Thompson et al. identify the dual specificity tyrosine-regulated kinase 1A (DYRK1A) as a new player in the control of lymphopoiesis. Loss of DYRK1A in mice results in Cyclin D3 stabilization and failure to repress E2F target genes, thus impairing cell cycle exit and proper pre–B and pre–T cell differentiation.
Abstract
Pre–B and pre–T lymphocytes must orchestrate a transition from a highly proliferative state to a quiescent one during development. Cyclin D3 is essential for these cells’ proliferation, but little is known about its posttranslational regulation at this stage. Here, we show that the dual specificity tyrosine-regulated kinase 1A (DYRK1A) restrains Cyclin D3 protein levels by phosphorylating T283 to induce its degradation. Loss of DYRK1A activity, via genetic inactivation or pharmacologic inhibition in mice, caused accumulation of Cyclin D3 protein, incomplete repression of E2F-mediated gene transcription, and failure to properly couple cell cycle exit with differentiation. Expression of a nonphosphorylatable Cyclin D3 T283A mutant recapitulated these defects, whereas inhibition of Cyclin D:CDK4/6 mitigated the effects of DYRK1A inhibition or loss. These data uncover a previously unknown role for DYRK1A in lymphopoiesis, and demonstrate how Cyclin D3 protein stability is negatively regulated during exit from the proliferative phases of B and T cell development.
B- and T-lymphocyte precursors follow analogous paths as they differentiate in the bone marrow and thymus, respectively: both progress through a defined sequence of developmental stages, during which entry into and exit from the cell cycle must be tightly and dynamically regulated (Rothenberg, 2014). A critical step in both pre–B and pre–T cell development is a clonal proliferative expansion after transient surface expression of a pre–B cell receptor (pre–BCR) or pre–T cell receptor (pre–TCR), indicating successful gene rearrangements at μ heavy chain or TCR-β loci, respectively (Muljo and Schlissel, 2000). After this burst of proliferation, pre–B and pre–T cells must then exit the cell cycle to allow further differentiation, namely the rearrangement of κ light or TCR-α chains en route to expressing a functional antigen receptor (Michie and Zuñiga-Pflucker, 2002; Clark et al., 2014).
One of the primary effectors of these processes is Cyclin D3, which plays essential and nonredundant roles in the proliferation of both pre–B and pre–T cells (Sicinska et al., 2003; Cooper et al., 2006; Sawai et al., 2012). The precise molecular mechanisms by which these cells transition from a proliferative state to a quiescent one are still being dissected. Transcriptional repression of Cyclin D3 (Mandal et al., 2009) and other cell cycle–associated genes (Hoffmann et al., 2002) occurs; however, little is known about the regulation of Cyclin D3 protein stability during this transition. The ubiquitin–proteasome system allows cells to rapidly diminish the quantity of certain proteins available for cell cycle progression. To initiate this mechanism, proteins must first be phosphorylated at specific residues within phosphodegrons (Ye et al., 2004). This phosphorylation facilitates polyubiquitylation of the proteins by ubiquitin ligases, which targets them for swift degradation by the proteasome (Teixeira and Reed, 2013). All three D-type Cyclins (D1, D2, and D3) contain phosphodegrons that can be targeted by various kinases to initiate protein turnover (Casanovas et al., 2004; Naderi et al., 2004; Låhne et al., 2006; Barbash et al., 2009); however, the identities and relative contributions of the kinases that specifically regulate Cyclin D3 stability during lymphoid development remain unclear.
Dual specificity tyrosine-regulated kinase 1A (DYRK1A) has been shown to phosphorylate more than 30 proteins to regulate diverse biological functions, including synaptic transmission (Xie et al., 2012; Chen et al., 2014), neurodegeneration (Wegiel et al., 2011), transcription (Gwack et al., 2006), mRNA splicing (de Graaf et al., 2006), proliferation (Hämmerle et al., 2011; Litovchick et al., 2011; Chen et al., 2013), and survival (Guo et al., 2010; Barallobre et al., 2014). DYRK1A phosphorylates Cyclin D1 on threonine 286 (T286) to promote its degradation and subsequent cell cycle arrest in developing neurons (Yabut et al., 2010; Soppa et al., 2014) and fibroblasts (Chen et al., 2013). Recent work in our laboratory uncovered a tumor-promoting role for DYRK1A in the megakaryocytic leukemia associated with Down syndrome (Malinge et al., 2012); this was the first report of DYRK1A’s importance in a hematopoietic cell type. To understand how DYRK1A functions during hematopoiesis, we conditionally inactivated the gene using the Mx1-Cre, Lck-Cre, and CD19-Cre LoxP systems. Here, we reveal that DYRK1A phosphorylates Cyclin D3 to decrease its stability in pre–B and pre–T cells and promote quiescence during the large-to-small pre–B, and double negative-to-double positive thymocyte transitions. Loss of DYRK1A results in Cyclin D3 stabilization and failure to repress E2F target genes, which ultimately impairs cell cycle exit and proper differentiation of pre–B and pre–T cells.
RESULTS
Dyrk1a is selectively required for lymphopoiesis
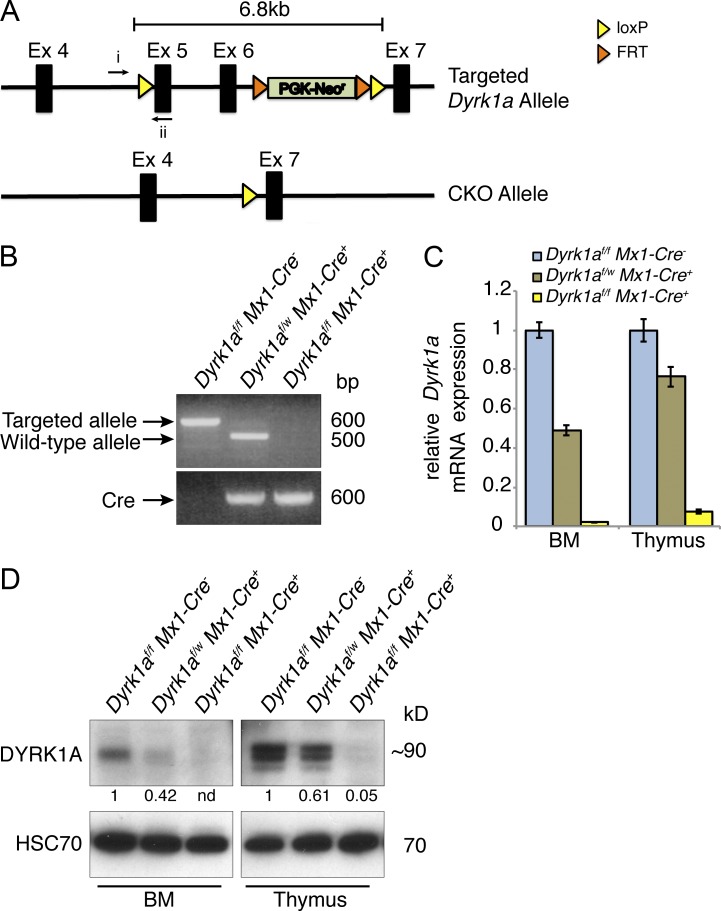
To achieve conditional inactivation of Dyrk1a, we generated a Dyrk1a allele with loxP sites flanking (floxed) exons 5 and 6, which encode an essential portion of the protein’s kinase domain (Fig. 1 A). The frameshift caused by loss of exons 5 and 6 allows for potential expression of a truncated 12.5-kD protein; however, if expressed it would lack most of the essential functional domains of DYRK1A.
Figure 1.
Conditional inactivation of the Dyrk1a gene. (A) Exons 5 and 6 were floxed in the targeted allele and excised in the conditional knockout (CKO) allele. (B) PCR from thymocyte genomic DNA was performed 2 wk after pI:pC treatment using the indicated primers in A (i and ii) and assessing the presence or loss of the targeted allele in Dyrk1af/f Mx1-Cre−, Dyrk1af/w Mx1-Cre+, and Dyrk1af/f Mx1-Cre1+,mice. (C) Dyrk1a mRNA expression measured by qRT-PCR using primers within the excised gene segment in bone marrow and thymus for the indicated mice. (D) Western blot shows DYRK1A protein expression in bone marrow and thymus after loss of 0, 1, or 2 Dyrk1a alleles. Densitometry values were normalized to HSC70. Loss of DYRK1A expression was verified for all subsequent experiments by qPCR, Western blot, or both. Data are derived from 1 litter and are representative of over 20 cohorts that were analyzed by RT-PCR and/or Western blot.
Mice with the floxed allele were crossed to Mx1-Cre transgenic mice to generate Dyrk1a conditional knockout (Dyrk1af/f Mx1-Cre+, referred to hereafter as CKO) animals. To induce Cre expression and subsequent excision at the Dyrk1a locus in all hematopoietic cells, we treated animals with four injections of polycytidylic:polyinosinic acid (pI:pC). Loss of DYRK1A expression in the bone marrow and thymus 2 wk after pI:pC was confirmed at the genomic (Fig. 1 B), mRNA (Fig. 1 C), and protein levels (Fig. 1 D).
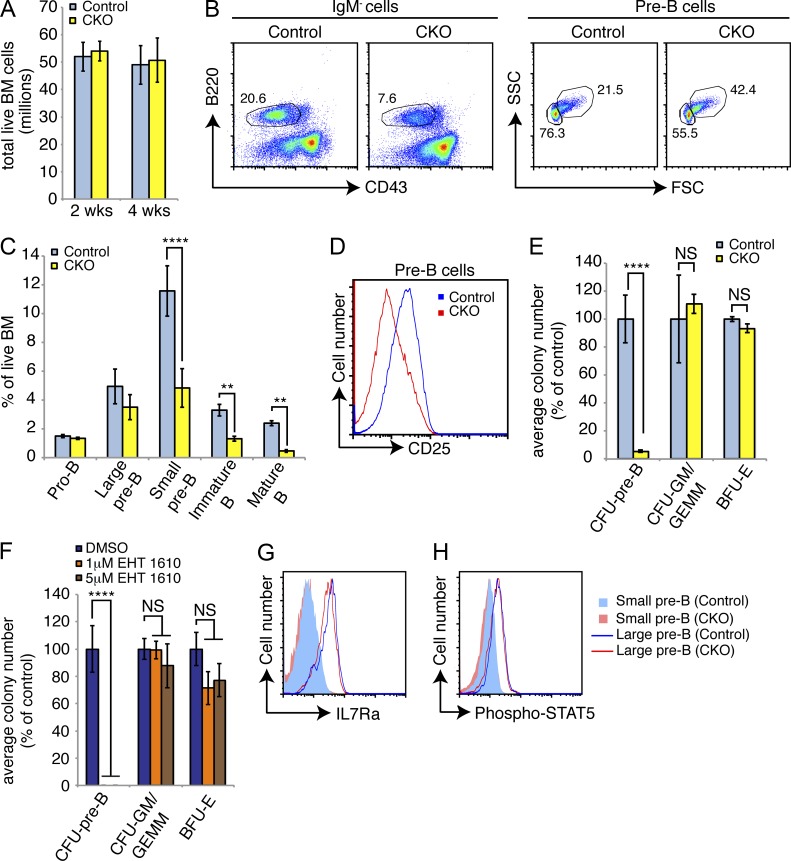
Peripheral blood B cell, but not neutrophil, numbers were reduced in CKO mice at 2 and 4 wk after pI:pC (unpublished data). In the bone marrow, total cellularity in the CKO mice was unchanged compared with controls at 2 and 4 wk after pI:pC (Fig. 2 A). Within the B cell lineage, the percentage of pro–B cells (IgM−/B220+/CD43+/cKit+) was unchanged; however, we observed a two- to threefold reduction in the percentage and absolute number of pre–B cells (IgM−/B220+/CD43low/−; Fig. 2, B and C). This population contains both rapidly cycling large pre–B cells (IgM−/B220+/CD43low/FSChigh) and quiescent small pre–B cells (IgM−/B220+/CD43−/FSClow). The reduced number of pre–B cells in CKO bone marrow primarily reflected a loss of small pre–B cells (Fig. 2, B and C), suggesting a defect in the large-to-small pre–B transition. Consistent with this prediction, CKO pre–B cells expressed lower surface levels of the differentiation marker CD25 (Fig. 2 D). Later stages of B cells were also reduced in numbers in CKO bone marrow, including immature B (IgM+/IgD−/B220+) and mature recirculating B cells (IgM+/IgD+/B220high; Fig. 2 C). In contrast, myeloid cells were not decreased after loss of Dyrk1a; in fact, the numbers of Gr1+/Mac1+ granulocytes modestly increased with time (unpublished data).
Figure 2.
Loss of Dyrk1a adversely affects B cell development. (A) Mean bone marrow cellularity was assessed in Dyrk1af/f Mx1-Cre− (Control) and Dyrk1af/f Mx1-Cre+ (CKO) mice 2 and 4 wk after pI:pC treatment; n = 6 mice per genotype, pooled from 3 independent cohorts of 2 mice per genotype. (B) Representative flow cytometry analysis of bone marrow for total pre–B (IgM− gate, left) and large/small pre–B (pre–B gate, right) cells 4 wk after pI:pC treatment is shown. Numbers indicate percentages in each gate; mean percentages of each population among total bone marrow cells were quantified in (C); n = 6 mice per genotype, pooled from 3 independent cohorts of 2 mice per genotype. (D) Representative flow cytometry analysis of pre–B cell surface CD25 expression in bone marrow from Control and CKO mice 2 wk after pI:pC treatment is shown; n = 6 mice per genotype, pooled from 3 independent cohorts of 2 mice per genotype. (E) Mean number of colony forming units from total bone marrow 2 wk after pI:pC treatment as percent of Control for each colony type was calculated; n = 3 mice per genotype, with duplicate plates for each mouse. Data are representative of three independent experiments. (F) The mean numbers and phenotypes of colonies formed by WT bone marrow in the presence of indicated concentrations of EHT 1610 were calculated. Results depict duplicate plates for each condition, and represent 3 independent experiments. (G) Surface IL7R-α expression (H) and intracellular STAT5 phosphorylation (Y694, C) in small and large pre–B cells from the bone marrow of Control and CKO mice 2 wk after pI:pC treatment were assessed by flow cytometry. Data are representative of 3three mice per genotype. For all graphs, error bars depict SD. **, P < 0.01; ****, P < 0.0001.
In accord with the lack of an effect on myeloid development, ex vivo myeloid colony-forming ability in cytokine-supplemented methylcellulose was similar in both control and CKO bone marrow in terms of colony number and morphology (Fig. 2 E). However, loss of Dyrk1a severely impaired pre–B colony formation in cultures containing IL-7 (Fig. 2 E). We observed the same phenomenon when we cultured WT bone marrow in the presence of the small molecule DYRK1A/B inhibitor, EHT 1610 (Foucourt et al., 2014a,b); pre–B colony formation was completely blocked at inhibitor concentrations that did not affect myeloid colony formation (Fig. 2 F). IL-7 signaling, which drives pre–B cell expansion both in vivo and in ex vivo colony assays, was intact, as loss of Dyrk1a did not markedly change surface IL7Rα expression (Fig. 2 G) or intracellular STAT5 activation (Fig. 2 H).
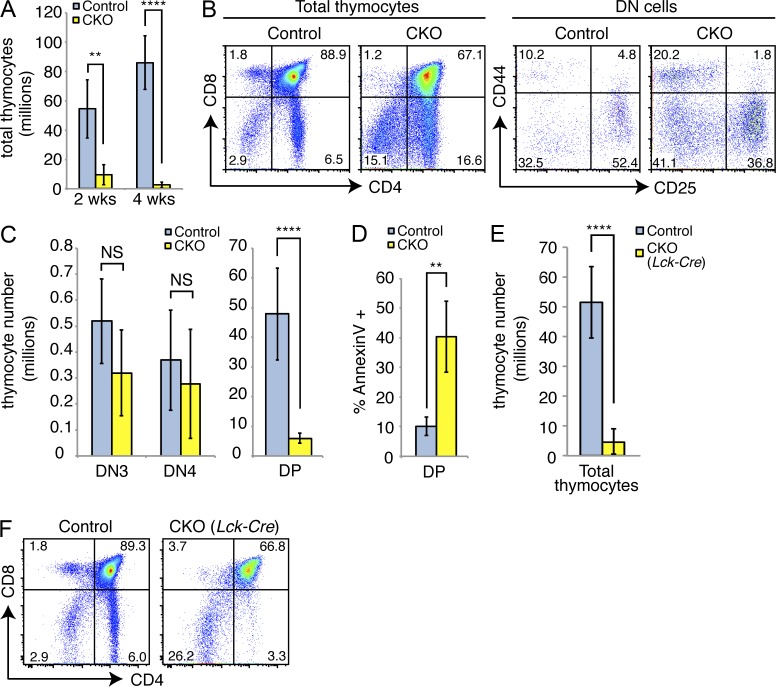
In addition to a loss of small pre–B cells in the bone marrow, thymic pre–T cells were also adversely affected in the CKO mice. Total thymocyte cellularity was reduced 5–10 fold after loss of Dyrk1a (Fig. 3 A). CD4−/CD8− double-negative (DN) cells were relatively enriched among the remaining CKO thymocytes compared with controls (Fig. 3 B). Similar to what we observed for the B lineage, proliferating DN3 (CD44−/CD25+) and DN4 (CD44−/CD25−) thymocytes were present in similar numbers to controls, but a substantial loss in cellularity occurred at the next stage of development, as thymocytes become CD4+/CD8+ double-positive (DP) cells and exit the cell cycle (Fig. 3 C). This loss was accompanied by a fourfold increase in apoptosis of DP thymocytes (Fig. 3 D). Apoptosis in pre–B cells was not affected, as measured by either Annexin V or cleaved Caspase 3 staining (unpublished data). Thus, the loss of Dyrk1a affects similar developmental stages within the B and T lineages, namely, the transition from a highly proliferative state to a quiescent one.
Figure 3.
Loss of Dyrk1a adversely affects T cell development. (A) Mean thymocyte numbers in Control and CKO mice were assessed 2 and 4 wk after pI:pC treatment; n = 6 mice per genotype, pooled from 3 independent cohorts of 2 mice per genotype. (B) Representative flow cytometry analysis of total (left) and CD4−/CD8− double-negative (DN, right) thymocytes is shown. Numbers indicate the percentages in each gate; mean absolute numbers were quantified in (C); n = 6 mice per genotype, pooled from 3 independent cohorts of 2 mice per genotype. (D) Mean percentage of Annexin V+ cells was assessed from CD4+CD8+ double-positive (DP) thymocytes 2 wk after pI:pC treatment; n = 4 mice per genotype. Data are representative of 2 independent cohorts of n = 4 mice per genotype. (E) Mean total thymocyte numbers and representative flow cytometry analysis (F) of thymi from Dyrk1awt/wt Lck-Cre+ (Control) and Dyrk1af/f Lck-Cre+ (CKO) mice are shown; n = 6 mice per genotype, pooled from 3 independent cohorts of 2 mice per genotype. **, P < 0.01; ****, P < 0.0001.
To validate our observations using a different Cre driver, we crossed Dyrk1af/f mice to the Lck-Cre strain, which induces gene excision beginning at the DN3 stage of thymic development and leads to nearly complete excision by the DP stage (Hennet et al., 1995). The thymi of Dyrk1af/f Lck-Cre+ mice mirrored those from Dyrk1af/f Mx1-Cre+ mice, showing marked hypocellularity and loss of DP thymocytes (Fig. 3, E and F). Separately, we crossed Dyrk1af/f mice to CD19-Cre mice to delete DYRK1A exclusively in the B lineage. Because this system is known to give inefficient excision at the pre–B cell stage (Hobeika et al., 2006), we crossed in a ROSA26lox-STOP-lox-EGFP allele (Mao et al., 2001) to follow the cells that have undergone Cre-mediated excision. Surprisingly, flow-purified EGFP+ pre–B cells from Dyrk1af/f CD19-Cre+/− EGFP+/− mice still expressed considerable levels of Dyrk1a (40–50% of that in control cells; unpublished data). The fact that Cre-expressing pre–B cells excised one locus (i.e., became EGFP+), but not another (i.e., retained Dyrk1a expression) indicates a selective pressure against loss of Dyrk1a in this system. In contrast, we typically achieved 95–99% reduction in Dyrk1a mRNA in CKO cells using Mx1-Cre or Lck-Cre (Fig. 1 C and not depicted).
The requirement for Dyrk1a is cell autonomous
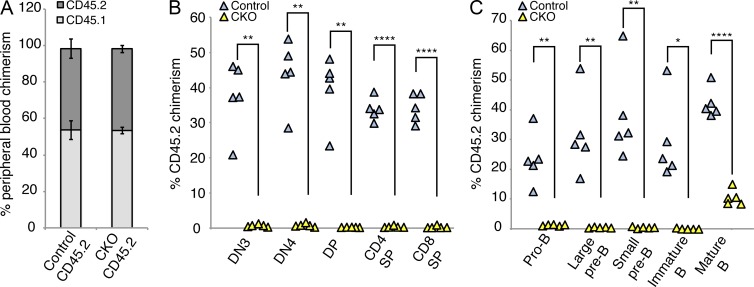
Because the Mx1-Cre system also results in deletion of target sequences within bone marrow stromal cells, we next investigated whether the phenotype of Dyrk1a loss is cell autonomous. To address this point, we performed competitive bone marrow transplantation experiments. CD45.2-expressing Dyrk1af/f Mx1-Cre− (Control) or Dyrk1af/f Mx1-Cre+ (CKO) bone marrow cells before pI:pC treatment were mixed 1:1 with congenic WT CD45.1 bone marrow cells and then transplanted into lethally irradiated CD45.1 recipient mice. Hematopoietic reconstitution of peripheral blood was confirmed 4 wk after transplant (Fig. 4 A). Subsequently, mice were treated with pI:pC to inactivate Dyrk1a in CKO cells, and analyzed 2 wk later. Whereas control CD45.2 cells provided 30–50% donor chimerism in the bone marrow and thymus, we observed no contribution to B or T cell development from CKO cells (Fig. 4, B and C). Transplantation of CKO cells at fourfold excess provided variable donor chimerism in pre–B cells and thymocytes, but total pre–B cell and thymocyte numbers were significantly reduced in these recipients, recapitulating the phenotypes described in Figs. 2 and 3 (unpublished data). These data confirm that loss of Dyrk1a causes a cell-autonomous defect in lymphoid development.
Figure 4.
Genetic deletion of Dyrk1a results in cell autonomous loss of competitiveness in lymphoid cells. Total bone marrow cells from either Dyrk1af/f Mx1-Cre− (Control CD45.2) or Dyrk1af/f Mx1-Cre+ (CKO CD45.2) were mixed with an equal number WT CD45.1 bone marrow cells and transplanted into lethally irradiated recipients, which were treated with pI:pC 4 wk later. (A) Mean peripheral blood chimerism (pre–pI:pC treatment) of transplanted mice analyzed by flow cytometry for surface CD45 isoform expression 4 wk after transplant is shown; n = 5 mice per group; error bars depict SD. (B and C) Donor chimerism in thymocytes (B) and bone marrow B cell populations (C) 2 wk after pI:pC treatment was assessed. Each triangle represents one mouse. n = 5 mice per group; error bars depict SD. Data represent two independent transplant experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Dyrk1a-deficient pre–B and pre–T cells fail to become quiescent
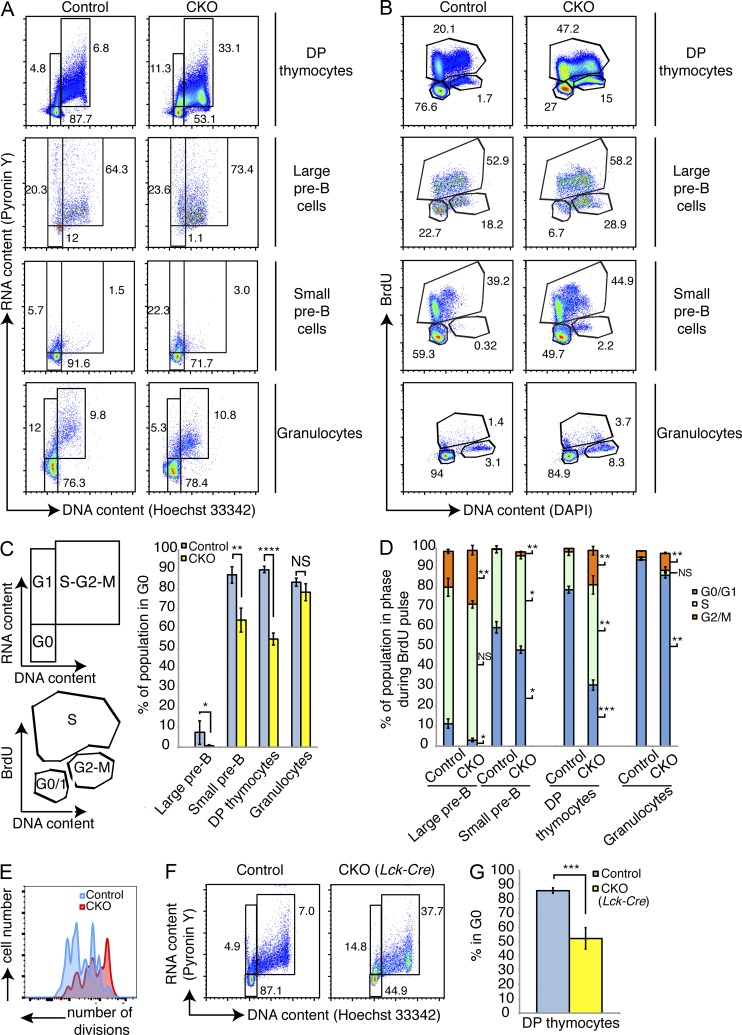
Given that Dyrk1a loss affected pre–B and pre–T cells at developmental stages that are characterized by cell cycle exit after a burst of proliferation, we wondered whether Dyrk1a regulates quiescence in these populations. Cell cycle analysis revealed that, although the vast majority of control pre–B and DP thymocytes were in the G0 phase, a substantial fraction of both populations remained in S-G2-M in CKO mice (Fig. 5, A–C). However, not all hematopoietic cell populations were similarly affected: for example, the G0 fraction of Gr1high/Mac1high mature granulocytes was not significantly changed after loss of Dyrk1a (Fig. 5, A and C). We confirmed these findings using intracellular Ki67 staining as a G0/G1 discriminator (unpublished data). Furthermore, BrdU incorporation experiments revealed multiple cell cycle abnormalities. CKO small pre–B cells and DP thymocytes were significantly enriched for cells that had entered S-phase (i.e., incorporated BrdU) during the pulse (Fig. 5 D). In addition, every population analyzed, including granulocytes, contained significantly more cells in G2-M at the end of the 24-h pulse (i.e., 4N, did not incorporate BrdU), indicating a delay in the completion of mitosis in DYRK1A-defcient cells (Fig. 5 D). We tested the ability of CKO pre–B cells to undergo progressive rounds of cell division by labeling FACS-purified large pre–B cells with CellTrace dye and monitoring dye dilution over time in cultures containing IL-7. Despite the increased steady-state fraction of cycling CKO large pre–B cells, they completed fewer cell divisions than control large pre–B cells (Fig. 5 E). We obtained similar results when culturing WT pre–B cells in the presence of EHT 1610 (unpublished data). Finally, thymocytes from Dyrk1af/f Lck-Cre+ mice (Fig. 5, F and G) displayed similar cell cycle abnormalities, demonstrating that the cell cycle defects we observed in the Mx1-Cre model were not simply due to an artifact of that system.
Figure 5.
Loss of Dyrk1a results in failed cell cycle exit during lymphoid development. Cell cycle status of bone marrow and thymocytes from Dyrk1af/f Mx1-Cre− (Control) and Dyrk1af/f Mx1-Cre+ (CKO) was assessed 4 wk after pI:pC treatment. (A and B) Representative flow cytometry plots depicting steady-state cell cycle status using DNA versus RNA content (A) and in vivo BrdU incorporation 24 h after BrdU injection (B) are shown. Numbers depict percentages in each gate. (C) The corresponding defined cell cycle phases and mean percentages in G0 are shown from the indicated Control and CKO cell types shown in A; n = 6 mice per genotype, pooled from 2 independent cohorts of 3 mice per genotype. (D) The mean percentages of cells in each phase of the cell cycle from the indicated Control and CKO cell types in B are shown; n = 4 mice per genotype. Data are representative of 2 independent experiments. Significant differences are denoted for each phase to the right of each CKO bar. (E) Flow cytometry was performed using CellTrace dye dilution in cultured FACS-purified Control and CKO large pre–B cells. Data are representative of two independent experiments. (F) Thymocytes from Control and CKO mice (Lck-Cre) were stained for DNA and RNA content to assess cell cycle status as in A. Representative flow cytometry plots for DP thymocytes from Control and CKO mice are shown; n = 4 mice per genotype, pooled from 2 independent cohorts of 2 mice per genotype. (G) Mean percentages of DP thymocytes in G0 are depicted; n = 4 mice per genotype, pooled from 2 independent cohorts of 2 mice per genotype. For all graphs, error bars depict SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant.
DYRK1A regulates expression of E2F target genes
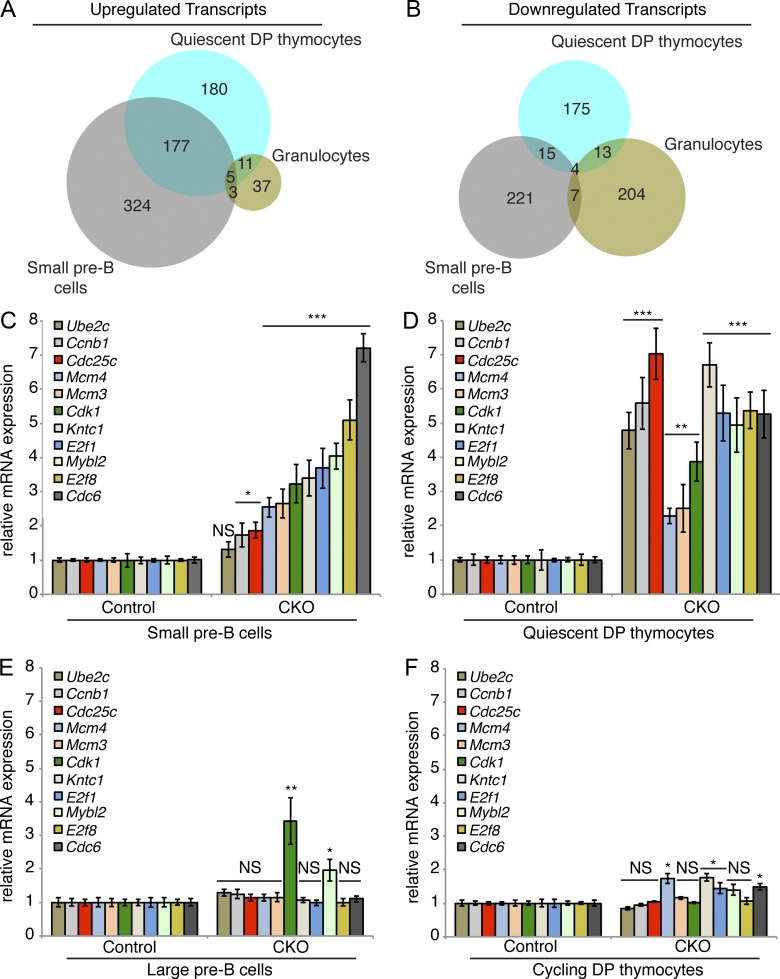
To gain insights into the mechanism by which Dyrk1a loss affects lymphopoiesis, we performed RNA-sequencing (RNA-seq) of multiple hematopoietic cell populations, including B cells, thymocytes, and mature granulocytes. We used fluorescence-activated cell sorting (FACS) to isolate five highly purified populations for RNA-seq analysis: quiescent DP thymocytes, cycling DP thymocytes, quiescent small pre–B cells, cycling large pre–B cells, and mature granulocytes. We generated lists of transcripts that were up- or down-regulated in CKO versus control cells, and then performed gene functional enrichment analysis using DAVID bioinformatics resources (Huang et al., 2007). Among the lists of up- and down-regulated transcripts in each of the five cell populations, only two populations showed enrichment of cell cycle, proliferative, and mitotic related terms: up-regulated transcripts in small pre–B cells, and up-regulated transcripts in quiescent DP thymocytes. In contrast, no significant enrichment of proliferative, mitotic, or cell cycle terms was observed in either the cycling cell populations or the granulocytes.
Among the up-regulated transcripts specific to small pre–B cells and quiescent DP thymocytes, 177 were shared (Fig. 6 A). In contrast, there were only 15 commonly down-regulated transcripts (Fig. 6 B). DAVID analysis of the 177 shared up-regulated transcripts revealed highly significant enrichments for terms associated with cell cycle, DNA replication, and mitosis. Furthermore, the top hit for known transcription factor binding sites within the promoters of the shared up-regulated genes was E2F. E2F factors are considered master regulators of cell cycle–dependent transcription: before cell cycle entry, Cyclin–CDK complexes phosphorylate the retinoblastoma protein (Rb), which releases activator E2Fs to induce transcription of genes necessary for cell cycle progression (Lees et al., 1993; Wu et al., 2001).
Figure 6.
Dyrk1a-deficient quiescent DP thymocytes and small pre–B cells fail to repress E2F target gene transcription. (A and B) Up- and down-regulated transcripts were identified by RNA-sequencing of FACS-purified quiescent DP thymocytes (CD4+/CD8+/DNA content2N/Pyronin Ylow), cycling DP thymocytes (CD4+/CD8+/DNA content>2N/Pyronin Yhigh), small pre–B cells (IgM−/B220+/CD43−/FSClow), large pre–B cells (IgM−/B220+/CD43low/FSChigh), and granulocytes (Gr-1high/Mac-1/CD11bhigh) from Dyrk1af/f Mx1-Cre− (Control) and Dyrk1af/f Mx1-Cre+ (CKO) mice. Venn diagrams depict shared up-regulated (A) and down-regulated (B) transcripts identified by RNA-sequencing in the three quiescent cell populations. (C–F) mRNA expression of E2F target genes was assessed by qRT-PCR in FACS-purified small pre–B cells (C), quiescent DP thymocytes (D), large pre–B cells (E), and cycling DP thymocytes (F) from Control and CKO mice 2 wk after pI:pC treatment. Transcript levels were normalized to Actb expression. PCRs were performed using 3 independently sorted pairs of samples (each with 1 mouse per genotype) for pre–B cells, and pooled samples from 3 mice per genotype for thymocytes. Error bars depict SD of triplicate wells for representative samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (CKO vs. Control).
The RNA-seq data indicated that E2F target genes are expressed at aberrantly high levels in Dyrk1a-deficient quiescent small pre–B cells and DP thymocytes. We validated these findings in independent cohorts of mice by sorting identical populations and interrogating the expression levels of E2F target genes by qRT-PCR. As expected, all E2F targets tested were expressed at higher levels in WT cycling cells compared with WT quiescent ones (unpublished data). Strikingly, E2F targets were de-repressed in CKO small pre–B cells (Fig. 6 C) and quiescent DP thymocytes (Fig. 6 D) compared with controls. In contrast, expression levels of these genes in CKO large pre–B cells (Fig. 6 E) and cycling DP thymocytes (Fig. 6 F) were similar to controls. Thus, loss of Dyrk1a leads to a failure to repress cell cycle–dependent gene transcription as pre–B and pre–T cells attempt to enter quiescence.
Loss of DYRK1A activity alters a subset of differentiation makers without affecting pre–BCR signaling
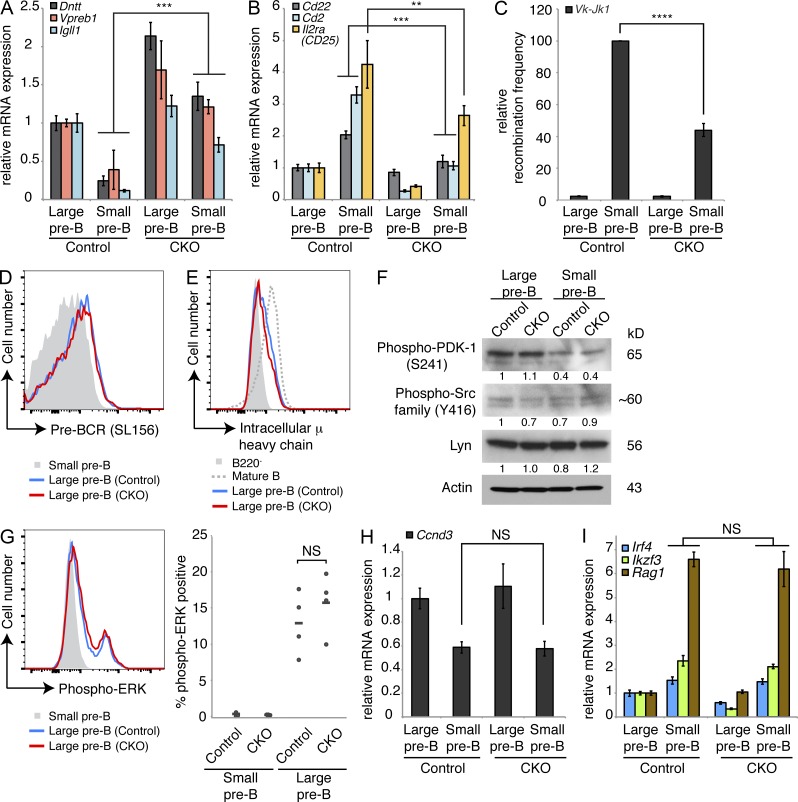
Cell cycle exit is tightly coupled to the differentiation of pre–B and pre–T cells. Thus, we next investigated whether failed cell cycle exit would coincide with failure to initiate transcriptional hallmarks of B cell differentiation in the absence of DYRK1A activity. Through our RNA-seq study, we noted that several transcripts that are normally repressed at the large-to-small pre–B transition (Parker et al., 2005; Schuh et al., 2008), including Dntt and the pre–BCR components Vpre–B1 and Igll1, remained elevated in CKO small pre–B cells (Fig. 7 A). Likewise, markers normally up-regulated during this transition (Schuh et al., 2008), such as Cd2, Cd22, and Il2ra (CD25), remained low in CKO small pre–B cells (Fig. 7 B). Quantitative PCR using genomic DNA from these cells also revealed lower amounts of Vκ-Jκ1 light chain rearrangement, indicating a failure to differentiate properly (Fig. 7 C).
Figure 7.
Loss of Dyrk1a alters pre–B cell differentiation markers without affecting pre–BCR signaling. (A and B) mRNA expression was analyzed by qRT-PCR to assess transcripts dynamically regulated during pre–B differentiation in FACS-purified large and small pre–B cells from the bone marrow of Dyrk1af/f Mx1-Cre− (Control) and Dyrk1af/f Mx1-Cre+ (CKO) 4 wk after pI:pC treatment. PCRs were performed using cDNA from 2–3 independently sorted pairs of mice, with 1 mouse per genotype in each pair. Error bars depict SD of triplicate wells for representative samples. (C) Vκ-Jκ1 light chain gene rearrangement was assessed by qPCR using genomic DNA from the same purified cell populations as in A and B; graph depicts mean recombination frequency as a percentage of Control small pre–B from two independent cohorts of mice, with cells pooled from two mice per genotype in each cohort; error bars depict SD. (D and E) Representative flow cytometry plots depicting surface pre–BCR (D) and intracellular μ heavy chain expression (E) in pre–B cells from the bone marrow of Control and CKO mice are shown. Data are representative of three independent cohorts of mice, each with two to three mice per genotype. (F) Western blots were performed to assess the levels of signaling proteins in FACS-purified large and small pre–B cells from the bone marrow of Control and CKO mice as indicated. Data are representative of two independently sorted samples, each pooled from three mice per genotype. Densitometry values were normalized to Actin. (G) ERK activation was assessed by intracellular staining for phospho-ERK in large and small pre–B cells from the bone marrow of Control and CKO mice as indicated in the flow cytometry plot (left); (right) individual (dots) and mean (bars) percentages of phospho-ERK+ cells in each population are shown; n = 4 mice per genotype. Data are representative of three independent experiments. (H and I) qRT-PCR analysis of Ccnd3 mRNA expression (H) and pre–BCR induced transcripts (I) from the same purified populations as in A and B were measured. All qRT-PCRs were performed using cDNA from two to three independently sorted pairs of mice, with one mouse per genotype in each pair. Error bars depict SD of triplicate wells for representative samples. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant.
Key signals that drive the large-to-small pre–B cell transition emanate from the pre–BCR (Clark et al., 2014). Therefore, it remained possible that impaired differentiation and cell cycle exit in CKO cells might result from altered pre–BCR signaling. We did not detect any differences in pre–BCR expression at the large pre–B stage in CKO mice, either by staining for surface pre–BCR (Fig. 7 D) or intracellular μ heavy chain (Fig. 7 E). Furthermore, we did not detect any changes in the activation of downstream Src family kinases or PDK-1 (activated via PI3K; Fig. 7 F). The ERK pathway—also activated by pre–BCR—was previously shown to promote cell cycle exit at this stage by repressing Cyclin D3 transcription (Mandal et al., 2009). ERK activation in CKO large pre–B cells was similar to, if not greater than, controls (Fig. 7 G), and Ccnd3 (Cyclin D3) transcript levels were repressed to the same extent as controls in CKO small pre–B cells (Fig. 7 H). Additionally, the pre–BCR–dependent up-regulation of Irf4, Ikzf3 (Aiolos), and Rag1 transcripts occurred normally in CKO small pre–B cells (Fig. 7 I). Thus, despite functional pre–BCR signaling, certain hallmarks of the differentiation program it induces cannot be fully enforced without prior DYRK1A-mediated cell cycle exit.
DYRK1A promotes quiescence via phosphorylation of Cyclin D3
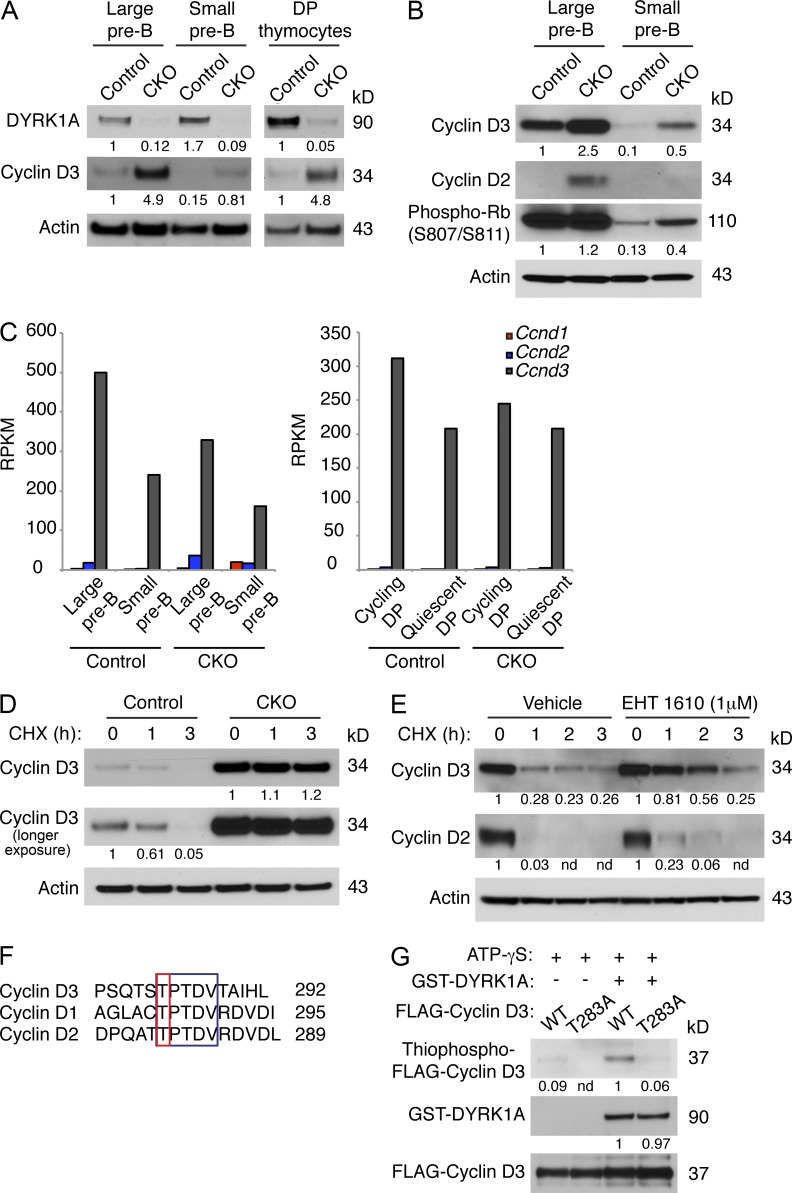
We next asked whether E2F deregulation in CKO cells might stem from increased Cyclin D–CDK4/6 activity. Cyclin D1, although regulated by DYRK1A (Chen et al., 2013; Soppa et al., 2014), is expressed at very low or undetectable levels during lymphoid development (Cooper et al., 2006; Fig. 8 C). Cyclin D3, however, is a critical mediator of proliferation in both pre–B and pre–T cells, and is highly expressed in both cell types (Sicinska et al., 2003; Cooper et al., 2006). We found a dramatic increase in Cyclin D3 protein level in CKO large and small pre–B cells, as well as in CKO thymocytes (Fig. 8 A), and this correlated with increased phosphorylation of Rb at S807/S811 (Fig. 8 B). Cyclin D2 protein, which is dispensable for pre–B cell development (Cooper et al., 2006) and not expressed in thymocytes after the DN3 stage (Sicinska et al., 2003), also accumulated in CKO large pre–B cells (Fig. 8 B), although its mRNA expression levels were much lower than for Cyclin D3 (Fig. 8 C). Despite the increases in Cyclin D3 protein, Ccnd3 transcript levels were not elevated in CKO pre–B cells or thymocytes (Figs. 7 H and 8 C). Accumulation of Cyclin D3 protein, but not transcript, suggests that it might be stabilized in the absence of DYRK1A. Indeed, cycloheximide pulse-chase experiments showed increased Cyclin D3 protein stability in CKO thymocytes (Fig. 8 D).
Figure 8.
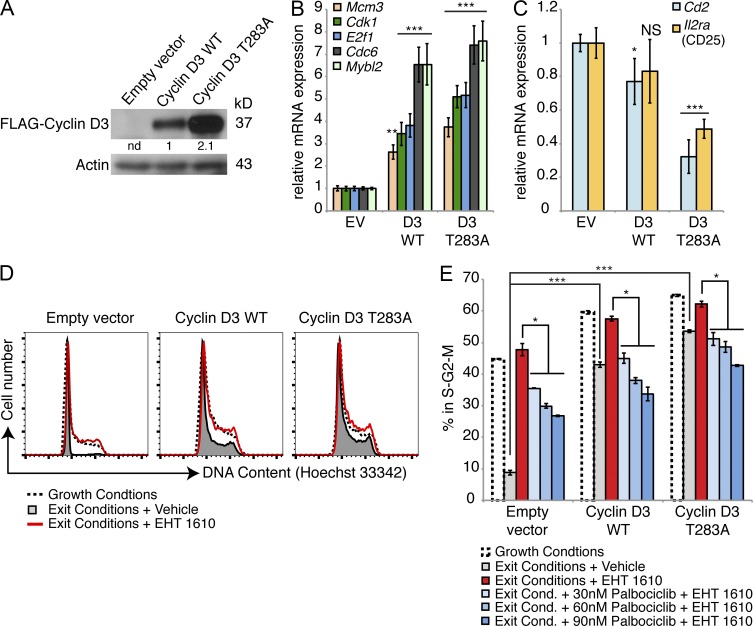
DYRK1A destabilizes Cyclin D3 by phosphorylating T283. (A and B) Western blots of FACS-purified large and small pre–B cells from Dyrk1af/f Mx1-Cre− (Control) and Dyrk1af/f Mx1-Cre+ (CKO) mice 4 wk after pI:pC treatment were assessed for the indicated proteins as shown. The same membrane (cut into segments according to molecular weight) from Fig. 7 F was stripped and reprobed, hence the same loading control is shown here. Data are representative of three independent experiments. Densitometry values were normalized to actin. (C) The expression levels of the D-type Cyclin transcripts from RNA-seq data (from Fig. 6) are shown for Control and CKO mice. (D and E) Control and CKO thymocytes (D), or WT cultured pre–B cells ± EHT 1610 (E) were treated with cycloheximide (25 µg/ml) for the indicated times before Western blot analysis. Data are representative of three independent experiments. Densitometry values were normalized to actin. (F) An amino acid alignment of C-terminal phosphodegrons in the D-type Cyclins is shown. Conserved residues are highlighted in the blue box; phosphorylated threonines are highlighted in the red box. (G) FLAG-tagged Cyclin D3 (WT or T283A) was immunoprecipitated from transfected 293T cells and used for in vitro kinase assays with recombinant active DYRK1A and ATP-γS. The reaction products were alkylated with PNBM and analyzed for thiophosphate esters by Western blotting. Data are representative of three independent assays. Densitometry values were normalized to FLAG-Cyclin D3.
Because the CKO cells contained much higher steady-state amounts of Cyclin D3, we also performed these experiments with WT pre–B cells treated acutely with EHT 1610. Here, the Cyclin D3 half-life was similarly prolonged in the presence of the inhibitor (Fig. 8 E). Cyclin D2 protein stability also increased after DYRK1A inhibition, but to a much lesser extent than Cyclin D3 (Fig. 8 E). As Cyclin D3 harbors a phosphodegron at its C terminus, centered on T283 (Casanovas et al., 2004), that is nearly identical to the one phosphorylated by DYRK1A in Cyclin D1 (Fig. 8 F), we asked whether DYRK1A destabilizes Cyclin D3 by phosphorylating this critical residue. Indeed, in vitro kinase assays demonstrated that DYRK1A can phosphorylate Cyclin D3 but not when T283 is mutated to alanine (Fig. 8 G). Together, these results indicate that DYRK1A destabilizes Cyclin D3 in pre–B and pre–T cells by phosphorylating T283.
To test whether enhanced Cyclin D3 stability was sufficient to prevent cell cycle exit and differentiation in pre–B cells, we transduced cultured pre–B cells with retroviral constructs encoding the empty vector, WT Cyclin D3, or Cyclin D3 T283A. As expected, the T283A mutant generated much higher steady-state Cyclin D3 expression levels than the WT gene in transduced pre–B cells (Fig. 9 A). We sorted transduced small pre–B cells to assess the levels of the transcripts that were aberrantly expressed in CKO small pre–B cells. Compared with empty vector-transduced cells, Cyclin D3 T283A-expressing small pre–B cells contained three- to sevenfold higher levels of the same E2F target transcripts that were de-repressed in CKO small pre–B cells (Fig. 9 B), and likewise had reduced levels of the differentiation markers that failed to be up-regulated in CKO small pre–B cells (Figs. 9 C and 7 B). Expression of the T283A mutant also resulted in increased cell size (unpublished data) and an increased proportion of cells in the S–G2–M phases of the cell cycle compared with cells expressing WT Cyclin D3 or empty vector (Fig. 9, D and E).
Figure 9.
Expression of Cyclin D3 T283A mimics loss of DYRK1A activity. (A) Protein expression levels of FLAG-tagged Cyclin D3 retroviral constructs in cultured WT pre–B cells 2 d after transduction were assessed. Densitometry values were normalized to Actin. (B and C) mRNA expression levels of E2F target genes (B) and differentiation markers (C) were assessed by qRT-PCR in FACS-purified WT small pre–B cells after transduction with the indicated constructs. RT-PCRs were performed from two independently sorted samples for each construct; error bars depict SD of triplicate wells for representative samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant (indicated condition vs. empty vector). (D) Cell cycle analysis was performed in cultured WT pre–B cells after 2 d of growth conditions (5 ng/ml IL-7 and 10 ng/ml SCF) (dotted line), cell cycle exit conditions (0.05 ng/ml IL-7 and 0.1 ng/ml SCF; shaded gray), or exit conditions in the presence of 500 nM EHT 1610 (red line). (E) Percentages of cells from (D) in S-G2-M under the indicated conditions are shown; error bars depict SD of duplicate wells from a representative of four independent experiments, two of which included Palbociclib treatment. *, P < 0.05; ***, P < 0.001.
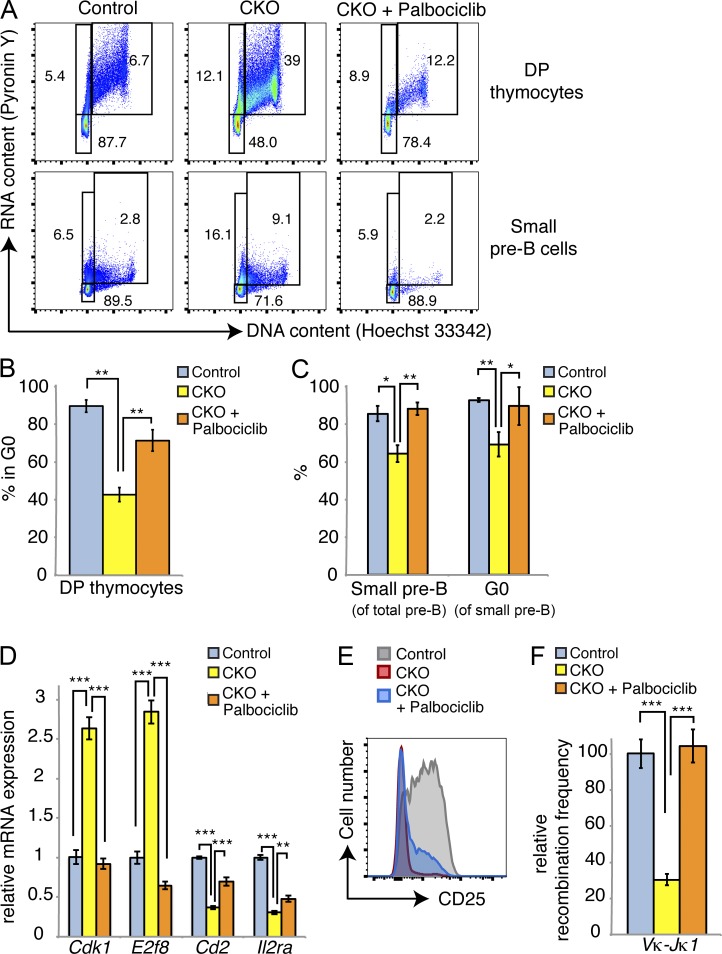
To induce cell cycle exit in our culture system, we replated transduced pre–B cells in medium containing 100-fold lower cytokine concentrations for 2 d. In empty vector–transduced cells, this yielded a population of 90% small pre–B cells that had exited the cell cycle (Fig. 9, D and E). In stark contrast, Cyclin D3 T283A-expressing cells failed to exit the cell cycle, with ∼40% of cells remaining in S-G2-M. We observed a similar block in cell cycle exit among empty vector–transduced cells treated with EHT 1610 during cytokine withdrawal; however, EHT 1610 treatment had much less of an effect on Cyclin D3 T283A-expressing cells, indicating that DYRK1A inhibition only minimally potentiates retention in S–G2–M within cells that already express stabilized Cyclin D3 (Fig. 9, D and E). Concomitant treatment with Palbociclib, a CDK4/6 inhibitor, mitigated the effects of EHT 1610 on cell cycle exit in both empty vector- and Cyclin D3 T283A-expressing cells (Fig. 9 E). This result confirms that the inability to exit the cell cycle after loss of DYRK1A activity occurs, at least in part, via a Cyclin D–CDK4/6-dependent mechanism. To extend these results to our in vivo model, we treated cohorts of Control and CKO mice with either 30 or 150 mg/kg Palbociclib daily for 1 wk, beginning 1 wk after the last pI:pC injection. We observed minimal differences in the mice treated with 30 mg/kg (not depicted), but saw significant restoration of cell cycle exit in CKO DP thymocytes and small pre–B cells at the 150 mg/kg dose (Fig. 10, A–C). The percentage of small pre–B cells relative to large ones was also restored to control levels in Palbociclib-treated CKO mice (Fig. 10 C). This coincided with decreased expression of E2F target genes (e.g., Cdk1 and E2f8) and increased expression of Cd2 and Il2ra in FACS-purified small pre–B cells (Fig. 10 D). Thus, Palbociclib treatment allowed CKO cells to both exit the cell cycle as well as initiate a small pre–B differentiation program, further supported by partially restored surface CD25 expression (Fig. 10 E) and completely restored frequency of κ light chain rearrangement (Fig. 10 F). Collectively, these data suggest that DYRK1A couples cell cycle exit to differentiation in developing lymphoid cells by targeting Cyclin D3 for degradation.
Figure 10.
Inhibition of CDK4/6 can rescue cell cycle exit and differentiation in Dyrk1a-defcient cells. (A) Mice of the indicated genotypes were treated with vehicle or 150 mg/kg Palbociclib (oral gavage) for 1 wk after completion of pI:pC treatment. Cell cycle analysis of DP thymocytes and bone marrow small pre–B cells was assessed. Flow cytometry plots represent two independent experiments, each with two to three mice per condition. (B and C) Quantifications of mean quiescent DP thymocyte and small pre–B cell populations are shown; n = 2–3 mice per genotype; error bars depict SD. Data represent two independent experiments, each with two to three mice per condition. (D) qRT-PCR analysis was performed for the indicated transcripts in FACS-purified small pre–B cells from the mice shown in B and C. Data show two pooled mice per condition; error bars depict SD of triplicate wells. Data are representative of two independent Palbociclib treatment experiments. (E) Flow cytometry plot depicts surface CD25 expression on bone marrow small pre–B cells from a representative mouse from each condition as indicated. Data represent two independent experiments, each with two to three mice per condition. (F) Light chain recombination frequencies were measured by qPCR from genomic DNA in FACS-purified bone marrow small pre–B cells from the indicated mice; each sample was pooled from 2 mice; data represent two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
An essential transition in lymphocyte development is the switch from a proliferative phase to quiescence. Our data uncover a previously unknown role for DYRK1A during this step of pre–B and pre–T cell development, wherein it directs cell cycle exit by phosphorylating T283 on Cyclin D3 to mark the protein for degradation. Although p38 (Casanovas et al., 2004) and GSK3β (Naderi et al., 2004), have been previously shown to phosphorylate T283 in cell lines, and T283-indpendent degradation of Cyclin D3 has also been reported (Låhne et al., 2006), ours is the first in vivo evidence that DYRK1A plays a major role in murine pre–B and pre–T development. A previous report found enhanced Cyclin D3 stability in cells that express a pre–BCR compared with those that do not (Cooper et al., 2006); here, we describe a novel regulatory mechanism by which stabilizing signals are antagonized by DYRK1A during these cells’ transit from proliferation to quiescence.
This study adds to a growing body of evidence that DYRK1A—and the DYRK family in general (Becker, 2012)—broadly influences cell cycle entry and exit in multiple cell types by phosphorylating both positive and negative cell cycle regulators, including Cyclin D1 (Chen et al., 2013), LIN-52 (Litovchick et al., 2011), p27 (Soppa et al., 2014), and now Cyclin D3. Underscoring its involvement in cell cycle regulation, the human DYRK1A promoter is itself bound and activated by E2F1 (Maenz et al., 2008), but whether this mechanism exists to initiate negative feedback on cell cycle entry or to facilitate cell cycle progression remains to be determined.
Given that DYRK1A phosphorylates multiple cell cycle regulators, we considered an alternative mechanism by which loss of DYRK1A activity might result in de-repression of E2F targets: a complex named DREAM (DP1, Rb-like, E2F, and MuvB) has been shown to silence E2F targets during quiescence using the repressor E2F4, and its assembly depends on phosphorylation of a MuvB subunit, LIN-52, on S28. DYRK1A can phosphorylate that site to promote DREAM assembly (Litovchick et al., 2011). Overexpression of WT LIN-52, nonphosphorylatable S28A, and phosphomimetic S28D mutants did not affect the cell cycle status of pre–B cells in our culture system in the presence or absence of EHT 1610 (unpublished data). Additionally, the phosphomimetic S28D mutant could not rescue the block in cell cycle exit caused by DYRK1A inhibition. Although our study cannot formally exclude the possibility that DYRK1A also phosphorylates LIN-52 and/or p27 during lymphoid development, it provides strong evidence that Cyclin D3 is the primary DYRK1A target in these cells. Nonetheless, it is interesting to consider that DYRK1A could potentially promote cell cycle exit via at least three distinct yet ultimately convergent mechanisms: by destabilizing D-type Cyclins, stabilizing p27, and promoting assembly of the DREAM complex via LIN-52 phosphorylation.
DYRK family kinases also phosphorylate NFAT transcription factors, triggering their export from the nucleus and thereby antagonizing NFAT activity (Gwack et al., 2006). Calcium signaling and NFAT activation play important roles in lymphocyte activation and differentiation (Peng et al., 2001); however, we did not detect any differences in NFATc2 phosphorylation in CKO pre–B cells (unpublished data). Furthermore, known NFAT target genes were not significantly enriched among up-regulated transcripts in any of the populations we analyzed by RNA-seq (unpublished data). Still, it remains possible that DYRK1A regulates NFATs at other stages of lymphoid development.
It is interesting that some outcomes of pre–BCR-induced differentiation seemed to occur normally amid failed cell cycle exit, yet others did not. For example, several studies have shown that pre–BCR signaling initiates a negative feedback loop, through which transcriptional up-regulation of Irf4 and Ikzf3 (Aiolos) leads to transcriptional silencing of the pre–BCR surrogate light chain components Vpre–B and λ5 (Parker et al., 2005; Thompson et al., 2007; Johnson et al., 2008). In Dyrk1a-deficient small pre–B cells, Vpre–B1 and Igll1 (λ5) transcript levels remained high, despite normal up-regulation of Irf4 and Ikzf3 mRNA. Likewise, these cells had less κ light chain gene rearrangement, despite normal up-regulation of Rag1 transcript. Thus, our data suggest that a portion of the pre–B cell differentiation program, such as down-regulation of surrogate light chain components, up-regulation of CD2 and CD25, and initiation of light chain rearrangements, requires E2F target gene repression and cell cycle exit to be fully enforced, whereas another subset of pre–BCR–induced changes, including up-regulation of IRF4, Aiolos, and Rag1, does not. We have demonstrated that inhibition of Cyclin D3–CDK4/6 activity in CKO pre–B and pre–T cells not only restores cell cycle exit by facilitating E2F target gene repression, but it also restores aspects of differentiation that require cell cycle exit, including expression of markers like CD2 and CD25, and κ light chain rearrangement. Thus, DYRK1A activity couples cell cycle exit to differentiation at these stages of development.
However, an interesting and somewhat paradoxical outcome of DYRK1A inactivation is our finding that, despite high levels of Cyclin D3, Dyrk1a-deficient pre–B cells show severely impaired proliferative capacity: they complete fewer cell divisions than control cells and fail to form colonies ex vivo, and cannot compete with WT cells in vivo. These data, combined with the fact that earlier stages of B and T cell differentiation are also impaired by loss of DYRK1A in mixed bone marrow chimera experiments (Fig. 4), suggest that DYRK1A regulates multiple aspects of lymphocyte differentiation, including some that are independent of Cyclin D3 phosphorylation, but nonetheless affect growth, mitotic progression, or both.
A recent study found an intriguing novel role for DYRK1A, in which it is recruited to specific promoters, including those of many growth-associated genes, to phosphorylate RNAP II and promote transcription (Di Vona et al., 2015). Thus, a potential connection to our study could be that DYRK1A performs a similar function in developing lymphocytes to facilitate growth and proliferation; indeed, that study found high expression levels of genes regulated by DYRK1A in this manner within lymphocyte populations. Additionally, DYRK1A might facilitate the mechanics of mitosis via interactions with cytoskeletal components: in accord with that idea, DYRK1A associates with multiple components of the cytoskeleton (Scales et al., 2009; Dowjat et al., 2012), and its overexpression can result in abnormal mitotic spindle formation and centrosome amplification (Funakoshi et al., 2003). Although the phosphorylation targets that mediate these effects of DYRK1A are unknown, it remains possible that such a mechanism underlies the delay in G2-M progression we observed in DYRK1A-deficient cells.
Finally, our study revisits a persistent question about the general nature of DYRK1A in cell growth and proliferation: is it an oncogene, a tumor suppressor, or both? Several studies have reported that DYRK1A promotes normal cell proliferation (Rachdi et al., 2014) and tumor growth (Malinge et al., 2012), prevents apoptosis (Laguna et al., 2013; Barallobre et al., 2014), and enables growth factor signaling (Aranda et al., 2008; Pozo et al., 2013). However, equally compelling studies have shown that DYRK1A suppresses proliferation (Hämmerle et al., 2011; Liu et al., 2014; Soppa et al., 2014), lengthens the time to cell cycle reentry (Chen et al., 2013), and promotes quiescence (Litovchick et al., 2011; Forristal et al., 2014). Our present work indicates that, even within the same cell type, DYRK1A can suppress as well as facilitate proliferation. The oncogene versus tumor suppressor question is particularly relevant to this study because we have shown that DYRK1A promotes cell cycle exit and differentiation during stages of lymphoid development that are susceptible to malignant transformation in acute lymphoblastic leukemia (ALL; Inaba et al., 2013). However, several lines of evidence suggest that DYRK1A is not a tumor suppressor, per se, in pre–B or pre–T cells. First, we have not detected any hematologic malignancies in our CKO mice that have been monitored for (thus far) 12 mo after loss of Dyrk1a. Second, our preliminary work has shown that ALL cells undergo growth inhibition and apoptosis when treated with DYRK1 inhibitors (unpublished data). Third, loss-of-function mutations in DYRK1A have not been reported in ALL; we have directly sequenced the gene in 12 ALL cell lines and did not detect any mutations, and our searches of public databases failed to turn up any DYRK1A mutations or silencing in ALL. In fact, among 3,505 sequenced primary hematopoietic tumor samples in the COSMIC database, only two DYRK1A mutations were found; both were in chronic lymphocytic leukemia (CLL), a mature B cell neoplasm. SIFT and PolyPhen software predict that both mutations (L261R and R300P) could be deleterious to protein function, but we are unable to say definitively how they might affect DYRK1A-mediated Cyclin D3 turnover without functional studies. Still, the overall scarcity of DYRK1A mutations in lymphoid leukemias suggests that DYRK1A activity is required for multiple aspects of lymphocyte biology, and that proliferative advantages afforded by loss of DYRK1A (via decreased Cyclin D3 turnover) might be offset by impairment of other DYRK1A-dependent pathways. Our mouse data are consistent with this idea: despite high levels of Cyclin D3 and increased cell cycle entry, CKO lymphoid cells proliferate less efficiently than control counterparts. Thus, one must consider not only cell type–specific functions of DYRK1A, but also the integration of multiple and potentially disparate functions within individual cell types.
Our study adds Cyclin D3 to a growing list of DYRK1A targets. We have demonstrated that normal lymphocyte development depends on DYRK1A activity and that its loss results in Cyclin D3 stabilization, failure to fully repress E2F target genes, and inability to enter quiescence. These findings enhance our understanding of cell cycle regulation during lymphocyte differentiation and identify a previously unknown mechanism by which developing lymphocytes enter quiescence.
MATERIALS AND METHODS
Mice.
Construction of the targeting vector to generate Dyrk1a conditional knockout (CKO) mice was performed as described previously (Liu et al., 2003). In brief, a 12,273-bp genomic region obtained from the C57BL/6 RP23 BAC library was introduced in the PL253 retrieval vector through homologous recombination. The loxP sites flanking exons 5 and 6, as well as a PGK-Neomycin cassette flanked by two FRT sites, were then subcloned in this retrieval vector. After electroporation, C57BL/6 ES clones resistant to G418 were screened for the targeted integration of the recombined allele by PCR (primer sequences available upon request). ES-recombined clones were then injected into Albino C57BL/6 blastocysts to generate chimeras. Two founding chimeric males yielded germline transmission and Mendelian proportions were verified by genotyping using primers forward, 5′-ATTACCTGGAGAAGAGGGCAAG-3′ and reverse 5′-TTCTTATGACTGGAATCGTCCC-3′ (primers i and ii in Fig. 1). For inducible pan-hematopoietic and B or T lymphocyte-specific inactivation of Dyrk1a gene, CKO mice were crossed with Mx1-Cre, CD19-Cre, and Lck-Cre strains, respectively; CD19-Cre CKO mice were subsequently crossed to ROSA26lox-STOP-lox-EGFP mice. Mx1-Cre, CD19-Cre, Lck-Cre, and ROSA26lox-STOP-lox-EGFP mice were obtained from The Jackson Laboratory. Mice were maintained in specific pathogen–free conditions. All in vivo experiments were approved by the Northwestern University Institutional Animal Care and Use Committee.
For all experiments, mice were analyzed between 5–10 wk of age. Littermate controls were used for each analysis cohort.
pI:pC administration.
To induce Cre expression in Mx1-Cre mice, animals received intraperitoneal injections of pI:pC (20 µg/gram of body weight; InvivoGen) every other day for a total of 4 injections. Injections began when mice were 3–6 wk of age.
Flow cytometry and cell sorting.
Single-cell suspensions of bone marrow and thymocytes were prepared and stained in FACS buffer (PBS + 3% FBS) containing fluorochrome-conjugated antibodies for surface markers, including B220 (RA3-6B2; eBioscience), CD43 (S7; BD), IgM (II/41; eBioscience), IgD (11-26c-2a; BD), cKit (2B8; eBioscience), IL7Ra (A7R34; eBioscience), CD25 (PC61.5; eBioscience), Gr1 (RB6-8C5; BD), Mac1/CD11b (M1/70; eBioscience), and pre–BCR (SL156; BD). Apoptotic cells were quantified using Annexin V (BD) and cleaved Caspase 3 (BioLegend) staining kits.
For intracellular staining, cells were stained for surface markers, washed with FACS buffer, fixed for 30 min with Cytofix fixation buffer (BD), washed, permeabilized with ice-cold Perm Buffer iii (BD), washed again, and then stained with fluorochrome-conjugated antibodies against intracellular proteins, including: phospho-STAT5 Y694 (D47E7; Cell Signaling Technology), phospho-ERK1/2 T202/Y204 (197G2; Cell Signaling Technology), or IgM (II/41; eBioscience).
For cell cycle analysis, cells were first incubated with 10 µg/ml Hoechst 33342 (Life Technologies) for 1 h at 37°C, washed, then stained for surface markers on ice for 30 min, washed, fixed for 30 min on ice, washed, and finally resuspended in FACS buffer containing 1 µg/ml Pyronin Y (Sigma-Aldrich) before analysis. Cells were analyzed on a LSRII cytometer (BD).
For cell cycle–based sorting, cells were first incubated with Hoechst 33342 (2 µg/ml) for 1 h at 37°C, washed, stained for surface markers on ice for 30 min, washed, and resuspended in FACS buffer containing 1 µg/ml Pyronin Y and 2 µg/ml Hoechst 33342. Quiescent (Pyronin Ylow, 2N DNA content) and cycling (Pyronin Yhigh, >2N DNA content) cells were purified using BD FACS Aria cell sorters. Gating strategies were similar to those depicted in Figs. 2 and 4 A.
BrdU incorporation.
Mice were given intraperitoneal injections of BrdU (150 mg) and sacrificed 24 h later. Bone marrow and thymocytes were stained for surface markers, and then analyzed for BrdU incorporation and DNA content using BrdU Flow kits (BD).
Colony assays.
For pre–B colony assays, 200,000 total bone marrow cells were plated in M3630 medium; for myeloid colony assays, 25,000 total bone marrow cells were plated in M3434 medium (both media from Stem Cell Technologies) and incubated at 37°C, 5% CO2. For inhibitor treatments, EHT 1610 or vehicle was added directly to the medium and vortexed vigorously before plating the cells. Colony numbers and phenotypes were scored 7 d later.
Library construction and sequencing.
Library construction and sequencing (100 bp, paired-end) were performed by Beijing Genomics Institute. The total RNA samples were first treated with DNase I and then enriched for mRNA with oligo(dT) magnetic beads. After fragmentation, first strand of cDNA was synthesized using random hexamer primer. Double-stranded cDNA was purified with magnetic beads and end reparation and 3′-end single nucleotide A (adenine) addition were then performed. Finally, sequencing adaptors were ligated to the fragments and DNA fragments were enriched by PCR amplification. Library products were sequenced on the Illumina HiSeq2000.
Transcriptome analysis.
The quality of DNA reads, in Fastq format, was evaluated using FastQC. Adapters were removed and reads of poor quality were filtered. The data were processed largely following the procedure described in Trapnell et al. (2012). In brief, the reads were aligned to the Mus musculus genome (mm10) using TopHat (v2.0.8b). Subsequently, the aligned reads, in conjunction with a gene annotation file for mm10 obtained from the UCSC website, were used to determine the expression of known genes using Cufflinks (v2.1.1). The individual transcript files generated by Cufflinks for each sample were merged into a single gene annotation file, which was then used to calculate RPKM values. Transcripts with greater than 5 RPKM in either CKO or control conditions were kept, and the log2 ratio (CKO:control) was calculated. Finally, transcripts with an absolute value of log2 ≤ 0.9 were removed. Sequencing data are available in the Gene Expression Omnibus under accession no. GSE67052.
Functional enrichment analysis.
All mRNA transcripts classified as up and down-regulated mRNA from each of the five cell populations were examined independently using DAVID (Huang et al., 2007). Additionally, to compare sets, the top 47 up or down-regulated transcripts were submitted to DAVID for each population and enriched terms were compared (47 was used because it was the minimum number of transcripts that were expressed at log2 > ±0.9).
qRT-PCR.
Total RNA was isolated using RNeasy kits (QIAGEN) and reverse transcribed using SuperScript III First Strand Synthesis kits (Life Technologies). Real-time PCR was performed using PerfeCTa SYBR Green (Quanta Biosciences) and gene-specific primers. Data were quantified using the standard curve method. Quantification of Vκ-Jκ1 gene rearrangements from genomic DNA was done as previously described (Johnson et al., 2008).
Western blotting.
Cells were lysed in TENT buffer (50 mM Tris, pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100) supplemented with 2 mM NaF, 2 mM NaVO3, 2 mM PMSF, and 1× cOmplete protease inhibitor cocktail (Roche) for 30 min on ice. Insoluble debris was pelleted by centrifugation at 21,000 g for 10 min at 4°C. Lysates were denatured in LDS sample loading buffer (Life Technologies) at 100°C for 5 min, and electrophoresed on 4–12% Bis-Tris gradient gels (Life Technologies). Proteins were transferred to PVDF membranes and probed with primary antibodies for: DYRK1A (7D10; Abnova), Cyclin D3 (C-16; Santa Cruz Biotechnology, Inc.), Cyclin D2 (M-20; Santa Cruz Biotechnology, Inc.), phospho-RB S807/811 (D20B12; Cell Signaling Technology), phospho-Src family Y461 (D49G4; Cell Signaling Technology), phospho-PDK1 S241 (C49H2; Cell Signaling Technology), Lyn (5G2, Cell Signaling Technology), phospho-NFATc2 S326 (sc-32994; Santa Cruz Biotechnology, Inc.), and FLAG (M2; Sigma-Aldrich) and detected with HRP-conjugated secondary antibodies and ECL substrate (GE Healthcare). β-Actin was detected using an HRP-conjugated primary antibody (C4; Santa Cruz Biotechnology, Inc.). Band densitometry values were calculated using ImageJ software.
Plasmids and site-directed mutagenesis.
Coding sequences for Cyclin D3 and LIN-52 were amplified by PCR from mouse pre–B cell cDNA using high-fidelity Pfu Ultra polymerase (Agilent Technologies) and ligated into the p3XFLAG-CMV7.1 expression vector (Sigma-Aldrich). Site-directed mutagenesis was performed using QuikChange kits (Agilent Technologies), and sequences were verified by direct DNA sequencing. 3xFLAG-tagged Cyclin D3 and LIN-52 cDNAs were amplified by PCR and subcloned into the MIGR1 retroviral vector.
Kinase assays.
Nonradioactive assays were performed as described previously (Allen et al., 2007). In brief, 293T cells were transfected with p3xFLAG-CMV7.1-Cyclin D3 WT or T283A and lysed 2 d later for immunoprecipitation with anti-FLAG affinity gel (Sigma-Aldrich). Bead-bound Cyclin D3 immunoprecipitates were washed extensively, and then added to kinase reactions containing a recombinant active fragment of hDYRK1A (Millipore), ATP-γS (Abcam), and kinase assay buffer (40 mM Tris, pH 7.5, 10 mM MgCl2, and 50 mM NaCl). Reactions were incubated on a rotator at room temperature for 45 min, before adding 25 mM p-nitrobenzyl mesylate (PNBM; Abcam) for 2 h to alkylate thiophosphates. Reaction products were subjected to Western blot and probed with an anti-thiophosphate ester antibody (51–8; Abcam) to visualize phosphorylated proteins.
Retrovirus production.
293T cells were grown to 70–80% confluence in 10-cm dishes and transfected with 10 µg of MIGR1 plasmid and 10 µg of ψEco packaging plasmid using Xtreme Gene 9 (Roche). Fresh medium was replaced after 12 h. Retrovirus-containing supernatants were collected 24-48 h after transfection, 0.45-µm filtered to remove cell debris, and frozen at −80°C until use. Thawed supernatants were used at a 2:1 ratio with fresh medium for transductions.
Pre–B cell cultures.
CD19+ cells were positively selected from suspensions of total bone marrow mononuclear cells using EasySep kits (Stem Cell Technologies), and expanded in DMEM (supplemented with 10% FBS [Hyclone], 2 mM l-glutamine, 10 mM Hepes, 1 mM sodium pyruvate, 1× Primocin [InvivoGen], and 55 µM β-mercaptoethanol) containing 5 ng/ml IL-7 and 10 ng/ml SCF (PeproTech) for 4–5 d, replating in fresh medium every 2 d. Retroviral transductions were performed by centrifuging the cells at 700 g for 1 h in the presence of retroviral supernatant and 8 µg/ml polybrene (Millipore), and then incubated for an additional 4–6 h before replating in fresh medium. Experiments were performed 2 d after transduction. For cell cycle exit experiments, cells were replated in either the above medium (growth conditions), or the same base medium containing 0.05 ng/ml IL-7 and 0.1 ng/ml SCF (exit conditions) for 2 additional days. Palbociclib was purchased from Selleckchem.
In vivo Palbociclib treatments.
Control and CKO mice were treated with pI:pC as described above; then, Palbociclib treatment was initiated one week after the last pI:pC injection. Mice received 30 or 150 mg/kg Palbociclib (in 50 mM sodium lactate) via oral gavage once daily for 7 d. Mice were analyzed 1 d after completion of the treatment protocol.
Statistical analysis.
Data were analyzed by Student’s t test (unpaired, two-tailed). Statistically significant differences are indicated in each figure, with corresponding p-values in each figure legend.
Acknowledgments
The authors wish to thank Drs. Barbara Kee and Alex Minella for helpful discussions.
This work was supported by grants from the National Institutes of Health (R01 CA101774), the Samuel Waxman Cancer Research Foundation, the Leukemia and Lymphoma Society, the Rally Foundation, and the Bear Necessities Foundation.
B. Leblond, L. Désiré, and A.S. Casagrande are current or former employees of Diaxonhit. The authors declare no additional competing financial interests.
Footnotes
Abbreviations used:
- CKO
- conditional KO
- CDK
- cyclin dependent kinase
- DN
- double negative
- DP
- double positive
- DYRK1A
- dual specificity tyrosine-regulated kinase 1A
- FACS
- fluorescence-activated cell sorting
References
- Allen J.J., Li M., Brinkworth C.S., Paulson J.L., Wang D., Hübner A., Chou W.H., Davis R.J., Burlingame A.L., Messing R.O., et al. 2007. A semisynthetic epitope for kinase substrates. Nat. Methods. 4:511–516. 10.1038/nmeth1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda S., Alvarez M., Turró S., Laguna A., and de la Luna S.. 2008. Sprouty2-mediated inhibition of fibroblast growth factor signaling is modulated by the protein kinase DYRK1A. Mol. Cell. Biol. 28:5899–5911. 10.1128/MCB.00394-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barallobre M.J., Perier C., Bové J., Laguna A., Delabar J.M., Vila M., and Arbonés M.L.. 2014. DYRK1A promotes dopaminergic neuron survival in the developing brain and in a mouse model of Parkinson’s disease. Cell Death Dis. 5:e1289 10.1038/cddis.2014.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash O., Egan E., Pontano L.L., Kosak J., and Diehl J.A.. 2009. Lysine 269 is essential for cyclin D1 ubiquitylation by the SCF(Fbx4/alphaB-crystallin) ligase and subsequent proteasome-dependent degradation. Oncogene. 28:4317–4325. 10.1038/onc.2009.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W. 2012. Emerging role of DYRK family protein kinases as regulators of protein stability in cell cycle control. Cell Cycle. 11:3389–3394. 10.4161/cc.21404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanovas O., Jaumot M., Paules A.B., Agell N., and Bachs O.. 2004. P38SAPK2 phosphorylates cyclin D3 at Thr-283 and targets it for proteasomal degradation. Oncogene. 23:7537–7544. 10.1038/sj.onc.1208040 [DOI] [PubMed] [Google Scholar]
- Chen J.Y., Lin J.R., Tsai F.C., and Meyer T.. 2013. Dosage of Dyrk1a shifts cells within a p21-cyclin D1 signaling map to control the decision to enter the cell cycle. Mol. Cell. 52:87–100. 10.1016/j.molcel.2013.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.K., Bregere C., Paluch J., Lu J.F., Dickman D.K., and Chang K.T.. 2014. Activity-dependent facilitation of Synaptojanin and synaptic vesicle recycling by the Minibrain kinase. Nat. Commun. 5:4246 10.1038/ncomms5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.R., Mandal M., Ochiai K., and Singh H.. 2014. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 14:69–80. 10.1038/nri3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.B., Sawai C.M., Sicinska E., Powers S.E., Sicinski P., Clark M.R., and Aifantis I.. 2006. A unique function for cyclin D3 in early B cell development. Nat. Immunol. 7:489–497. 10.1038/ni1324 [DOI] [PubMed] [Google Scholar]
- de Graaf K., Czajkowska H., Rottmann S., Packman L.C., Lilischkis R., Lüscher B., and Becker W.. 2006. The protein kinase DYRK1A phosphorylates the splicing factor SF3b1/SAP155 at Thr434, a novel in vivo phosphorylation site. BMC Biochem. 7:7 10.1186/1471-2091-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Vona C., Bezdan D., Islam A.B., Salichs E., López-Bigas N., Ossowski S., and de la Luna S.. 2015. Chromatin-wide profiling of DYRK1A reveals a role as a gene-specific RNA polymerase II CTD kinase. Mol. Cell. 57:506–520. 10.1016/j.molcel.2014.12.026 [DOI] [PubMed] [Google Scholar]
- Dowjat K., Adayev T., Kaczmarski W., Wegiel J., and Hwang Y.W.. 2012. Gene dosage-dependent association of DYRK1A with the cytoskeleton in the brain and lymphocytes of down syndrome patients. J. Neuropathol. Exp. Neurol. 71:1100–1112. 10.1097/NEN.0b013e31827733c8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forristal C., Henley S.A., MacDonald J.I., Bush J.R., Ort C., Passos D.T., Talluri S., Ishak C.A., Thwaites M.J., Norley C.J., et al. 2014. Loss of the mammalian DREAM complex deregulates chondrocyte proliferation. Mol. Cell. Biol. 34:2221–2234. 10.1128/MCB.01523-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucourt A., Hédou D., Dubouilh-Benard C., Désiré L., Casagrande A.S., Leblond B., Loäec N., Meijer L., and Besson T.. 2014a. Design and synthesis of thiazolo[5,4-f]quinazolines as DYRK1A inhibitors, part I. Molecules. 19:15546–15571. 10.3390/molecules191015546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucourt A., Hédou D., Dubouilh-Benard C., Girard A., Taverne T., Casagrande A.S., Désiré L., Leblond B., and Besson T.. 2014b. Design and synthesis of thiazolo[5,4-f]quinazolines as DYRK1A inhibitors, part II. Molecules. 19:15411–15439. 10.3390/molecules191015411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi E., Hori T., Haraguchi T., Hiraoka Y., Kudoh J., Shimizu N., and Ito F.. 2003. Overexpression of the human MNB/DYRK1A gene induces formation of multinucleate cells through overduplication of the centrosome. BMC Cell Biol. 4:12 10.1186/1471-2121-4-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Williams J.G., Schug T.T., and Li X.. 2010. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J. Biol. Chem. 285:13223–13232. 10.1074/jbc.M110.102574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwack Y., Sharma S., Nardone J., Tanasa B., Iuga A., Srikanth S., Okamura H., Bolton D., Feske S., Hogan P.G., and Rao A.. 2006. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 441:646–650. 10.1038/nature04631 [DOI] [PubMed] [Google Scholar]
- Hämmerle B., Ulin E., Guimera J., Becker W., Guillemot F., and Tejedor F.J.. 2011. Transient expression of Mnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing NOTCH signaling. Development. 138:2543–2554. 10.1242/dev.066167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T., Hagen F.K., Tabak L.A., and Marth J.D.. 1995. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc. Natl. Acad. Sci. USA. 92:12070–12074. 10.1073/pnas.92.26.12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., and Reth M.. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794. 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann R., Seidl T., Neeb M., Rolink A., and Melchers F.. 2002. Changes in gene expression profiles in developing B cells of murine bone marrow. Genome Res. 12:98–111. 10.1101/gr.201501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., and Lempicki R.A.. 2007. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35(Web Server):W169-75 10.1093/nar/gkm415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba H., Greaves M., and Mullighan C.G.. 2013. Acute lymphoblastic leukaemia. Lancet. 381:1943–1955. 10.1016/S0140-6736(12)62187-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Hashimshony T., Sawai C.M., Pongubala J.M., Skok J.A., Aifantis I., and Singh H.. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 28:335–345. 10.1016/j.immuni.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Laguna A., Barallobre M.J., Marchena M.A., Mateus C., Ramírez E., Martínez-Cue C., Delabar J.M., Castelo-Branco M., de la Villa P., and Arbonés M.L.. 2013. Triplication of DYRK1A causes retinal structural and functional alterations in Down syndrome. Hum. Mol. Genet. 22:2775–2784. 10.1093/hmg/ddt125 [DOI] [PubMed] [Google Scholar]
- Låhne H.U., Kloster M.M., Lefdal S., Blomhoff H.K., and Naderi S.. 2006. Degradation of cyclin D3 independent of Thr-283 phosphorylation. Oncogene. 25:2468–2476. 10.1038/sj.onc.1209278 [DOI] [PubMed] [Google Scholar]
- Lees J.A., Saito M., Vidal M., Valentine M., Look T., Harlow E., Dyson N., and Helin K.. 1993. The retinoblastoma protein binds to a family of E2F transcription factors. Mol. Cell. Biol. 13:7813–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L., Florens L.A., Swanson S.K., Washburn M.P., and DeCaprio J.A.. 2011. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 25:801–813. 10.1101/gad.2034211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., and Copeland N.G.. 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 13:476–484. 10.1101/gr.749203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Liu N., Zang S., Liu H., Wang P., Ji C., and Sun X.. 2014. Tumor suppressor DYRK1A effects on proliferation and chemoresistance of AML cells by downregulating c-Myc. PLoS ONE. 9:e98853 10.1371/journal.pone.0098853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenz B., Hekerman P., Vela E.M., Galceran J., and Becker W.. 2008. Characterization of the human DYRK1A promoter and its regulation by the transcription factor E2F1. BMC Mol. Biol. 9:30 10.1186/1471-2199-9-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinge S., Bliss-Moreau M., Kirsammer G., Diebold L., Chlon T., Gurbuxani S., and Crispino J.D.. 2012. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J. Clin. Invest. 122:948–962. 10.1172/JCI60455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M., Powers S.E., Ochiai K., Georgopoulos K., Kee B.L., Singh H., and Clark M.R.. 2009. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat. Immunol. 10:1110–1117. 10.1038/ni.1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Fujiwara Y., Chapdelaine A., Yang H., and Orkin S.H.. 2001. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 97:324–326. 10.1182/blood.V97.1.324 [DOI] [PubMed] [Google Scholar]
- Michie A.M., and Zúñiga-Pflücker J.C.. 2002. Regulation of thymocyte differentiation: pre-TCR signals and beta-selection. Semin. Immunol. 14:311–323. 10.1016/S1044-5323(02)00064-7 [DOI] [PubMed] [Google Scholar]
- Muljo S.A., and Schlissel M.S.. 2000. Pre-B and pre-T-cell receptors: conservation of strategies in regulating early lymphocyte development. Immunol. Rev. 175:80–93. 10.1111/j.1600-065X.2000.imr017509.x [DOI] [PubMed] [Google Scholar]
- Naderi S., Gutzkow K.B., Låhne H.U., Lefdal S., Ryves W.J., Harwood A.J., and Blomhoff H.K.. 2004. cAMP-induced degradation of cyclin D3 through association with GSK-3beta. J. Cell Sci. 117:3769–3783. 10.1242/jcs.01210 [DOI] [PubMed] [Google Scholar]
- Parker M.J., Licence S., Erlandsson L., Galler G.R., Chakalova L., Osborne C.S., Morgan G., Fraser P., Jumaa H., Winkler T.H., et al. 2005. The pre-B-cell receptor induces silencing of VpreB and lambda5 transcription. EMBO J. 24:3895–3905. 10.1038/sj.emboj.7600850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.L., Gerth A.J., Ranger A.M., and Glimcher L.H.. 2001. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity. 14:13–20. 10.1016/S1074-7613(01)00085-1 [DOI] [PubMed] [Google Scholar]
- Pozo N., Zahonero C., Fernández P., Liñares J.M., Ayuso A., Hagiwara M., Pérez A., Ricoy J.R., Hernández-Laín A., Sepúlveda J.M., and Sánchez-Gómez P.. 2013. Inhibition of DYRK1A destabilizes EGFR and reduces EGFR-dependent glioblastoma growth. J. Clin. Invest. 123:2475–2487. 10.1172/JCI63623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachdi L., Kariyawasam D., Aiello V., Herault Y., Janel N., Delabar J.M., Polak M., and Scharfmann R.. 2014. Dyrk1A induces pancreatic beta cell mass expansion and improves glucose tolerance. Cell Cycle. 13:2221–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E.V. 2014. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 32:283–321. 10.1146/annurev-immunol-032712-100024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai C.M., Freund J., Oh P., Ndiaye-Lobry D., Bretz J.C., Strikoudis A., Genesca L., Trimarchi T., Kelliher M.A., Clark M., et al. 2012. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer Cell. 22:452–465. 10.1016/j.ccr.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales T.M., Lin S., Kraus M., Goold R.G., and Gordon-Weeks P.R.. 2009. Nonprimed and DYRK1A-primed GSK3 beta-phosphorylation sites on MAP1B regulate microtubule dynamics in growing axons. J. Cell Sci. 122:2424–2435. 10.1242/jcs.040162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh W., Meister S., Herrmann K., Bradl H., and Jäck H.M.. 2008. Transcriptome analysis in primary B lymphoid precursors following induction of the pre-B cell receptor. Mol. Immunol. 45:362–375. 10.1016/j.molimm.2007.06.154 [DOI] [PubMed] [Google Scholar]
- Sicinska E., Aifantis I., Le Cam L., Swat W., Borowski C., Yu Q., Ferrando A.A., Levin S.D., Geng Y., von Boehmer H., and Sicinski P.. 2003. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 4:451–461. 10.1016/S1535-6108(03)00301-5 [DOI] [PubMed] [Google Scholar]
- Soppa U., Schumacher J., Florencio Ortiz V., Pasqualon T., Tejedor F.J., and Becker W.. 2014. The Down syndrome-related protein kinase DYRK1A phosphorylates p27(Kip1) and Cyclin D1 and induces cell cycle exit and neuronal differentiation. Cell Cycle. 13:2084–2100. 10.4161/cc.29104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira L.K., and Reed S.I.. 2013. Ubiquitin ligases and cell cycle control. Annu. Rev. Biochem. 82:387–414. 10.1146/annurev-biochem-060410-105307 [DOI] [PubMed] [Google Scholar]
- Thompson E.C., Cobb B.S., Sabbattini P., Meixlsperger S., Parelho V., Liberg D., Taylor B., Dillon N., Georgopoulos K., Jumaa H., et al. 2007. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 26:335–344. 10.1016/j.immuni.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J., Gong C.X., and Hwang Y.W.. 2011. The role of DYRK1A in neurodegenerative diseases. FEBS J. 278:236–245. 10.1111/j.1742-4658.2010.07955.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Timmers C., Maiti B., Saavedra H.I., Sang L., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., et al. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 414:457–462. 10.1038/35106593 [DOI] [PubMed] [Google Scholar]
- Xie W., Adayev T., Zhu H., Wegiel J., Wieraszko A., and Hwang Y.W.. 2012. Activity-dependent phosphorylation of dynamin 1 at serine 857. Biochemistry. 51:6786–6796. 10.1021/bi2017798 [DOI] [PubMed] [Google Scholar]
- Yabut O., Domogauer J., and D’Arcangelo G.. 2010. Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J. Neurosci. 30:4004–4014. 10.1523/JNEUROSCI.4711-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Nalepa G., Welcker M., Kessler B.M., Spooner E., Qin J., Elledge S.J., Clurman B.E., and Harper J.W.. 2004. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 279:50110–50119. 10.1074/jbc.M409226200 [DOI] [PubMed] [Google Scholar]