Insight from Chao Zhong (left) and Jinfang Zhu (right)
Effective immune responses to pathogens require the activation and differentiation of both innate and adaptive lymphocytes, such as innate lymphoid cells (ILCs) and T cells, respectively. ILCs consist of distinct helper-like subsets representing innate versions of CD4+ T helper subsets, whereas natural killer cells represent the innate counterpart of CD8+ cytotoxic T cells. By producing effector cytokines, T helper subsets and helper-like ILCs are also involved in the pathogenesis of many inflammatory diseases. Strikingly, the development of ILCs and T cells seems to depend on a shared transcriptional network. For example, GATA3 and Tcf7, which play essential roles during T cell development, are also indispensable for the development of multiple ILC subsets. Bcl11b is another critical component of a T cell lineage fate-specifying transcriptional network. In this issue, Yu et al. and Walker et al. independently describe the role of Bcl11b in specifying ILC2 development.
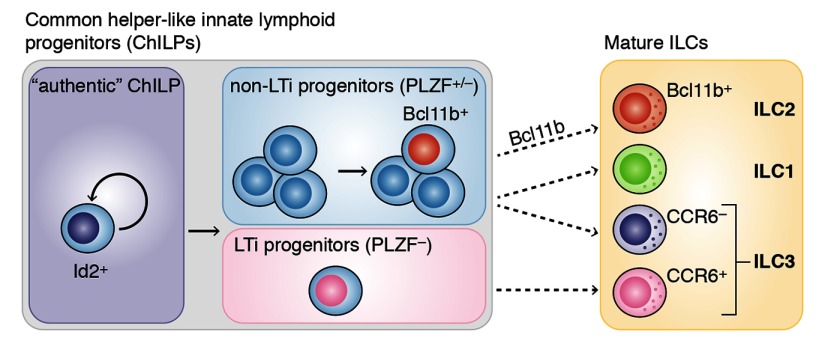
One would have expected that, similar to GATA3 and Tcf7, Bcl11b would be critical for the development of multiple ILC lineages. However, the two papers in this issue report that Bcl11b appears to be essential for ILC2 development only. Consequently, Bcl11b deficiency leads to impaired immune responses to either papain treatment, influenza virus infection (Yu et al.), or Nippostrongylus brasiliensis helminth infection, without affecting the ability of mice to clear bacterial Citrobacter rodentium infection (Walker et al.). The authors show that Bcl11b is expressed in mature ILC2s as well as in a subset of Id2+ common helper-like innate lymphoid progenitors (ChILPs); Walker et al. but not Yu et al. also show that a proportion of other mature ILC subsets, such as CCR6− ILC3s, express Bcl11b. The discrepancy in Bcl11b expression by mature ILC3s between these two studies has not been resolved. Nevertheless, the Bcl11b-expressing ChILPs only give rise to ILC2s but not other ILCs, suggesting that the previously identified ChILPs contain a mixture of distinct committed ILC progenitors. This is consistent with another report in Nature suggesting that within the ChILP compartment, the PLZF+ progenitors are largely committed to specific ILC subsets and fail to generate lymphoid tissue inducers, which represent another subset of ChILP-derived ILCs.
Scheme of helper-like innate lymphoid cell (ILC) development. LTi: hematopoietic lymphoid tissue inducer lineage.
Both reports also show that Bcl11b-expressing ILC2 progenitors express high levels of GATA3. However, other studies have reported that GATA3hi progenitors also give rise to other ILCs. In addition, GATA3 is constantly required for the maintenance of ILC2s, whereas inducible deletion of Bcl11b in mature ILC2s does not result in loss of functional ILC2s. Thus, Bcl11b either functions independently of GATA3 or collaborates with GATA3 during ILC2 development, but the detailed mechanism requires further investigation. Moreover, it is not known whether Bcl11b is critical for ILC2 development in humans.
It is intriguing that Bcl11b, which is critical for the development of all T cell subsets, appears to be dispensable for the development of ILC1s and ILC3s. Does this mean that T cells have preferentially adopted the ILC2 transcriptional program during their early development in the thymus? If so, one would predict that T cell progenitors, particularly CD4−CD8− thymocytes, might display certain features of ILC2s. This idea is consistent with the notion that T cells carry an endogenous Th2-prone program that makes Th2 lymphocyte differentiation a default cellular fate.
The studies of Yu et al. and Walker et al. not only identify Bcl11b as an ILC2 lineage-specifying factor, they also provide important general insights into the mechanisms of ILC development. Furthermore, this work raises the possibility that dysregulation of Bcl11b in ILC progenitors may contribute to human diseases involving ILC2 function.
Acknowledgments
Acknowledgments: C. Zhong and J. Zhu are supported by the Division of Intramural Research (DIR), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), USA.
References
- Constantinides, M.G., et al. 2014. Nature. 508:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.A., et al. 2015. J. Exp. Med. 10.1084/jem.20142224 [DOI] [Google Scholar]
- Yu, Y., et al. 2015. J. Exp. Med. 10.1084/jem.20142318 [DOI] [Google Scholar]