–965. doi: 10.1038/nrc1506. [DOI] [PubMed] [Google Scholar]
- Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, 3rd, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Zhou HM, Kelly K, Manova K, Blobel CP. Evaluation of the contributions of ADAMs 9, 12, 15, 17, and 19 to heart development and ectodomain shedding of neuregulins β1 and β2. Dev. Biol. 2005;283:459–471. doi: 10.1016/j.ydbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Huang S, Armstrong EA, Benavente S, Chinnaiyan P, Harari PM. Dual-agent molecular targeting of the epidermal growth factor receptor (EGFR): combining anti-EGFR antibody with tyrosine kinase inhibitor. Cancer Res. 2004;64:5355–5362. doi: 10.1158/0008-5472.CAN-04-0562. [DOI] [PubMed] [Google Scholar]
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J. Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- Kim HH, Sierke SL, Koland JG. Epidermal growth factor-dependent association of phosphatidylinositol 3-kinase with the erbB3 gene product. J. Biol. Chem. 1994;269:24747–24755. [PubMed] [Google Scholar]
- Liu X, Fridman JS, Wang Q, Caulder E, Yang G, Covington M, Liu C, Marando C, Zhuo J, Li Y, et al. Selective inhibition of ADAM metalloproteases blocks HER-2 extracellular domain (ECD) cleavage and potentiates the anti-tumor effects of Trastuzumab. Cancer Biol. Ther. 2006;5:648–656. doi: 10.4161/cbt.5.6.2707. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Matar P, Rojo F, Cassia R, Moreno-Bueno G, Di Cosimo S, Tabernero J, Guzman M, Rodriguez S, Arribas J, Palacios J, Baselga J. Combined epidermal growth factor receptor targeting with the tyrosine kinase inhibitor gefitinib (ZD1839) and the monoclonal antibody cetuximab (IMC-C225): superiority over single-agent receptor targeting. Clin. Cancer Res. 2004;10:6487–6501. doi: 10.1158/1078-0432.CCR-04-0870. [DOI] [PubMed] [Google Scholar]
- Mendelsohn J. Epidermal growth factor receptor inhibition by a monoclonal antibody as anticancer therapy. Clin. Cancer Res. 1997;3:2703–2707. [PubMed] [Google Scholar]
- Merlos-Suarez A, Ruiz-Paz S, Baselga J, Arribas J. Metalloprotease-dependent protransforming growth factor-α ectodomain shedding in the absence of tumor necrosis factor-α-converting enzyme. J. Biol. Chem. 2001;276:48510–48517. doi: 10.1074/jbc.M103488200. [DOI] [PubMed] [Google Scholar]
- Montero JC, Yuste L, Diaz-Rodriguez E, Esparis-Ogando A, Pandiella A. Differential shedding of transmembrane neuregulin isoforms by the tumor necrosis factor-α-converting enzyme. Mol. Cell. Neurosci. 2000;16:631–648. doi: 10.1006/mcne.2000.0896. [DOI] [PubMed] [Google Scholar]
- Mosesson Y, Yarden Y. Oncogenic growth factor receptors: implications for signal transduction therapy. Semin. Cancer Biol. 2004;14:262–270. doi: 10.1016/j.semcancer.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature. 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- Mukohara T, Engelman JA, Hanna NH, Yeap BY, Kobayashi S, Lindeman N, Halmos B, Pearlberg J, Tsuchihashi Z, Cantley LC, et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J. Natl. Cancer Inst. 2005;97:1185–1194. doi: 10.1093/jnci/dji238. [DOI] [PubMed] [Google Scholar]
- Normanno N, Bianco C, De Luca A, Salomon DS. The role of EGF-related peptides in tumor growth. Front. Biosci. 2001;6:D685–D707. doi: 10.2741/normano. [DOI] [PubMed] [Google Scholar]
- Normanno N, Campiglio M, De LA, Somenzi G, Maiello M, Ciardiello F, Gianni L, Salomon DS, Menard S. Cooperative inhibitory effect of ZD1839 (Iressa) in combination with trastuzumab (Herceptin) on human breast cancer cell growth. Ann. Oncol. 2002;13:65–72. doi: 10.1093/annonc/mdf020. [DOI] [PubMed] [Google Scholar]
- O-Charoenrat P, Rhys-Evans P, Eccles S. A synthetic matrix metalloproteinase inhibitor prevents squamous carcinoma cell proliferation by interfering with epidermal growth factor receptor autocrine loops. Int. J. Cancer. 2002;100:527–533. doi: 10.1002/ijc.10531. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Arteaga CL. Autocrine and paracrine growth regulation of breast cancer: clinical implications. Breast Cancer Res. Treat. 1990;15:3–11. doi: 10.1007/BF01811884. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- Riemenschneider MJ, Bell DW, Haber DA, Louis DN. Pulmonary adenocarcinomas with mutant epidermal growth factor receptors. N. Engl. J. Med. 2005;352:1724–1725. doi: 10.1056/NEJM200504213521622. [DOI] [PubMed] [Google Scholar]
- Roghani M, Becherer JD, Moss ML, Atherton RE, Erdjument-Bromage H, Arribas J, Blackburn RK, Weskamp G, Tempst P, Blobel CP. Metalloprotease-disintegrin MDC9: intracellular maturation and catalytic activity. J. Biol. Chem. 1999;274:3531–3540. doi: 10.1074/jbc.274.6.3531. [DOI] [PubMed] [Google Scholar]
- Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit. Rev. Oncol. Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- Sporn MB, Todaro GJ. Autocrine secretion and malignant transformation of cells. N. Engl. J. Med. 1980;303:878–880. doi: 10.1056/NEJM198010093031511. [DOI] [PubMed] [Google Scholar]
- Sunnarborg SW, Hinkle CL, Stevenson M, Russell WE, Raska CS, Peschon JJ, Castner BJ, Gerhart MJ, Paxton RJ, Black RA, Lee DC. Tumor necrosis factor-α converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J. Biol. Chem. 2002;277:12838–12845. doi: 10.1074/jbc.M112050200. [DOI] [PubMed] [Google Scholar]
- Tateishi M, Ishida T, Mitsudomi T, Kaneko S, Sugimachi K. Immunohistochemical evidence of autocrine growth factors in adenocarcinoma of the human lung. Cancer Res. 1990;50:7077–7080. [PubMed] [Google Scholar]
- Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson BE, Janne PA. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- Umekita Y, Ohi Y, Sagara Y, Yoshida H. Co-expression of epidermal growth factor receptor and transforming growth factor-α predicts worse prognosis in breast-cancer patients. Int. J. Cancer. 2000;89:484–487. doi: 10.1002/1097-0215(20001120)89:6<484::aid-ijc3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Groffen J. Four human carcinoma cell lines with novel mutations in position 12 of c-K-ras oncogene. Nucleic Acids Res. 1986;14:843–852. doi: 10.1093/nar/14.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst EH, Murgia M, Treder M, Ullrich A. Anti-HER-3 MAbs inhibit HER-3-mediated signaling in breast cancer cell lines resistant to anti-HER-2 antibodies. Int. J. Cancer. 2005;115:519–527. doi: 10.1002/ijc.20867. [DOI] [PubMed] [Google Scholar]
- Wong ST, Winchell LF, McCune BK, Earp HS, Teixido J, Massague J, Herman B, Lee DC. The TGF-α precursor expressed on the cell surface binds to the EGF receptor on adjacent cells, leading to signal transduction. Cell. 1989;56:495–506. doi: 10.1016/0092-8674(89)90252-3. [DOI] [PubMed] [Google Scholar]
- Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J. Cell Biol. 2002;158:221–226. doi: 10.1083/jcb.200112026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zhou B-B, Fridman JS, Liu X, Friedman SM, Newton RC, Scherle PA. ADAM proteases, ErbB pathways and Cancer. Expert Opin. Investig. Drugs. 2005;14:591–606. doi: 10.1517/13543784.14.6.591. [DOI] [PubMed] [Google Scholar]
- Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.