Wilson et al. show that individuals with loss-of-function mutations in STAT3 have reduced numbers of peripheral blood MAIT and NKT cells, but not γδ T cells. Residual MAIT cells had normal expression of RORγt, but displayed impaired secretion of IL-17A and IL-17F.
Abstract
Unconventional T cells such as γδ T cells, natural killer T cells (NKT cells) and mucosal-associated invariant T cells (MAIT cells) are a major component of the immune system; however, the cytokine signaling pathways that control their development and function in humans are unknown. Primary immunodeficiencies caused by single gene mutations provide a unique opportunity to investigate the role of specific molecules in regulating human lymphocyte development and function. We found that individuals with loss-of-function mutations in STAT3 had reduced numbers of peripheral blood MAIT and NKT but not γδ T cells. Analysis of STAT3 mosaic individuals revealed that this effect was cell intrinsic. Surprisingly, the residual STAT3-deficient MAIT cells expressed normal levels of the transcription factor RORγt. Despite this, they displayed a deficiency in secretion of IL-17A and IL-17F, but were able to secrete normal levels of cytokines such as IFNγ and TNF. The deficiency in MAIT and NKT cells in STAT3-deficient patients was mirrored by loss-of-function mutations in IL12RB1 and IL21R, respectively. Thus, these results reveal for the first time the essential role of STAT3 signaling downstream of IL-23R and IL-21R in controlling human MAIT and NKT cell numbers.
Unconventional T cells possess unique properties that set them apart from conventional T cells. These include limited TCR diversity and a preactivated phenotype allowing them to respond rapidly upon activation and recognition of antigens (Ags) that are not presented by classical MHC molecules. Unconventional T cells include γδ T cells, mucosal-associated invariant T cells (MAIT cells), and NKT cells. These cells, particularly MAIT and γδ T cells, are a significant component of the human immune system, comprising up to 15% of peripheral blood lymphocytes, and in mouse models can provide protection against multiple pathogens (Gold and Lewinsohn, 2013; Chien et al., 2014). However, our knowledge of their precise role in protective immunity in humans and the molecular mechanisms regulating their development and function is limited. This is particularly true for MAIT cells, which until recently have been largely overlooked in studies of human disease and difficult to detect in mice because of a lack of appropriate reagents (Le Bourhis et al., 2013; Reantragoon et al., 2013). As a result, almost nothing is known of the obligate signals required for their development and function.
Several distinct populations of NKT cells have been identified, including type I NKT cells (also called invariant NKT [iNKT] cells) that in humans typically express an invariant Vα24-Jα18 TCR α-chain paired with Vβ11 TCR β-chain. These iNKT cells recognize self, synthetic, and bacterial glycolipids presented by CD1d (Rossjohn et al., 2012). Other NKT cell populations (type II and atypical) express a more diverse TCR repertoire (Rossjohn et al., 2012) and do not respond to the prototypical type I NKT cell ligand, α-galactosylceramide, making them more difficult to identify and thus study. MAIT cells are enriched in the mucosa. They typically express the invariant Vα7.2-Jα33 TCRα chain and are restricted by the MHC-related molecule 1 (MR1; Gold and Lewinsohn, 2013). MR1 binds vitamin B metabolites, produced by multiple microorganisms including Candida albicans and Staphylococcus aureus, and presents them to MAIT cells (Kjer-Nielsen et al., 2012). γδ T cells are more diverse than type I NKT or MAIT cells, with humans possessing 8 Vδ and 6 Vγ functional TCR chains. γδ TCRs bind a range of Ags and Ag-presenting molecules, including CD1 and MHC-I stress–induced molecules (Bonneville et al., 2010; Chien et al., 2014). Certain chains are preferentially expressed and paired. Thus, in the blood ∼70% of γδ T cells express Vγ9Vδ2 and the remainder are mostly Vδ1+ (Parker et al., 1990; Bonneville et al., 2010; Chien et al., 2014). These different δ-chain–expressing subsets have been associated with immune responses to different pathogens. For example, δ2+ cells respond to Mycobacterium tuberculosis (Li et al., 1996), whereas δ1+ cells are involved in responses to C. albicans (Fenoglio et al., 2009). Despite differences in TCR gene usage and mode of recognition of distinct Ags, a common feature of these unconventional T cell populations is their ability to promptly produce a broad range of effector cytokines, IFNγ, IL-4, IL-17, and IL-21, after activation (Bonneville et al., 2010; Dusseaux et al., 2011; Rossjohn et al., 2012; Gold and Lewinsohn, 2013; Chien et al., 2014).
Monogenic primary immunodeficiencies (PIDs) provide a unique opportunity to establish the nonredundant functions of specific molecules in regulating human lymphocyte development and function. Indeed, studies of PIDs have provided valuable insights into the molecular mechanisms that control conventional T and B cells. However, little analysis of unconventional T cells in these conditions has been performed. Autosomal-dominant hyper IgE syndrome (AD-HIES) is a PID characterized by elevated serum IgE, eczema, and susceptibility to a well-defined spectrum of pathogens. Patients suffer from recurrent skin and lung abscesses caused by S. aureus and chronic mucocutaneous infections caused by C. albicans (Chandesris et al., 2012). AD-HIES results from heterozygous loss of function mutations in STAT3 (Holland et al., 2007; Minegishi et al., 2007). STAT3 signals downstream of many cytokine receptors, including those for IL-6, IL-10, IL-21, and IL-23, as well as growth hormones and IFNγ (Kane et al., 2014). Studies of AD-HIES patients have revealed multiple roles for STAT3 in the adaptive immune system. For example, STAT3 signaling is crucial for the differentiation of naive CD4+ T cells into Th17 cells (de Beaucoudrey et al., 2008; Ma et al., 2008; Milner et al., 2008). This deficiency in Th17 cells partly explains the susceptibility of AD-HIES patients to C. albicans and S. aureus as IL-17 is crucial for host defense against these pathogens (Puel et al., 2011; Cypowyj et al., 2012).
Human unconventional T cells have been reported to recognize C. albicans and S. aureus, and mouse models support a role for these cells in immunity against these pathogens. Furthermore, unconventional T cells produce IL-17 and express many cytokine receptors that signal through STAT3 (Cua and Tato, 2010; Constantinides and Bendelac, 2013; Gold and Lewinsohn, 2013). However, the role of STAT3 in the development and function of human unconventional T cells, and how they may contribute to the clinical phenotype of AD-HIES, has not previously been assessed. Thus, we investigated the development and function of MAIT, NKT, and γδ T cells in patients with mutations in STAT3-dependent cytokine signaling pathways.
RESULTS AND DISCUSSION
Functional STAT3 deficiency causes a decrease in MAIT and NKT cells in vivo
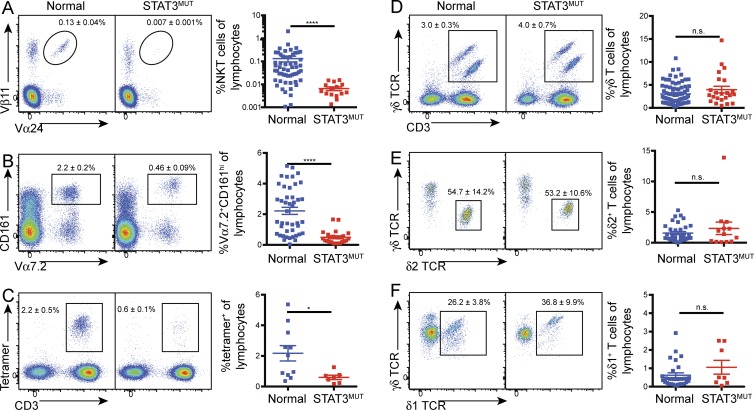
To determine whether functional STAT3 deficiency affects the numbers of circulating unconventional T cells, we analyzed PBMCs from AD-HIES patients and normal controls. This showed a 20-fold reduction in iNKT (CD3+Vα24+Vβ11+) cells in STAT3 mutant individuals (Fig. 1 A). Similarly, we observed a fourfold decrease in the percentage of MAIT cells as identified both by expression of the invariant Vα7.2 TCRα chain and high levels of CD161 (Fig. 1 B) or by using MR1 tetramers loaded with 5-OP-RU, the riboflavin metabolites recognized by MAIT cells (Fig. 1 C; Reantragoon et al., 2013; Corbett et al., 2014). We assessed the phenotype of the MAIT cells and observed no difference in the percentages of cells that had down-regulated CD45RA (control: 94.1 ± 1.6% [n = 11] vs. STAT3MUT: 93.8 ± 1.6% [n = 8]), nor a selective loss of any particular MAIT cell subset in the STAT3 mutant individuals based on CD8α and CD4 expression (CD8+: control 84.5 ± 2.4%, STAT3MUT 85.4 ± 2.3%; CD4+: control 2.1 ± 0.5%, STAT3MUT 3.7 ± 1.6%; DN: control 12.0 ± 2.3% [n = 12], STAT3MUT 8.6 ± 1.4% [n = 9]). This established that the reduction in MAIT cells caused by STAT3 deficiency was not caused by the loss of a particular subset, but rather by a global reduction in all subsets, at least as defined by these phenotypic characteristics. This dramatic decrease in NKT and MAIT cells suggests that STAT3 regulates the generation and/or survival of both of these unconventional T cell populations.
Figure 1.
Mutations in STAT3 result in decreased NKT and MAIT cell numbers. (A–F) PBMCs from normal controls or STAT3 mutant patients (STAT3MUT) were stained for iNKT cells (TCRVα24+ Vβ11+; A), MAIT cells [CD3+Vα7.2+ CD161+ (B); CD3+ cells binding MR1–rRL-6-CH2OH tetramers (C)], and total γδ T cells (D), as well as δ2+ (E) and δ1+ (F) subsets. Representative staining of total lymphocytes (A, C, and D), CD3+ cells (B), or γδ T cells (E and F) is shown on the left. Numbers represent mean percentage (±SEM) of lymphocytes (A–D) or γδ T cells (E and F). Graphs show combined data with each symbol representing a single control (n = 11–78) or patient (n = 7–23); error bars indicate SEM; *, P < 0.05; ****, P < 0.0001.
In contrast, the frequency of γδ T cells was not significantly different between normal controls and STAT3-deficient individuals (Fig. 1 D). As the different TCRδ chains are associated with responses to different pathogens (Li et al., 1996; Fenoglio et al., 2009; Chien et al., 2014), we also examined the relative proportions of δ2+ and δ1+ T cells to ascertain whether STAT3 deficiency selectively affects a particular γδ T cell subpopulation. However, our analysis showed that STAT3 deficiency had no significant effect on the percentage of δ1- or δ2-expressing cells (Fig. 1, E and F), suggesting STAT3 is not critical for development or survival of γδ T cells in vivo.
A cell-intrinsic requirement for STAT3 in unconventional T cells
Although our results clearly show that STAT3 mutations result in a decrease in iNKT and MAIT cells, it does not necessarily follow that there is a cell-intrinsic requirement for STAT3 in their survival or development. Indeed, the reduction in these cells could result from a defect in supporting cells that require STAT3 or as a result of the chronic infections associated with AD-HIES.
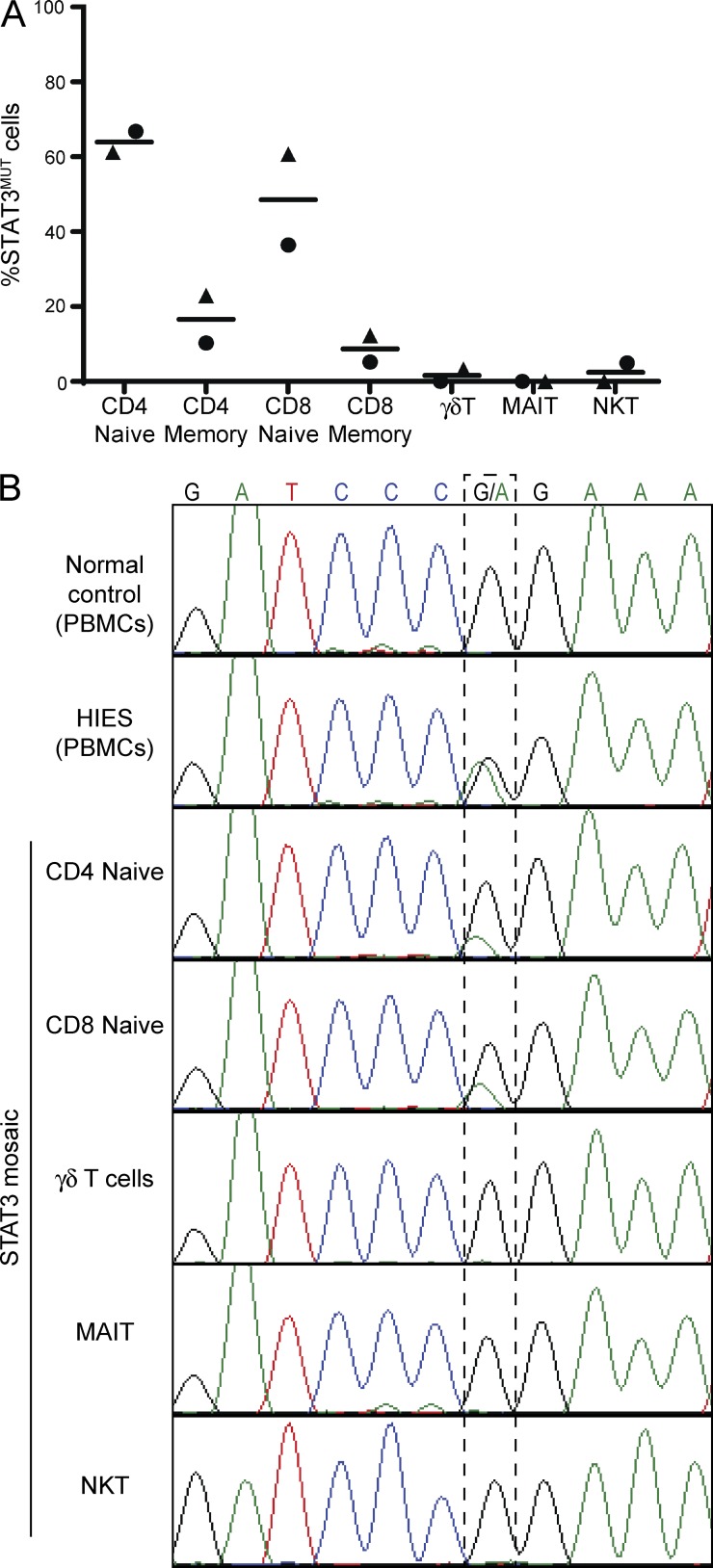
To delineate whether the paucity of iNKT and MAIT cells in AD-HIES is caused by a cell-intrinsic function of STAT3, we took advantage of two individuals who have been identified to have an intermediate AD-HIES phenotype caused by somatic mosaicism at one STAT3 allele (Siegel et al., 2011; Hsu et al., 2013). In these individuals ∼50% of their immune cells express mutant STAT3, whereas the other half express the WT genotype and thus have normal functioning STAT3 (Siegel et al., 2011; Hsu et al., 2013). Samples from these individuals offer the unique opportunity to examine normal and STAT3-deficient unconventional T cells in the same environment, thereby allowing the comparison of cell-extrinsic and -intrinsic effects of STAT3 mutations. Thus, if STAT3 has a critical intrinsic role in development or survival of unconventional T cells, cells with the WT genotype will have a distinct selective advantage and would therefore comprise a far greater proportion of the circulating T cells. To test this hypothesis, unconventional T cells were sorted from PBMCs from two STAT3 mosaic individuals, and the frequency of expression of the WT and mutant STAT3 alleles was determined by quantitative PCR (qPCR). As it has previously been observed that conventional CD4+ and CD8+ memory, but not naive, T cells have a cell-intrinsic dependency on STAT3 (Siegel et al., 2011), we included these cell types as controls. The populations of naive T cells were composed of approximately equal numbers of cells expressing each genotype, whereas the memory populations were composed almost entirely of cells with the WT genotype (Fig. 2 A). Strikingly, the unconventional T cell populations comprised almost entirely WT cells (Fig. 2 A). We confirmed the enrichment of the WT allele in these populations from one mosaic individual by Sanger sequencing of genomic DNA at the position of the 1145G>A SNP (Fig. 2 B). Traces from the healthy control showed only the WT G at position 1145, whereas an AD-HIES patient harboring this same mutation in the germline showed equal G and A peaks consistent with the heterozygous mutations in AD-HIES. Although traces from the unconventional T cells showed the same genotype as the WT control with no A peak visible at the mutation point, naive CD4+ and CD8+ T cells were clearly mosaic at this nucleotide, confirming the results of the qPCR. Together, these results unequivocally demonstrate that STAT3 functions as a crucial intrinsic mediator of NKT and MAIT cell frequency.
Figure 2.
Circulating unconventional T cells in individuals mosaic for STAT3 mutations all express the WT genotype. (A) qPCR was performed on DNA from sorted T cell populations from STAT3 mosaic individuals using probes specific for the WT or disease-causing genotype. Different symbols represent the two individuals examined. Horizontal bars indicate the mean. (B) Exons 12–14 of STAT3 were amplified from DNA from PBMCs from a normal donor (WBC) or a STAT3-deficient patient (AD-HIES), or the indicated populations of sorted cells from a mosaic individual with the 1145G>A STAT3 mutation. Chromatograms depict heterozygosity of the STAT3-deficient patient at this SNP and different degrees of mosaicism in distinct cell lineages in the mosaic individual.
STAT3-deficient MAIT cells express normal levels of RORγt and PLZF
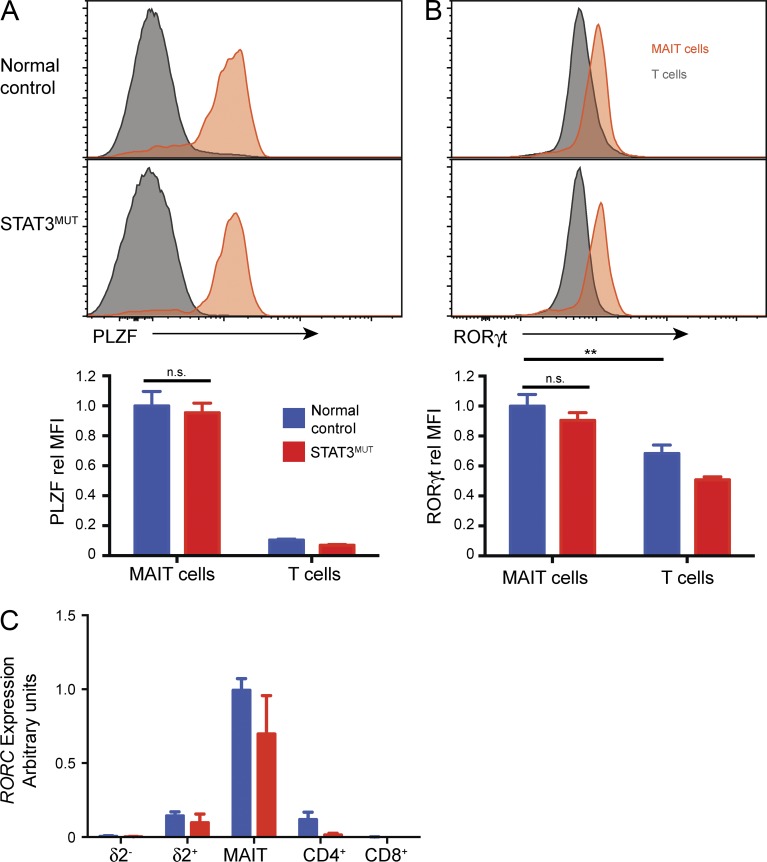
Given the role of STAT3 in regulating the numbers of iNKT cells and MAIT cells, we considered whether STAT3 might control expression of transcription factors important for development of these unconventional T cells. PLZF is a transcription factor highly expressed by both MAIT and NKT cells, which controls the development and preactivated phenotype of these cells (Kovalovsky et al., 2008; Savage et al., 2008; Eidson et al., 2011). Thus, we determined whether the levels of this molecule were altered in STAT3-deficient MAIT cells. Consistent with previous results (Savage et al., 2008), MAIT cells from normal controls expressed significantly more PLZF than other T cells (Fig. 3 A). Strikingly, STAT3-deficient MAIT cells expressed similar levels of PLZF as normal controls. As a result of the paucity of iNKT cells in STAT3-deficient patients, we were not able to determine the levels of PLZF in these cells.
Figure 3.
STAT3-deficient MAIT cells express normal levels of RORγt and PLZF. (A and B) PBMCs from normal controls or STAT3MUT patients were stained with mAbs to identify MAIT cells or total T cells, and then expression of PLZF (A) and RORγt (B) was determined. Representative histogram plots of MAIT or T cells (CD3+Vα7.2-γδ−). Graphs show mean ± SEM (n = 6–10) of the MFI normalized to MFI of MAIT cells from controls in each experiment; **, P < 0.01. (C) RORC transcripts were determined by qPCR of sorted T cell populations (n = 5–9). Error bars indicate SEM.
RORγt is a transcription factor key to the differentiation of Th17 cells and IL-17 production (Ivanov et al., 2006); however, it is also highly expressed by MAIT cells (Dusseaux et al., 2011). Furthermore, induction of RORγt in CD4+ T cells requires STAT3 such that patients with AD-HIES lack CD4+ Th17 cells in vivo and naive CD4+ T cells from these individuals fail to up-regulate RORγt and produce Th17-type cytokines after in vitro Th17-polarizing conditions (de Beaucoudrey et al., 2008; Ma et al., 2008; Milner et al., 2008). Thus, we reasoned that STAT3 deficiency might also ablate RORγt expression in MAIT cells, leading to their decrease in AD-HIES patients. However, RORγt was similarly expressed by normal and STAT3-deficient MAIT cells, as determined both by intracellular staining (Fig. 3 B) and qPCR for RORC transcripts (Fig. 3 C). We also observed no significant difference in expression of other transcriptional regulators of T cell differentiation, such as Eomesodermin and Tbet, between STAT3-deficient and control MAIT cells (not depicted). Thus, it is unlikely that the reduction in MAIT cells (and by extension NKT cells) is caused by a deregulation in either PLZF or RORγt. Rather it is likely that the deficiency of NKT and MAIT cells stems from defects in survival or expansion of these cells.
Furthermore, these results suggest that, unlike CD4+ Th17 cell differentiation, RORγt is acquired independently of STAT3 in MAIT cells. In mice, populations of RORγt+ IL-17+ NKT and γδ T cells have been described that appear to be preprogrammed in the thymus (Coquet et al., 2008; Rachitskaya et al., 2008; Shibata et al., 2011), and at least for γδ T cells, this has been shown to occur independently of STAT3 signaling (Shibata et al., 2011). Thus, it is possible that a similar STAT3-independent RORγt up-regulation occurs during thymic development of human MAIT cells.
STAT3 is required for IL-17 secretion by unconventional T cells
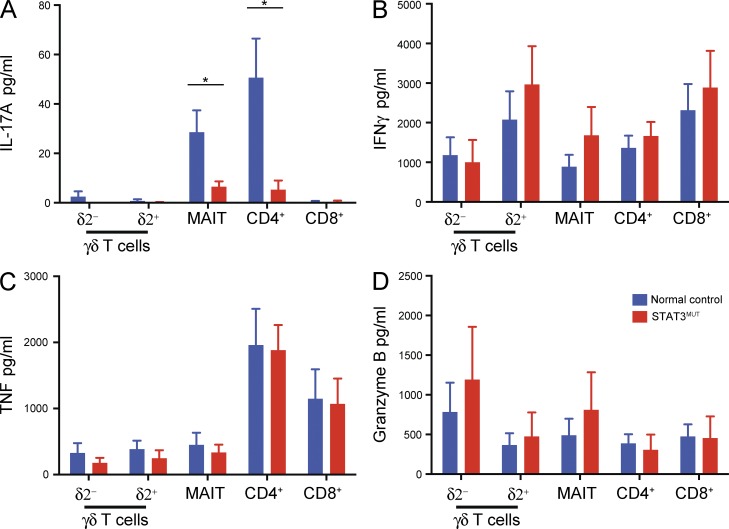
The normal expression of RORγt by STAT3-deficient MAIT cells, coupled with normal levels of other markers of IL-17–producing cells such as CD161 and CCR6 (Fig. 1 B and not depicted) led us to investigate whether these cells were also capable of expressing normal levels of IL-17. To address this, δ2− and δ2+ γδ T cells, MAIT cells and total CD4+ and CD8+ T cells were sorted from patients and normal controls and stimulated with PMA/ionomycin. iNKT cells could not be analyzed because of their rarity both in normal controls but more strikingly in AD-HIES patients. Consistent with a previous study (Dusseaux et al., 2011), both MAIT cells and CD4+ T cells from normal donors produced low levels of IL-17A. Importantly, this IL-17A production was abrogated by STAT3 deficiency (Fig. 4 A), suggesting STAT3 plays a crucial role in inducing IL-17 not only in conventional CD4+ T cells but also in MAIT cells. This reflected a specific defect in producing IL-17A, rather than a general dysfunction in cytokine secretion because production of IFNγ, TNF, Granzyme A and B, and IL-2 (Fig. 4, B–D; and not depicted) was comparable for STAT3 mutant and normal T cell subsets. We also analyzed γδ T cell populations to determine whether a human equivalent of the thymically programmed IL-17–producing population could be identified. However, we detected little IL-17 production from either γδ T cell population (Fig. 4 A).
Figure 4.
STAT3-deficient T cells are impaired in their ability to produce IL-17A. (A–D) Unconventional T cell populations (δ2− and δ2+ γδ T cells and MAIT cells) and total CD4+ and CD8+ T cells were isolated from control or STAT3MUT PBMCs and stimulated with PMA/ionomycin for 72 h. Culture supernatants were then harvested and secretion of IL-17A (A), IFNγ (B), TNF (C), and Granzyme B (D) determined. Graphs show mean ± SEM (n = 5–7); *, P < 0.05.
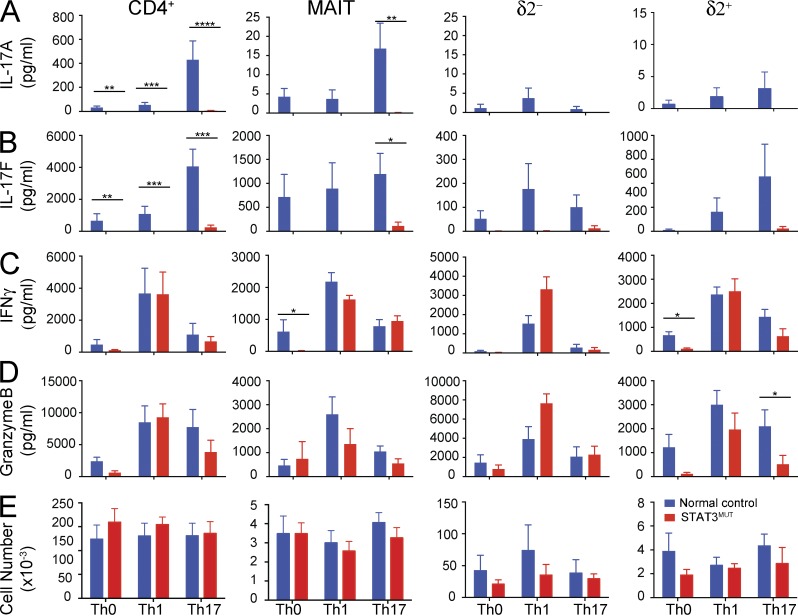
Although we found that all T cells secreted high levels of IFNγ and Granzyme B (Fig. 4, B and D) in response to PMA/ionomycin, secreted levels of IL-17A and IL-17F were very low to absent (Fig. 4 A and not depicted). The STAT3-activating cytokines IL-6 and IL-23, together with TGFβ and IL-1β, induce differentiation of CD4+ and γδ T cells to an IL-17–secreting phenotype (Acosta-Rodriguez et al., 2007; Wilson et al., 2007; Ness-Schwickerath et al., 2010). To determine whether these cytokines promoted IL-17 production from human MAIT cells, and whether STAT3 plays a role in this process in unconventional T cells, sorted T cells (δ2− and δ2+ γδ T, MAIT, and as controls total CD4+) were cultured for 5 d with T cell activation and expansion (TAE) beads (beads expressing anti-CD3, anti-CD28, and anti-CD2 mAb) alone or together with Th17-polarizing stimuli. The cells were also cultured under Th1-polarizing conditions as a control for intact responsiveness to alternative differentiation stimuli. As shown previously (de Beaucoudrey et al., 2008; Ma et al., 2008; Milner et al., 2008), Th17 culture conditions induced high levels of IL-17A and IL-17F secretion from CD4+ T cells, and this response was ablated in STAT3-deficient cells (Fig. 5, A and B). The Th17 culture also stimulated high levels of IL-17F and a lower level of IL-17A by MAIT cells, and this response was severely reduced in STAT3-deficient cells (Fig. 5, A and B). The level of IL-17 secretion by γδ T cells also appeared to be reduced in STAT3-deficient cells; however, this difference was not statistically significant, possibly because of the large variability among γδ T cells even from normal donors.
Figure 5.
STAT3-deficient MAIT cells do not respond to Th17-polarizing stimuli. Sorted δ2+ and δ2− γδ T, MAIT, and total CD4+ T cells from normal controls or STAT3MUT patients were cultured under Th0 (TAE beads), Th1 (TAE + IL-12), and Th17 (TAE + IL-1β/IL-6/IL-23/TGFβ) conditions. (A–D) After 5 d, secretion of IL-17A (A), IL-17F (B), IFNγ (C), and Granzyme B (D) was determined. (E) Absolute cell numbers in each culture were also determined at this time. Graphs show mean ± SEM (n = 4–9); *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We also examined secretion of IFNγ and Granzyme B (Fig. 5, C and D). Interestingly, in contrast to normal secretion of IFNγ by STAT3-deficient δ2+ γδ T and MAIT cells in response to PMA/ionomycin stimulation (Fig. 4 B), both of these cell types displayed decreased IFNγ secretion in the nonpolarizing culture of TAE beads alone (Fig. 5 C). However, this impairment was rescued in the Th1 and Th17 cultures. The δ2+ γδ T also showed decreased Granzyme A and B expression (Fig. 5 D and not depicted) in Th0 cultures, which again was rescued by addition of IL-12. It is not clear what the mechanism underlying this defect in IFNγ and Granzyme secretion is as the normal production in response to PMA/ionomycin (Fig. 4) coupled with normal expression of T-bet (not depicted) suggests that the in vivo differentiation of these cells to a Th1-like phenotype is intact. Instead it may reflect the requirement for a STAT3-dependent response to endogenously produced STAT3-activating cytokines in the Th0 cultures. To determine whether STAT3 deficiency also affected T cell survival or expansion, we enumerated the number of live cells after 5 d of culture (Fig. 5 E). We saw no differences in cell recovery between controls and STAT3-deficient cultures for any cell type (Fig. 5 E). However, although we observed a significant increase in cell numbers over the starting 20,000 cells in cultures of CD4+ and δ2− γδ T cells, consistent with substantial proliferation, very few cells were recovered from cultures of δ2+ T and MAIT cells. Flow cytometric analysis of forward scatter of cells from these cultures also demonstrated that only CD4+ and δ2− γδ T cells showed characteristics of cells undergoing blastogenesis and proliferation (not depicted). Indeed, CFSE labeling of the different populations confirmed that CD4+ and δ2− γδ T cells, but neither MAIT nor δ2+ γδ T cells, underwent proliferation after 5 d in culture (not depicted). Remarkably, although we recovered ∼30–40-fold fewer cells from cultures of MAIT cells compared with CD4+ T cells, the level of IFNγ and IL-17F secretion was only two- to fourfold lower. This may reflect the greater heterogeneity of CD4+ T cells. For example, IFNγ production is largely restricted to Th1 cells, whereas the majority of MAIT cells can secrete IFNγ after in vitro stimulation (not depicted).
Together these results reveal that STAT3 deficiency leads to a loss of IL-17 production by all T cell populations. It is of particular interest that although STAT3-deficient MAIT cells expressed many features of IL-17–producing cells, including CD161, CCR6, and high levels of RORγt, they were still unable to secrete appreciable levels of IL-17. This suggests that although RORγt can be up-regulated independently of STAT3 in these cells, possibly during thymic development, the action of RORγt alone is not sufficient to drive transcription of the IL17A and IL17F genes and thus induce their secretion. This is presumably the result of a requirement for STAT3 to act directly at the promoters of these genes (Durant et al., 2010).
These experiments also provide further insight into the clinical phenotype of AD-HIES, which is characterized by recurrent infection with C. albicans and S. aureus (Kane et al., 2014). Control of these infections has been associated with the actions of IL-17A and IL-17F (Puel et al., 2011; Cypowyj et al., 2012). Indeed, previous work demonstrated a lack of IL-17–producing CD4+ T cells in STAT3-deficient individuals (de Beaucoudrey et al., 2008; Ma et al., 2008; Milner et al., 2008), suggesting this defect in CD4+ T cell differentiation contributed to poor control of pathogen infection. Importantly, MAIT cells can be activated by C. albicans and S. aureus as both of these microorganisms generate the riboflavin metabolites specifically recognized by these cells (Kjer-Nielsen et al., 2012). Furthermore, γδ T cells are also implicated in responses to these organisms (Cho et al., 2010; Cua and Tato, 2010; Ness-Schwickerath et al., 2010). Thus, our results that STAT3-deficient MAIT and γδ T cells are also unable to produce IL-17 broaden these findings and suggest that functional and numerical defects in unconventional T cells contribute to the infectious susceptibility in AD-HIES. Together, these data reinforce the significance of unconventional T cells and IL-17 in the immune system and underscore the essential role STAT3 plays in their action.
IL-21 and IL-23 activate STAT3 in human MAIT cells and are required for the maintenance of innate-like T cells
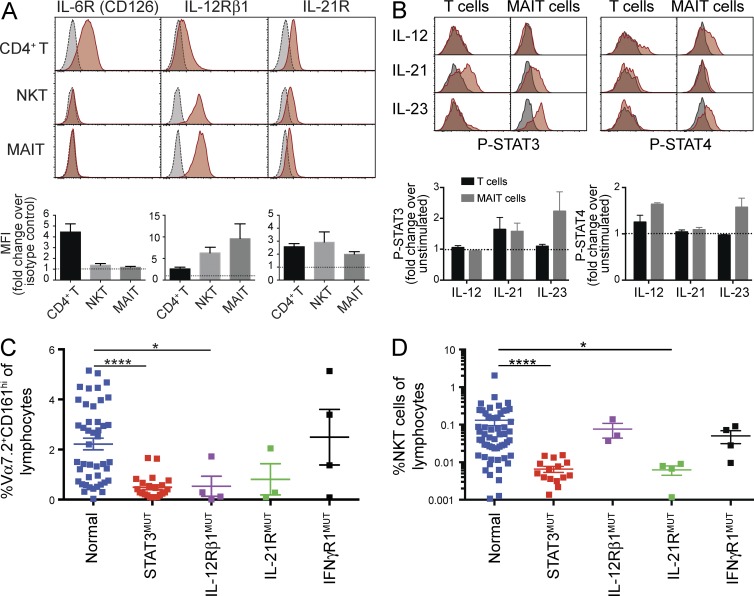
STAT3 can be activated by many cytokines, including IL-6, IL-10, IL-21, and IL-23 (Kane et al., 2014). Thus, we sought to identify which cytokine signaling pathways may be active in MAIT and NKT cells. We found prominent expression of IL-12Rβ1 and IL-21R on unstimulated MAIT and NKT cells; however, these cells lacked detectable expression of IL-6R (Fig. 6 A). This suggested these cells could potentially respond to IL-12, IL-23 (both of which share IL-12Rβ1), and IL-21. To test this, we examined STAT phosphorylation in sort-purified MAIT cells that were preactivated for 48 h. IL-12 induced phosphorylation of only STAT4, and IL-21 induced phosphorylation of STAT3 (Fig. 6 B). However, IL-23 induced robust activation of both STAT3 and STAT4, implying IL-12Rβ1 is used by IL-23, rather than IL-12, to initiate STAT3-dependent signaling in human MAIT cells. To determine whether signaling through IL-21R/STAT3 or IL-23R/STAT3 was important for controlling MAIT and iNKT cells numbers, we enumerated these populations in patients with loss-of-function mutations in IL21R or IL12RB1 (Fig. 6, C and D). We also examined IFNGR1 mutant individuals as a control for immune-deficient patients. This revealed intact numbers of MAIT and iNKT cells in IFNGR1 mutant patients, but striking deficiencies of MAIT cells in individuals with IL12RB1 mutations (Fig. 6 C) and of iNKT cells in IL-21R–deficient patients (Fig. 6 D). Indeed, the proportions of MAIT and iNKT cells in these patients approximated those in STAT3-deficient individuals, implicating IL-23 and IL-21 as the major cytokines responsible for STAT3-dependent maintenance of human MAIT and iNKT cells, respectively (Fig. 6, C and D). Importantly, ∼25% of patients with IL12RB1 mutations present with Candida infections (de Beaucoudrey et al., 2010), consistent with a role for MAIT cells in protective immunity to Candida. Together, these data define a novel requirement for specific cytokine-induced STAT3 signaling pathways that control MAIT and iNKT cell numbers in humans. Importantly, this represents the first description of a cell-intrinsic regulator of human MAIT cells. Identification of these pathways potentially paves the way for manipulating the frequencies of these cell types to modulate immune responses in the setting of infection with specific pathogens.
Figure 6.
IL-21 and IL-23 activate STAT3 and are required for the maintenance of iNKT and MAIT cells. (A) PBMCs from normal controls were stained for IL-6R, IL-12β1, and IL-21R on CD4+ T, iNKT, or MAIT cells (gray: isotype control, red: cytokine receptor). Graphs show fold increase in MFI over isotype control (mean ± SEM; n = 4–9). (B) Sorted MAIT (CD3+Vα7.2+CD161+) and non-MAIT (CD3+Vα7.2−CD161−) cells were stimulated with TAE beads, rested, and then stimulated with various cytokines for 20 min. After this time, cells were stained for phospho-STAT. Histograms show representative staining (gray: unstimulated, red: cytokine stimulated). Graphs show fold increase in MFI over unstimulated control (mean ± SEM; n = 3). (C and D) PBMCs from patients with IL12RB1, IL21R, or IFNGR1 mutations were stained for MAIT (CD3+Vα7.2+ CD161+; C) or iNKT (TCRVα24+ Vβ11+; D) cells. Data from normal controls and STAT3MUT patients from Fig. 1 are included for comparison. Each point represents a different individual; error bars indicate SEM; *, P < 0.05; ****, P < 0.0001.
MATERIALS AND METHODS
Human blood samples.
Buffy coats from normal donors were from the Australian Red Cross Blood Service. Peripheral blood was also collected from patients with mutations in STAT3 (Holland et al., 2007; Chandesris et al., 2012; Deenick et al., 2013), IL12RB1 (de Beaucoudrey et al., 2010), IL21R (Deenick et al., 2013; Ives et al., 2013; Kotlarz et al., 2013; Stepensky et al., 2015), or IFNGR1 (Dorman et al., 2004) and individuals mosaic for STAT3 mutations (Siegel et al., 2011; Hsu et al., 2013). Approval for this study was obtained from the ethics committees of the St. Vincent’s Hospital and Sydney South West Area Health Service (Australia), Rockefeller University Institutional Review Board (New York), and National Institute of Allergy and Infectious Diseases Intramural Institutional Review Board (Bethesda, MD). All participants gave written informed consent in accordance with the Declaration of Helsinki.
Flow cytometry reagents.
PerCp-Cy5.5 anti-CD161, PE anti-RORγt, PE anti–γδ TCR, biotinylated anti–γδ TCR, PerCp-Cy5.5 anti-CD45RA, and APC anti–αβ TCR antibodies were from eBioscience. A647 anti-PLZF, APC-Cy7 anti-CD4, PE-Cy7 anti-CD8, BV421 anti-CD3, BV421 anti-CD161, PE anti-STAT4 (pY693), PE anti–IL-6R, PE anti–IL-12Rβ1, PE anti–IL-21R, PE mouse IgG1 isotype control, PerCP-Cy5.5 anti-STAT3 (pY705), and FITC anti–Vδ2 TCR antibodies were from BD. FITC anti–Vα24 TCR and PE and biotinylated anti-Vβ11 antibodies were from Beckman Coulter. FITC anti–Vδ1 TCR was from Thermo Fisher Scientific. APC or APC-Cy7 anti-Vα7.2 and PE-Cy7 anti–αβ TCR antibodies were from BioLegend. FITC anti-CCR7 antibody was from R&D Systems. MR1–5-OP-RU tetramers have been described previously (Reantragoon et al., 2013; Corbett et al., 2014).
Cell sorting.
PBMCs were stained with mAbs and sorted as below using a BD FACSAria or Aria III. For SNP analysis, MAIT cells were sorted as CD3+Vα7.2+CD161hiγδ−, NKT cells as CD3+Vα24+Vβ11+, γδ T cells as CD3+Vα7.2−γδ+ δ2+/−, and CD4+ and CD8+ naive T cell populations as CD45RA+CCR7+, with the remaining cells sorted as memory cells. For cell culture, MAIT cells were sorted as CD3+Vα7.2+CD161hiγδ−, γδ T cells as CD3+Vα7.2−γδ+ and either δ2+ or δ2−, total CD4+ T cells as CD3+Vα7.2−γδ−CD8−CD4+, and total CD8+ T cells as CD3+Vα7.2−γδ−CD4−CD8+.
Cell culture.
Cells were cultured at 20,000/well/100 µl media (Deenick et al., 2013; Ives et al., 2013). Cells were stimulated with either TAE beads (1 bead/2 cells; Miltenyi Biotec) alone (Th0 culture) or under Th1 (20 ng/ml IL-12; R&D Systems) or Th17 (2.5 ng/ml TGFβ, 20 ng/ml IL-1β [PeproTech], 50 ng/ml IL-6 [PeproTech], or 20 ng/ml IL-23 [eBioscience]) conditions for 5 d or 100 ng/ml PMA plus 750 ng/ml ionomycin (Sigma-Aldrich) for 3 d before harvesting supernatants. Cytokine secretion was measured by cytometric bead array (BD) as per the manufacturer’s instructions. The absolute number of cells in culture was determined by adding a known number of CaliBRITE beads (BD) before harvest, which can then be distinguished by flow cytometry based on forward and side scatter.
Intracellular transcription factor staining.
Cells were stained for surface markers to identify γδ T and MAIT cells and then permeabilized using the BD transcription factor buffer set and stained with PLZF- or RORγt-specific mAbs.
SNP assay.
Genomic DNA was extracted from sorted cells (QIAGEN). Percent mosaicism was determined in different cell populations using the TaqMan SNP genotyping assay (Life Technologies; Hsu et al., 2013). This qPCR-based assay used primers for the region of DNA containing the disease-causing mutation, together with specific probes recognizing either the WT or disease SNPs. The difference between the cycle threshold for each probe was used to determine the proportion of the two alleles in each sample. In each assay a normal control and AD-HIES sample with the same mutation were used to normalize the data, with the normal control taken as 0% and the AD-HIES sample as 100%.
DNA sequencing.
Exons 12–14 of STAT3 were amplified from genomic DNA (Forward: 5′-TAGTTTAAAGAAATGCCCAGGAGCACAGAG-3′ and Reverse: 5′-TTGGCCTAAGTGACTTTTTGGAATAACTACAGC-3′). Sanger sequencing of the amplified product was performed by Garvan Molecular Genetics.
qPCR.
RNA was isolated from sorted populations using the RNeasy kit (QIAGEN). For qPCR, total RNA was reverse transcribed with oligo-dT. Expression of genes was determined using real-time PCR with the LightCycler 480 Probe Master Mix and System (Roche) as previously described (Ives et al., 2013).
Expression of phospho-STATs.
Sorted MAIT (CD3+Vα7.2+CD161+) and non-MAIT (CD3+Vα7.2−CD161−) cells were stimulated with TAE beads for 2 d and then rested for 2 h in media before being stimulated with 50 ng/ml IL-12, 100 ng/ml IL-21, or 100 ng/ml IL-23 or media alone for 20 min. Cells were fixed, permeabilized, and stained with anti–phospho-STAT3 (pY705) and STAT4 (pY693) mAbs (Deenick et al., 2013; Ives et al., 2013).
Statistical analysis.
Significant differences between datasets were determined using either the unpaired Student’s t test when comparing two groups, one-way ANOVA with Dunnet post test for more than two groups, or two-way ANOVA with Šídák’s multiple comparison test for two variables (Prism; GraphPad Software).
Acknowledgments
We thank Rob Salomon, Eric Lam, and Vitri Dewi for cell sorting and all of the patients and their families for participating in this project.
This work was funded by project and program grants from the National Health and Medical Research Council (NHMRC) of Australia (1016953, 1066694, and 1027400 to E.K. Deenick and S.G. Tangye) and the Rockefeller University Center for 541 Clinical and Translational Science (5UL1RR024143 to J.-L. Casanova). C.S. Ma is a recipient of a Career Development Fellowship (1008820), and S.G. Tangye is a recipient of a Principal Research Fellowship (1042925) from the NHMRC of Australia.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AD-HIES
- autosomal-dominant hyper IgE syndrome
- Ag
- antigen
- iNKT cell
- invariant NKT cell
- MAIT cell
- mucosal-associated invariant T cell
- MR1
- MHC-related molecule 1
- PID
- primary immunodeficiency
- qPCR
- quantitative PCR
- TAE
- T cell activation and expansion
References
- Acosta-Rodriguez E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., and Napolitani G.. 2007. Surface phenotype and antigenic specificity of human interleukin 17–producing T helper memory cells. Nat. Immunol. 8:639–646. 10.1038/ni1467 [DOI] [PubMed] [Google Scholar]
- Bonneville M., O’Brien R.L., and Born W.K.. 2010. γδ T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10:467–478. 10.1038/nri2781 [DOI] [PubMed] [Google Scholar]
- Chandesris M.-O., Melki I., Natividad A., Puel A., Fieschi C., Yun L., Thumerelle C., Oksenhendler E., Boutboul D., Thomas C., et al. 2012. Autosomal dominant STAT3 deficiency and hyper-IgE syndrome: molecular, cellular, and clinical features from a French national survey. Medicine (Baltimore). 91:e1–e19. 10.1097/MD.0b013e31825f95b9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y.H., Meyer C., and Bonneville M.. 2014. γδ T cells: first line of defense and beyond. Annu. Rev. Immunol. 32:121–155. 10.1146/annurev-immunol-032713-120216 [DOI] [PubMed] [Google Scholar]
- Cho J.S., Pietras E.M., Garcia N.C., Ramos R.I., Farzam D.M., Monroe H.R., Magorien J.E., Blauvelt A., Kolls J.K., Cheung A.L., et al. 2010. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J. Clin. Invest. 120:1762–1773. 10.1172/JCI40891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M.G., and Bendelac A.. 2013. Transcriptional regulation of the NKT cell lineage. Curr. Opin. Immunol. 25:161–167. 10.1016/j.coi.2013.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquet J.M., Chakravarti S., Kyparissoudis K., McNab F.W., Pitt L.A., McKenzie B.S., Berzins S.P., Smyth M.J., and Godfrey D.I.. 2008. Diverse cytokine production by NKT cell subsets and identification of an IL-17–producing CD4−NK1.1− NKT cell population. Proc. Natl. Acad. Sci. USA. 105:11287–11292. 10.1073/pnas.0801631105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett A.J., Eckle S.B.G., Birkinshaw R.W., Liu L., Patel O., Mahony J., Chen Z., Reantragoon R., Meehan B., Cao H., et al. 2014. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 509:361–365. 10.1038/nature13160 [DOI] [PubMed] [Google Scholar]
- Cua D.J., and Tato C.M.. 2010. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 10:479–489. 10.1038/nri2800 [DOI] [PubMed] [Google Scholar]
- Cypowyj S., Picard C., Maródi L., Casanova J.-L., and Puel A.. 2012. Immunity to infection in IL-17-deficient mice and humans. Eur. J. Immunol. 42:2246–2254. 10.1002/eji.201242605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beaucoudrey L., Puel A., Filipe-Santos O., Cobat A., Ghandil P., Chrabieh M., Feinberg J., von Bernuth H., Samarina A., Jannière L., et al. 2008. Mutations in STAT3 and IL12RB1 impair the development of human IL-17–producing T cells. J. Exp. Med. 205:1543–1550. 10.1084/jem.20080321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beaucoudrey L., Samarina A., Bustamante J., Cobat A., Boisson-Dupuis S., Feinberg J., Al-Muhsen S., Jannière L., Rose Y., de Suremain M., et al. 2010. Revisiting human IL-12Rβ1 deficiency: a survey of 141 patients from 30 countries. Medicine (Baltimore). 89:381–402. 10.1097/MD.0b013e3181fdd832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deenick E.K., Avery D.T., Chan A., Berglund L.J., Ives M.L., Moens L., Stoddard J.L., Bustamante J., Boisson-Dupuis S., Tsumura M., et al. 2013. Naive and memory human B cells have distinct requirements for STAT3 activation to differentiate into antibody-secreting plasma cells. J. Exp. Med. 210:2739–2753. 10.1084/jem.20130323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman S.E., Picard C., Lammas D., Heyne K., van Dissel J.T., Baretto R., Rosenzweig S.D., Newport M., Levin M., Roesler J., et al. 2004. Clinical features of dominant and recessive interferon γ receptor 1 deficiencies. Lancet. 364:2113–2121. 10.1016/S0140-6736(04)17552-1 [DOI] [PubMed] [Google Scholar]
- Durant L., Watford W.T., Ramos H.L., Laurence A., Vahedi G., Wei L., Takahashi H., Sun H.W., Kanno Y., Powrie F., and O’Shea J.J.. 2010. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 32:605–615. 10.1016/j.immuni.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., and Lantz O.. 2011. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17–secreting T cells. Blood. 117:1250–1259. 10.1182/blood-2010-08-303339 [DOI] [PubMed] [Google Scholar]
- Eidson M., Wahlstrom J., Beaulieu A.M., Zaidi B., Carsons S.E., Crow P.K., Yuan J., Wolchok J.D., Horsthemke B., Wieczorek D., and Sant’Angelo D.B.. 2011. Altered development of NKT cells, γδ T cells, CD8 T cells and NK cells in a PLZF deficient patient. PLoS ONE. 6:e24441 10.1371/journal.pone.0024441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio D., Poggi A., Catellani S., Battaglia F., Ferrera A., Setti M., Murdaca G., and Zocchi M.R.. 2009. Vδ1 T lymphocytes producing IFN-γ and IL-17 are expanded in HIV-1–infected patients and respond to Candida albicans. Blood. 113:6611–6618. 10.1182/blood-2009-01-198028 [DOI] [PubMed] [Google Scholar]
- Gold M.C., and Lewinsohn D.M.. 2013. Co-dependents: MR1-restricted MAIT cells and their antimicrobial function. Nat. Rev. Microbiol. 11:14–19. 10.1038/nrmicro2918 [DOI] [PubMed] [Google Scholar]
- Holland S.M., DeLeo F.R., Elloumi H.Z., Hsu A.P., Uzel G., Brodsky N., Freeman A.F., Demidowich A., Davis J., Turner M.L., et al. 2007. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357:1608–1619. 10.1056/NEJMoa073687 [DOI] [PubMed] [Google Scholar]
- Hsu A.P., Sowerwine K.J., Lawrence M.G., Davis J., Henderson C.J., Zarember K.A., Garofalo M., Gallin J.I., Kuhns D.B., Heller T., et al. 2013. Intermediate phenotypes in patients with autosomal dominant hyper-IgE syndrome caused by somatic mosaicism. J. Allergy Clin. Immunol. 131:1586–1593. 10.1016/j.jaci.2013.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Ives M.L., Ma C.S., Palendira U., Chan A., Bustamante J., Boisson-Dupuis S., Arkwright P.D., Engelhard D., Averbuch D., Magdorf K., et al. 2013. Signal transducer and activator of transcription 3 (STAT3) mutations underlying autosomal dominant hyper-IgE syndrome impair human CD8+ T-cell memory formation and function. J. Allergy Clin. Immunol. 132:400–411: e9. 10.1016/j.jaci.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane A., Deenick E.K., Ma C.S., Cook M.C., Uzel G., and Tangye S.G.. 2014. STAT3 is a central regulator of lymphocyte differentiation and function. Curr. Opin. Immunol. 28:49–57. 10.1016/j.coi.2014.01.015 [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Patel O., Corbett A.J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., et al. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 491:717–723. [DOI] [PubMed] [Google Scholar]
- Kotlarz D., Ziętara N., Uzel G., Weidemann T., Braun C.J., Diestelhorst J., Krawitz P.M., Robinson P.N., Hecht J., Puchałka J., et al. 2013. Loss-of-function mutations in the IL-21 receptor gene cause a primary immunodeficiency syndrome. J. Exp. Med. 210:433–443. 10.1084/jem.20111229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S., et al. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9:1055–1064. 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L., Mburu Y.K., and Lantz O.. 2013. MAIT cells, surveyors of a new class of antigen: development and functions. Curr. Opin. Immunol. 25:174–180. 10.1016/j.coi.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Li B., Rossman M.D., Imir T., Oner-Eyuboglu A.F., Lee C.W., Biancaniello R., and Carding S.R.. 1996. Disease-specific changes in γδ T cell repertoire and function in patients with pulmonary tuberculosis. J. Immunol. 157:4222–4229. [PubMed] [Google Scholar]
- Ma C.S., Chew G.Y.J., Simpson N., Priyadarshi A., Wong M., Grimbacher B., Fulcher D.A., Tangye S.G., and Cook M.C.. 2008. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J. Exp. Med. 205:1551–1557. 10.1084/jem.20080218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J.D., Brenchley J.M., Laurence A., Freeman A.F., Hill B.J., Elias K.M., Kanno Y., Spalding C., Elloumi H.Z., Paulson M.L., et al. 2008. Impaired TH17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 452:773–776. 10.1038/nature06764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y., Saito M., Tsuchiya S., Tsuge I., Takada H., Hara T., Kawamura N., Ariga T., Pasic S., Stojkovic O., et al. 2007. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 448:1058–1062. 10.1038/nature06096 [DOI] [PubMed] [Google Scholar]
- Ness-Schwickerath K.J., Jin C., and Morita C.T.. 2010. Cytokine requirements for the differentiation and expansion of IL-17A– and IL-22–producing human Vγ2Vδ2 T cells. J. Immunol. 184:7268–7280. 10.4049/jimmunol.1000600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C.M., Groh V., Band H., Porcelli S.A., Morita C., Fabbi M., Glass D., Strominger J.L., and Brenner M.B.. 1990. Evidence for extrathymic changes in the T cell receptor gamma/delta repertoire. J. Exp. Med. 171:1597–1612. 10.1084/jem.171.5.1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A., Cypowyj S., Bustamante J., Wright J.F., Liu L., Lim H.K., Migaud M., Israel L., Chrabieh M., Audry M., et al. 2011. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 332:65–68. 10.1126/science.1200439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachitskaya A.V., Hansen A.M., Horai R., Li Z., Villasmil R., Luger D., Nussenblatt R.B., and Caspi R.R.. 2008. Cutting edge: NKT cells constitutively express IL-23 receptor and RORγt and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J. Immunol. 180:5167–5171. 10.4049/jimmunol.180.8.5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon R., Corbett A.J., Sakala I.G., Gherardin N.A., Furness J.B., Chen Z., Eckle S.B.G., Uldrich A.P., Birkinshaw R.W., Patel O., et al. 2013. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210:2305–2320. 10.1084/jem.20130958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossjohn J., Pellicci D.G., Patel O., Gapin L., and Godfrey D.I.. 2012. Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 12:845–857. 10.1038/nri3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., and Bendelac A.. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403. 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K., Yamada H., Sato T., Dejima T., Nakamura M., Ikawa T., Hara H., Yamasaki S., Kageyama R., Iwakura Y., et al. 2011. Notch-Hes1 pathway is required for the development of IL-17–producing γδ T cells. Blood. 118:586–593. 10.1182/blood-2011-02-334995 [DOI] [PubMed] [Google Scholar]
- Siegel A.M., Heimall J., Freeman A.F., Hsu A.P., Brittain E., Brenchley J.M., Douek D.C., Fahle G.H., Cohen J.I., Holland S.M., and Milner J.D.. 2011. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 35:806–818. 10.1016/j.immuni.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepensky P., Keller B., Abuzaitoun O., Shaag A., Yaacov B., Unger S., Seidl M., Rizzi M., Weintraub M., Elpeleg O., and Warnatz K.. 2015. Extending the clinical and immunological phenotype of human interleukin-21 receptor deficiency. Haematologica. 100:e72–e76. 10.3324/haematol.2014.112508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.J., Boniface K., Chan J.R., McKenzie B.S., Blumenschein W.M., Mattson J.D., Basham B., Smith K., Chen T., Morel F., et al. 2007. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat. Immunol. 8:950–957. 10.1038/ni1497 [DOI] [PubMed] [Google Scholar]