Summary
In the wild, bacteria are predominantly associated with surfaces as opposed to existing as free-swimming, isolated organisms. They are thus subject to surface-specific mechanics including hydrodynamic forces, adhesive forces, the rheology of their surroundings and transport rules that define their encounters with nutrients and signaling molecules. Here, we highlight the effects of mechanics on bacterial behaviors on surfaces at multiple length scales, from single bacteria to the development of multicellular bacterial communities such as biofilms.
Bacteria occupy a broad variety of ecological niches on Earth. Their long evolutionary history has exposed them to vastly different environments, and they have evolved remarkable plasticity in response to locally changing physicochemical conditions. In particular, bacteria can detect and respond to chemical, thermal, and mechanical cues, as well as to electric and magnetic fields. How do these cues influence bacterial behaviors in natural environments? Characterizing bacterial behavior in realistic contexts requires integrating a spectrum of environmental stimuli to which they respond, and doing so in physical configurations representative of their natural habitats. Such analyses are critical to comprehensively understand bacterial biology and to thereby make progress in promoting or restricting bacterial growth in medical, industrial, and agricultural realms.
Mechanics is an integral part of eukaryotic cell biology: numerous studies have demonstrated the importance of fluid flow and surface mechanics in mammalian cell growth and behavior at many different length scales (Fritton and Weinbaum, 2009; Hoffman et al., 2011; Pruitt et al., 2014). In contrast, microbiology has traditionally focused on the influence of the chemical environment on bacterial behavior. Hence, for decades, growth in well-mixed batch cultures and on agar plates were the methods of choice for studies of bacterial physiology. As a result, the community has only recently recognized that mechanics also play a significant role in microbial biology on surfaces: fluid flow and contact between cells and surfaces are two ubiquitous and influential features of bacterial existence in natural environments. Advances in microscale engineering and microscopy now provide us with powerful tools to explore, at the relevant spatial scales, the roles physical forces play in bacterial sensory perception and adaptation (Rusconi et al., 2014). These new experimental platforms have revealed that bacteria are attuned to mechanical forces and, indeed, can exploit mechanics to drive adaptive behavior.
Swimming motility provides an elegant example of how bacteria are influenced by the mechanical nature of their surroundings. As a consequence of their small size (~1 µm), bacteria live in environments dominated by viscosity, which stands in contrast to the meter-scale world of humans in which dynamics are dominated by inertia (Purcell, 1977). Fluid motion can be broadly characterized by the Reynolds number (Re), which compares the magnitudes of inertial forces and viscous forces in a given flow (Re = ρUL/µ where U is a typical fluid speed, L a typical length scale, ρ the density of the fluid and µ its viscosity). We humans live a high Reynolds number life (at least 104), as we are meter-scale organisms moving at speeds on the order of meters per second. But swimming microorganisms live at Reynolds numbers far below unity (at most 10−3). To self-propel in such a regime, bacteria use motorized flagella that convert mechanical actuation (rotation) into net displacement. Thus, many bacteria have evolved a biological machine - the flagellum and its associated motor - to adapt to the mechanical properties of their (purely viscous) environment. The biology and physics of swimming motility have been intensively investigated and are reviewed elsewhere (Berg, 2003; Guasto et al., 2012; Macnab, 2003). Here, we provide perspective on a more general but understudied aspect of mechanics in bacterial biology, namely the effects of surfaces and flow on bacterial behavior.
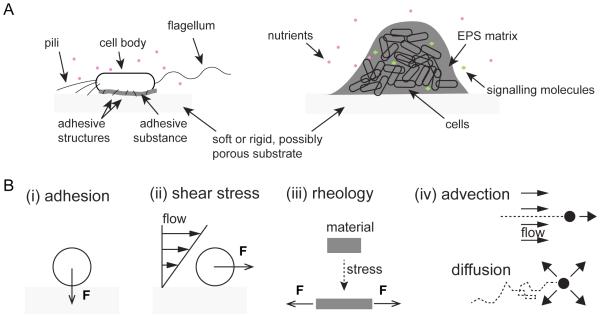
Outside of the oceans, most bacteria in nature exist on surfaces, rather than in the bulk liquid of their fluid environments (Costerton et al., 1995). Bacteria are equipped to live at the liquid-solid interface via the secretion of adhesive structures such as flagella, pili, exopolysaccharides, and other matrix components (Dunne, 2002) (Fig. 1A). The mechanical environment of surface-associated bacteria is remarkably different than that of their free-floating counterparts (Fig. 1B). From initial contact, a surface-attached bacterium will experience a local force that is normal to the surface, usually referred to as an adhesive force (Fig. 1B). In an environment with flow, the viscosity of the surrounding fluid generates a hydrodynamic (shear) force on the cell that is tangential to the surface in the direction of the flow (Fig. 1B). Surface motility may produce a friction force that is tangential to the cell wall and localized at the interface with the substrate. The principles of mechanics dictate that the forces on a stationary or steadily moving cell must balance, so that a local adhesive force toward the substrate at one point on the cell must be balanced by repulsive forces due to compression elsewhere.
Figure 1. Bacteria experience a variety of mechanical effects on surfaces.
(A) Flagella, pili, and adhesive substances are useful for attachment of individual bacterial cells to surfaces. Extracellular polymeric substances (EPS) aid in maintaining the integrity of community structures composed of multiple cells. Bacteria use diffusible signaling molecules, chemical weapons, and soluble public goods to interact within such communities. (B) A cell attaching to a surface is subject to a local adhesive force F in the direction normal to the surface. Shear stresses due to fluid flow generate a force F on the cell that is parallel to the surface. Bacteria experience the rheological properties of their surrounding extracellular matrix, which flows and/or deforms upon application of forces. Fluid flow (advection) and Brownian motion (diffusion) transport soluble compounds (black dots) that are released and/or internalized by bacteria.
Surface-attached bacterial cells can multiply to form large groups that develop into organized communities termed biofilms (Hall-Stoodley et al., 2004). At this multicellular scale, additional mechanical effects become relevant (Stewart, 2012). Attachment of a cell to a surface induces secretion of a mixture of proteins, polysaccharides, and DNA that form a surrounding matrix (EPS; extracellular polymeric substances) with both viscous and elastic properties (Fig. 1B). These extracellular polymers bind surface-attached cells and their progeny together in biofilm communities, and the rheology of the secreted matrix likely has important implications for the growth, spatial arrangement, and resilience of the resulting multicellular structures (Berk et al., 2012; Chew et al., 2014). The spatiotemporal distribution of small molecules internalized and/or released by bacterial cells residing within these communities can be strongly affected by the flow environment that the community experiences, with substantial and distinct consequences for individual and collective behaviors (Fig. 1B).
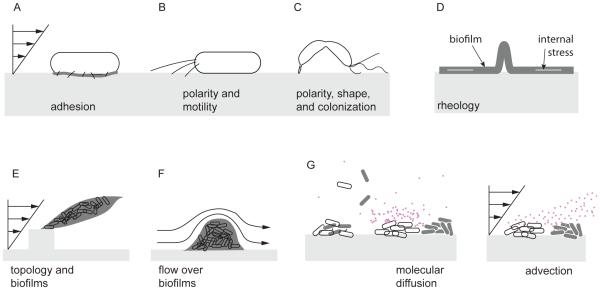
Here, we highlight how these mechanical effects play roles in bacterial behavior at the level of single cells and of multicellular structures. We discuss strategies that bacterial cells deploy specifically on surfaces, including enhanced adhesion under fluid flow, exploration via surface-specific motility, and control of cell shape to enhance colonization (Fig. 2A-C). At the level of multicellular structures, we discuss how the rheology of polymeric matrices affects populations growing in biofilms and how flow influences these structures (Fig. 2D-F). We also describe how fluid flow affects the transport of small molecules used in social interactions, e.g. quorum sensing, between individual bacterial cells (Fig. 2G). Finally, we provide insight into the scalability of the effects of mechanics on bacteria, i.e. how phenomena at the level of single cells influence emergent collective behaviors and group fitness consequences in multicellular communities.
Figure 2. Influences of environmental mechanics on individual cells and multicellular communities.
(A) On surfaces and in the presence of flow, individual bacterial cells use short appendages (fimbriae) and other adhesive structures or substances to remain strongly attached, and (B) they use motorized pili localized at their poles to move against the flow. (C) Bacteria exploit their shapes to orient in flow thereby enhancing surface colonization. (D) At the scale of multicellular communities, bacteria can alter the rheology of the extracellular polymeric substances (EPS) to optimize growth. (E, F) Flow modifies the architecture of bacterial biofilms by driving formation of filamentous structures called streamers, which can obstruct flow but also capture cells and metabolites suspended in the surrounding fluid. (G) Transport of nutrients and other solutes by diffusion and advection drives the growth of and interactions between surface-associated bacteria.
Mechanics at the level of single cells
To initiate and maintain intimate contact with solid surfaces, bacteria leverage a wide variety of adhesion strategies. Many bacteria, upon attaching to a surface, will secrete a mixture of EPS, which increases their affinity for porous, rough, and chemically heterogeneous surfaces (Flemming and Wingender, 2010). Bacteria also construct protein structures on their exteriors that enhance their adhesion to surfaces. For example, appendages such as pili and fimbriae aid cells in overcoming repulsive forces between the cell membrane and abiotic surfaces (Fig. 1A). EPS secretion and pilus formation are active areas of investigation and have been reviewed elsewhere (Burrows, 2012; Flemming and Wingender, 2010). Previous reviews have also highlighted surface-specific motility such as swarming and twitching (Harshey, 2003). Here we focus on strategies bacteria use – often employing fimbriae, pili and EPS –to maintain attachment to surfaces or to optimize surface colonization in flow environments.
Holding tight
The shear stress generated by flow at a solid-liquid interface can easily overcome the adhesive forces anchoring cells onto a surface, potentially detaching them from substrata (De La Fuente et al., 2007). In flow, a cell experiences a drag force that is well-estimated as Fdrag = Aσ s , where A is the area of the cell exposed to flow (approximately the product of length and width for a rod-shape bacterium) and σ s is the local shear stress at the surface (Berg, 1993). In a microfluidic channel with rectangular cross section of height h and width w , and given flow rate Q (in m3 per second), the shear stress at the center of the wall can be estimated by σ s = 6Qμ/wh2 where μ is the fluid viscosity. While shear stress depends highly on the geometry of the flow, it is generally larger in environments with higher flow speeds. Thus, the drag force on an attached cell typically increases with flow intensity, and the attachment strength required for a cell to resist removal by shear will depend on the flows that characterize its environmental niche (Bakker et al., 2004). Cell adhesion forces range from a few to hundreds of picoNewtons (pN), which is sufficient to maintain attachment in a variety of flow environments. These forces also strongly depend on chemistry (Garrett et al., 2008) and mechanical properties of the substrate (Lichter et al., 2008).
Some bacteria, like the prosthecate Caulobacter crescentus, stand out among microbial models of shear resistance with their extremely strong surface attachment. Single C. crescentus cells construct an adhesive holdfast, which is composed of a sticky substance that localizes at the cell poles, to withstand forces as large as 1 µN, which effectively renders them irreversibly surface-attached (Tsang et al., 2006). C. crescentus cells can withstand shear stresses as high as 1 MPa (their typical surface area being on the order of 1 µm2). It is not clear why C. crescentus evolved such extreme attachment strength, given that the typical shear stress in their natural freshwater environments is expected to be orders of magnitude lower. One hypothesis is that strong attachment prevents grazing by predators (Parry, 2004).
Catch bonds
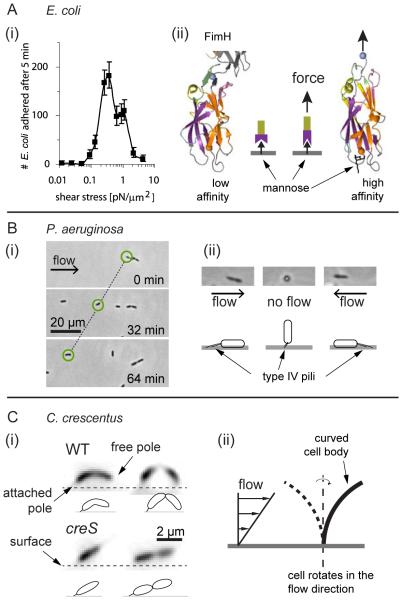
Paradoxically, multiple examples exist in which increasing shear stress enhances cell attachment to surfaces. For example, Escherichia coli is subject to flows spanning a wide range of intensities as it colonizes different host tissues (Thomas et al., 2004), and it has evolved adaptable fimbriae that counteract removal by flow to optimize colonization in these diverse environments (Thomas, 2008). Typical bacterial fimbriae fail to maintain adherence upon application of a sufficiently large force, whereas among many strains of E. coli, type I fimbriae attachment is enhanced under increasing tensile load (Thomas et al., 2008) (Fig. 3A). In these cases, type I fimbriae are capped with a tip protein called FimH that specifically binds the mannose that coats the surfaces of many tissues. Under tension, the mannose-bound FimH changes conformation, adopting a strong attachment state (Le Trong et al., 2010) (Fig. 3A). This force-dependent attachment is known as a catch bond, which increases the reliability of cell attachment to the surface in strong flow environments but also leads to a “stick and roll” adhesion where cells slowly continuously move in the direction of the flow while remaining attached to the surface (Thomas et al., 2004).
Figure 3. Bacteria leverage fluid flow at the scale of a single cell.
(A) E. coli use a catch bond mechanism to enhance attachment to surfaces in flow conditions. (left) Above a critical shear stress, cells are more likely to remain attached to a surface compared to cells that experience lower shear stress, as demonstrated by the peak in the number of adherent cells at finite shear stress (Nilsson et al., 2006). (right)The fimbrial capping protein FimH changes conformation when the fimbria is under tension, thereby increasing its affinity for surface-bound mannose (Le Trong et al., 2010). The mechanics of FimH are analogous to a finger trap toy, where extension enhances binding via twisting. (B) (left) P. aeruginosa attaches to surfaces with polar pili and cells migrate via twitching motility in the direction opposite to the flow. (right) Flow reorients cells in the direction opposite to the flow (Shen et al., 2012); successive pili extensions and retractions promote upstream migration. (C) (left) C. crescentus reorients in the direction of the flow when growing on surfaces (Persat et al., 2014). (right) Hydrodynamic forces act on the curved C. crescentus cell body, attached from one pole, to orient its free pole toward the surface. Thus, new cells are born close to the surface and can better attach immediately following separation from the mother cell. A straight cell dividing in flow has its pole oriented away from the surface, thereby reducing the likelihood of attachment to the surface after separation, which then reduces the rate of surface colonization.
Catch bonds thus may be beneficial during gastrointestinal colonization, allowing cells to remain in a beneficial microenvironment by anchoring to the epithelium and modulating their shear resistance in response to flow conditions. Notably, uropathogenic strains of E. coli possess mutations in FimH that reduce the dependence of adhesion on shear stress, indicating that the benefit of a catch bond may be lost in a low frequency pulsatile flow environment (Thomas et al., 2004). E. coli cells can further strengthen attachment by leveraging the mechanical deformation of type I fimbriae. These fibers extend under tension forces, so that the force applied to a single attached cell is distributed among multiple fimbriae, decreasing the load experienced by each fiber, and improving the ability of a cell to remain attached to a surface (Whitfield et al., 2014).
Shear stress also enhances the attachment properties of Pseudomonas aeruginosa, which exhibit longer residence times on surfaces when subjected to flow than under static conditions (Lecuyer et al., 2011). However, unlike E. coli, P. aeruginosa employs a mechanism that is independent of surface chemistry. Indeed, when subjected to shear, adhesion of P. aeruginosa increases on both glass and elastomeric substrata. The flow-dependent increase in residence time of P. aeruginosa is diminished in mutants lacking polar type I and IV pili (cupA1 and pilC), flagella (flgK), or the ability to synthesize certain EPS matrix components (pelA). While these observations do not entirely eliminate the possibility of attachment via catch bonds, the findings suggest an alternative mechanism of shear-dependent adhesion whereby multiple adhesive structures participate to increase surface attachment.
These two examples are not rare among bacteria, as shear-enhanced adhesion has been observed in a variety of other contexts. For example, other E .coli fimbral structures can form catch bonds with distinct ligands (Nilsson et al., 2006; Tchesnokova et al., 2010), and increased shear stress promotes the adhesion of Staphylococcus epidermis cell clusters to human fibrinogen-coated surfaces (Weaver et al., 2011) and of S. aureus to fibers of the mechanosensitive von Willebrand factor (Pappelbaum et al., 2013).
Upstream migration
Some bacterial surface interaction mechanisms simultaneously enable surface attachment and locomotion. Type IV pili, for example, are cell-surface structures that can be rapidly polymerized and depolymerized (Burrows, 2012), and cells use them to move over surfaces via successive pilus extension, tip attachment, and retraction, which altogether is termed twitching motility (Gibiansky et al., 2010; Mattick, 2002). P. aeruginosa and numerous other bacteria use twitching motility to explore surfaces prior to forming biofilms (Zhao et al., 2013). Pili extension and retraction also promote intimate contact between single cells and the host during infection (Comolli et al., 1999).
A striking architectural feature of type IV pili and other adhesive structures (e.g., flagella and holdfasts) is their strict localization to the poles of many rod-shaped cells. In environments with flow, a cell attached to a surface via a polar appendage will experience forces that tend to align the cell body with the vicinal flow field. In P. aeruginosa, which attaches to surfaces using its polar type IV pili, this phenomenon produces the surprising flow-driven behavior of upstream motion. Under flow, pilus-attached P. aeruginosa cells align in the direction of fluid movement with the piliated pole facing upstream. By successively retracting and extending pili, such cells migrate upstream, against the direction of the flow, despite the force oriented opposite to them generated by shear stress (Fig. 3B) (Shen et al., 2012). This behavior has also been observed in the plant pathogen Xyllela fastidiosa (Meng et al., 2005) and E. coli harboring type I pili (Rangel et al., 2013), and it may be a general feature of surface-attached species possessing motorized polar pili.
Cell shape
Bacteria possess a wide variety of cell morphologies, and each species robustly maintains a characteristic shape by precisely coordinating complex cell wall synthesis machineries (Typas et al., 2012). The function of cell shape is likely to depend on the typical environment of each species, but the underpinnings of this cell shape-niche relationship remain unknown in the vast majority of cases (Young, 2006). In some instances, however, there are hints about how cell shape may constitute an adaptation to specific environmental conditions. For example, the helical shape of Helicobacter pylori enhances swimming motility in hydrogels, a feature that aids cells in penetrating mucus layers during stomach infection (Sycuro et al., 2012). In contrast, the curved bacterium C. crescentus harnesses its shape and the mechanics of its hydrodynamic environment to enhance surface colonization (Persat et al., 2014). As mentioned above, C. crescentus cells attach to surfaces via a polar holdfast. In flow, surface-attached cells orient in the direction of the flow (Fig. 3C). Shear stress generates a torque on their curved cell bodies, which rotates them such that their unattached poles arc towards the substratum. Consequently, mother cells deposit newly born daughter cells onto the surface immediately downstream, which leads to the colonization of the downstream surface and the formation of a biofilm. Indeed, straight mutants of C. crescentus are less likely to have their progeny immediately attach to the surface following division, and such mutants are more frequently lost to the bulk flow (Persat et al., 2014). Thus C. crescentus may have evolved its curved shape to enhance surface colonization in environments with flow, indicating that bacterial morphology is potentially a result of adaptation to specific mechanical environments.
Touching down
As described above, bacteria adopt many phenotypes that can confer a fitness advantages when cells are associated with a substrate. Transitioning from a planktonic swimming state to surface attachment is presumably an expensive regulatory decision in terms of energetic and potential opportunity cost. In several notable cases, bacteria coordinate transitions between attachment to and detachment from surfaces, making specific use of mechanical cues transduced via cell-surface structures.
Swimming motility allows cells to explore the bulk of a fluid, but becomes largely unnecessary after surface attachment. Consequently, many flagellar systems possess a mechanism for disabling rotation in response to mechanical forces. In a low Reynolds number environment, an object moving very close to a boundary experiences a larger viscous force compared to that which it would experience far away from the surface (Goldman et al., 1967). Relative to that of a planktonic cell, the rotating flagellum of a surface-attached cell experiences a significantly larger drag force, increasing the load on the flagellar motors. E. coli harnesses this hydrodynamic effect and subsequently alters flagellar rotation (Lele et al., 2013). More generally, many bacterial species exhibit behavioral changes upon inhibition of flagellar rotation. EPS secretion, for example, is strongly modulated in response to the load on flagella by B. subtilis and V. parahaemolyticus (Belas, 2014; Guttenplan et al., 2010). Similarly, C. crescentus stimulates deployment of its holdfast using a flagellum-dependent mechanism when attaching to surfaces, strengthening adhesion when necessary (Li et al., 2012). Recent work has suggested that bacteria also possess the means to translate surface contact into physiological changes in gene expression independently of flagellar function (Siryaporn et al., 2014). However, the contributions of mechanics in all of the above examples remain to be determined quantitatively.
We note that other systems could potentially enable bacteria to mechanically sense surfaces. For example, the mechanosensitive channels protecting the integrity of the cell wall upon osmotic shock (Phillips et al., 2009) may also be sensitive to mechanical deformation of the cell wall and trigger surface-specific cellular responses, similar to those that occur among eukaryotes (Vogel and Sheetz, 2006). Analogous to flagella-surface interactions, type IV pili are mechanically actuated cellular structures that could represent ideal mechanosensors, since their function is highly dependent on surface contact (Skerker and Berg, 2001). Altogether, these mechanisms likely promote the colonization of surfaces and help to regulate the transition from the unicellular state to a multicellular lifestyle.
Mechanics at the level of multicellular structures
In favorable environments, single surface-associated cells grow and divide or aggregate, thus initiating the formation of sessile, multicellular structures known as biofilms (Hall-Stoodley et al., 2004). When nucleated from a single founder cell, biofilms are often genetically homogeneous (van Gestel et al., 2014), although their finite sizes and spatial organizations generate distinct microenvironments for individual cells within them. For example, some cells occupy space near the substratum while others localize to the biofilm exterior, adjacent to the surrounding fluid (O'Toole et al., 2000). The availability of growth-limiting nutrients and other solutes often varies along sharp concentration gradients as a function of biofilm depth; as a result, a genetically homogeneous biofilm population can exhibit strongly heterogeneous phenotypes (Stewart and Franklin, 2008). In this manner and many others, physical and mechanical constraints affect bacterial behavior within biofilms. There is currently only limited quantitative and qualitative understanding of the effects of mechanics on multicellular bacterial development. Here, we highlight recent work exploring the influence of mechanics on biofilms, with emphasis on matrix rheology, fluid flow, and molecular transport.
Rheology
The biofilm matrix is a complex, heterogeneous gel-like material whose components vary from one bacterial species to another. The biophysical properties of the matrix have implications for the bacteria residing within biofilms, including their spatial organization and ecological interactions. For example, the matrix is dense and thus only poorly permeable to the surrounding fluid, ensuring that flow primarily occurs around biofilms (de Beer et al., 1994; Stewart, 2012). A consequence of this structural feature is that bacterial cells residing deep inside of biofilms receive only those nutrients capable of diffusing through the matrix. Additionally, cells residing in the outermost biofilm layers often rapidly deplete these nutrients, further denying access to cells in the interior. The resulting gradients in nutrient availability (and other solute concentrations) commonly generate physiological heterogeneity within biofilms, even those that are monoclonal, as cells within them adjust to their distinct local microenvironments (Xu et al., 1998).
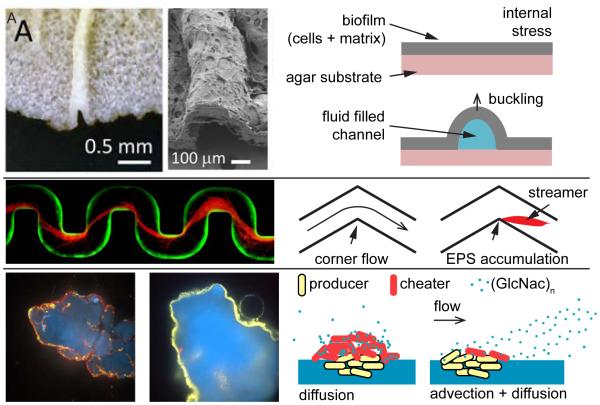
Macroscopically, biofilms are examples of living soft matter composed of cells and EPS matrix. The matrix displays elastic, plastic, and viscous properties, allowing biofilms to distort under mechanical forces. The effective mechanical response to forces on biofilms resembles that of a viscoelastic fluid (Wilking et al., 2011); biofilm stiffness is affected by the chemistry of the environment (Wloka et al., 2004) and the growth state of the resident bacterial population (Rogers et al., 2008). Biofilm expansion also generates mechanical stresses that can modify biofilm morphology and internal cellular organization. For example, in Bacillus subtilis biofilms growing at an air-liquid interface, compressive stresses generated by growth and expansion in a confined space leads to alteration of global biofilm morphology. In particular, B. subtilis biofilm sheets buckle to generate wrinkled pellicles at the surface of the fluid (Trejo et al., 2013). At solid-liquid interfaces, stresses generate wrinkles at locations of the weakest matrix stiffness. Cell death locally reduces biofilm thickness, thus focusing mechanical stress or reducing local biofilm adhesion, leading to vertical buckling of the matrix (Asally et al., 2012). These morphological changes can generate a network of fluid-filled channels that increase permeability and enable nutrient transport by flow, thus improving nutrient availability to cells within the biofilm compared to purely diffusive transport (Wilking et al., 2013). Similarly, the wrinkled morphology of P. aeruginosa biofilms increases their overall surface area, thereby improving oxygen uptake (Kempes et al., 2014).
Flow
The flow around biofilms can also strongly affect their morphology, deforming their structures. Shear stress applied to attached bacteria can wash away secreted compounds (Liu and Tay, 2002), affecting biofilm density, limiting growth, and as a result, reducing the overall biofilm size (Kostenko et al., 2010). Following the transition from single attached cells to larger multicellular biofilm communities, mechanical effects can underpin the spread of bacteria to new locations. Large shear stresses generated under strong flow conditions may lead to biofilm breakage, dislodging bacteria from the community (Purevdorj et al., 2002). Consequently, formerly biofilm-embedded bacterial cells are able to disperse to new territory and potentially colonize other environments that are more favorable (Stoodley et al., 1999).
One striking example of the effect of flow on biofilm architecture appears in irregular flow geometries. At corners and bends in curved channels, biofilms of some species develop as streamers – long, filamentous structures suspended in the fluid – nucleating at specific surface topological irregularities. Rusconi et al. showed that flow around corners promotes initiation of streamer formation (Rusconi et al., 2010). Biofilms nucleate at the downstream end of a bend and elongate in the direction of the flow, into the channel centerline (Fig. 4B). Numerical simulations suggest that the attachment point of a streamer co-localizes with flow moving fluid in the direction perpendicular to the main flow plane. The intensity of this secondary flow increases with sharper turns, which is consistent with the observation that streamers form more rapidly in such geometries (Rusconi et al., 2010).
Figure 4. Effect of mechanics on multicellular communities.
(A) Photograph and scanning electron micrographs show fluidic channels within biofilms generated by buckling of the EPS matrix (Wilking et al., 2013). Fluid evaporation through the biofilm generates flow within the channels leading to rapid advective transport of nutrients within the biofilm. In the absence of channels, nutrients only slowly diffuse into the biofilm. (B) Fluid flow promotes biofilm extrusions at channel bends that develop into fiber-like streamers extending into the channel centerline (Drescher et al., 2013). These streamers form as channel bends induce localized flow patterns potentially favoring the accumulation of EPS (Rusconi et al., 2011). (C) The interplay between diffusive and advective transport of a nutrient shapes the interactions between “producer” enzyme-secreting cells (yellow) that digest a chitin substrate (blue) and mutant “cheater” cells that do not secrete the chitinase enzyme (red) (Drescher et al., 2014). Without flow, diffusion of liberated chitin oligomers (GlcNac)n permits “cheater” cells to exploit populations of chitinase-producing cells. In contrast, flow rapidly removes the soluble (GlcNac)n released from the chitin surface, denying access to “cheaters” and rendering secreted chitinase-production evolutionarily stable.
The formation of streamers therefore depends on the characteristics of the flow coupled with the topology of the surface. A critical feature of streamer growth, in contrast to surface-associated biofilms, is their spatial extension far into the bulk of the fluid. While wall-associated biofilms only modestly impair flow, streamers obstruct flow channels near the centerline and thus slow flow much more dramatically than do biofilms on the walls. This effect generates a positive feedback loop in which the decrement of flow favors accumulation of EPS and other suspended materials, until streamers completely clog the channel and catastrophically stop the flow (Drescher et al., 2013). Because turns and bends are common within flow systems, such as an animal’s vasculature or an industrial cooling system, we anticipate that streamer-induced clogging is a frequent impediment in many fluidic systems and networks. Consistent with this idea, streamers form in flow elements common to various environmental systems, including filters, soil, and stents (Drescher et al., 2013). Many studies in this realm have focused on P. aeruginosa as a model organism (Stoodley et al., 2002); similarly S. aureus forms streamers that rapidly clog channels, and it does so in a manner that depends on the chemistry of the nucleating surface (Kim et al., 2014). Although only investigated recently, streamers are now predicted to be common when biofilms encounter bends and flow.
Transport
Bacteria survive, grow, and communicate by detecting and frequently importing compounds present in their environments. Nutrients, signaling molecules, and antimicrobials affect the behavior of individual bacteria and the development of their multicellular communities. These molecules generally reach cells by diffusion, but flow can dramatically modify the spatiotemporal distribution of such compounds and the effective concentrations that surface-attached cells experience (Berg and Purcell, 1977; Squires et al., 2008). In practice, the contribution of flow to the transport process can be estimated with the Péclet number, which measures the relative contribution to transport of a solute by advection (mediated by flow) compared to the contribution by diffusion (Pe = UL/D, where U is a typical value of flow speed, L is the length scale of the system, e.g. of the biofilm, and D is the diffusivity of the solute). For example, a Péclet number much larger than unity signifies that solutes mainly move with the flow. Conversely, the transport of a solute is dominated by diffusion when the flow is slow enough to yield a Péclet number much smaller than one.
In many instances, nutrients are not derived from the bulk fluid surrounding a biofilm, but rather from the substrata to which the cells are attached. In such cases, biofilm-dwelling cells often digest the substrata on which they reside by secreting extracellular enzymes, liberating nutrients that can freely diffuse away. This scenario generates a public goods conundrum: cells that secrete digestive enzymes can be exploited by other species that fail to produce the enzyme but nonetheless benefit from its production (West et al., 2006). Recent work has shown that a flow environment can help to overcome this evolutionary dilemma (Driscoll and Pepper, 2010). Outside of its human host, the pathogen Vibrio cholerae grows on solid particles of the biopolymer chitin, secreting chitinase enzymes that digest chitin and liberate the soluble product N-acetylglucosamine (GlcNac) and oligomers, which are excellent nutrients for growth. In the absence of flow, a “cheater” (a mutant that does not produce chitinase) can scavenge diffusing nutrients released by the producers’ chitinase activity and outcompete the producer because the cheater does not pay the cost of producing chitinase (Fig. 4C). In contrast, flow disperses nutrients generated by producers away from the chitin granules, limiting growth exclusively to chitinase-producers and their offspring who are residing on the chitin surface; cheater mutants are essentially starved out of the system (Drescher et al., 2014).
Similarly, flow transports signaling molecules and other secreted compounds that mediate social interactions within and between bacterial populations (Miller and Bassler, 2001). An important example is how flow affects quorum sensing, an intra- and inter-species bacterial communication system that is used to synchronize collective behaviors. Quorum sensing relies on the production, release, and detection of extracellular signal molecules called autoinducers. In well-mixed environments, cells assess cell density by measuring the local autoinducer concentration to initiate group behaviors. By contrast, in a heterogeneous environment, for example in a sessile population in flow, advection and diffusion affect local autoinducer concentrations. Bacteria may, in fact, use quorum-sensing systems to detect the flow in their surroundings, thus probing their vicinal surroundings for their growth potential or for other cues (Cornforth et al., 2014).
Connecting scales
Fully understanding bacterial behavior requires efforts to address the common and intimate association of cells with surfaces. We must consider individual cells, their morphology, and their responses to environmental cues and mechanical forces such as those described above: surface adhesion in the presence of flow, solute transport, and biofilm rheological properties. Cell division and matrix secretion combine to promote biofilm formation on the scale of many hundreds to millions of cells for which flow, transport, and rheology feed back onto the population dynamics within biofilm populations. Clarifying the consequences of mechanics for bacterial cells in isolation and as members of collectives is therefore central not only to understanding the transitions between individual and multicellular bacterial behavior but also to bacterial evolution in the broadest sense.
The phenomena discussed in the previous sections, which largely pertain to the behavior of single cells, influence transitions from surface occupation by individual bacteria to the formation of large multicellular communities. An adaptation that first appears to benefit only individual cells could influence the fitness of its descendants many generations later (Odling-Smee et al., 1996). A prime example of such adaptation is the increased ability of C. crescentus to colonize surfaces in flow as described above. Cell curvature increases the rate of surface attachment by daughter cells, thereby increasing the rate of surface colonization and ultimately enabling capture of increased niche space in a three-dimensional biofilm matrix that may extend many cell lengths away from the substratum. Populations of curved cells thus form robust biofilms more rapidly than straight cells, such that curvature may be viewed as adaptive not only at the scale of a single colonizing bacterium but at the scale of clonal populations descended from surface-associated founder cells. Similarly, the upstream motility of single P. aeruginosa cells provides a group-level advantage when competing with planktonic cells and other species during colonization of fluidic networks (Siryaporn et al., 2015). Phenotypes that evolved in response to mechanical forces experienced by single surface-attached cells could thus contribute to pathogenic infection by a large bacterial population.
When forming biofilms, bacteria implement individual gene expression programs that ultimately contribute to the properties of the entire community. As individual cells respond to or modify the chemical and mechanical features of their environment, they express phenotypes and generate cell group configurations that contribute to collective survival. For instance, impaired flagellar rotation upon surface contact has been proposed as a signal that induces biofilm formation, as motility and EPS secretion are negatively correlated (Blair et al., 2008; Krasteva et al., 2010). Thus, surface contact generates a mechanical cue triggering an intracellular signaling cascade that leads to many phenotypes, which culminate in stable adhesion to the substratum. Upon growth and division, bacteria modify their mechanical environment by secreting EPS that helps maintain the spatial coherence of clonal lineages within nascent biofilms (Millet et al., 2014; Nadell and Bassler, 2011) and discourages invasion by planktonic cells (Nadell et al., 2015). These and other recent studies together suggest that biofilms often emerge from the behavior of individual cells that are locally cooperative within their strain or clonal lineage and globally competitive with other strains and species with which they may be growing (Mitri and Foster, 2013).
Subsequent to the formation of multicellular communities, biofilm structure and mechanical properties feed back upon the forces and solute concentrations experienced by biofilm-dwelling bacteria to further influence their individual and collective behaviors. For example, because advection is negligible within the matrix, solute transport occurs primarily through diffusion, which in turn, leads to heterogeneous concentration gradients of soluble compounds secreted or absorbed by individual bacteria (Stewart, 2003). As we have described above, these solute distributions further influence the competitive dynamics within and among competing cell lineages inside biofilms. Biofilm structure and diffusive properties likewise influence the accumulation of autoinducers involved in the quorum-sensing-regulated activation of group-wide phenotypes such as virulence in P. aeruginosa (De Kievit et al., 2001). In V. cholerae, by contrast, quorum sensing feeds back into the structure and rheology of the biofilm to repress EPS at high cell density, thus initiating dispersal of the pathogen (McDougald et al., 2012).
Do single cells actively sense mechanical forces? What is the relevance of these features in realistic ecological contexts? How do biofilms form in complex and diverse environments such as the digestive tract? Do organ mechanics affect bacterial population development? These and many other questions are natural extensions of past and current experimental explorations of bacterial behavior. Resolving the connections between mechanics and biology in the bacterial world will require integrative approaches that combine genetics, biochemistry, chemistry, evolutionary biology, physics, and engineering principles.
Surface-specific mechanics are ubiquitous in the bacterial world, and the examples above highlight the essential role that they play in many elements of bacterial processes. The insights gained from a mechanics-informed view of bacteria will, in turn, improve our ability to control microbes in settings where they can be helpful or harmful to humans. For instance, a better understanding of how mechanics contribute to regulating virulence may provide alternative approaches to fight infections and help overcome the rise of antibiotic resistance. More generally, bacterial mechanics represent an exciting research direction for biologists aiming to understand bacterial physiology in realistic environments and for engineers and physicists aiming to develop new tools and models to interface with microbiology and develop a fully interdisciplinary understanding of bacterial behavior.
Acknowledgements
This work was supported by the Gordon and Betty Moore Foundation through Grant GBMF 2550.02 to the Life Sciences Research Foundation (A.P.), the NSF grant CBET-1330288 (Z.G. and H.A.S.), the NSF grant MCB-1119232 (B.L.B., H.A.S. and N.S.W.) the Howard Hughes Medical Institute (B.L.B. and C.D.N.), the National Institutes of Health Grant R01GM065859 (B.L.B.), the STX fellowship (M.K.K.), the Human Frontier Science Program (K.D.), and the Max Planck Society (K.D. and C.D.N.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asally M, Kittisopikul M, Rue P, Du Y, Hu Z, Cagatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Suel GM. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker DP, Postmus BR, Busscher HJ, van der Mei HC. Bacterial strains isolated from different niches can exhibit different patterns of adhesion to substrata. Appl Environ Microbiol. 2004;70:3758–3760. doi: 10.1128/AEM.70.6.3758-3760.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014;22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Berg HC. Random walks in biology. Princeton University Press; Princeton, N.J.: 1993. Expanded edn. [Google Scholar]
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- Berg HC, Purcell EM. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, Yildiz FH, Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- Chew SC, Kundukad B, Seviour T, van der Maarel JR, Yang L, Rice SA, Doyle P, Kjelleberg S. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio. 2014;5:e01536–01514. doi: 10.1128/mBio.01536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli JC, Hauser AR, Waite L, Whitchurch CB, Mattick JS, Engel JN. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect Immun. 1999;67:3625–3630. doi: 10.1128/iai.67.7.3625-3630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth DM, Popat R, McNally L, Gurney J, Scott-Phillips TC, Ivens A, Diggle SP, Brown SP. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4280–4284. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- de Beer D, Stoodley P, Lewandowski Z. Liquid flow in heterogeneous biofilms. Biotechnol Bioeng. 1994;44:636–641. doi: 10.1002/bit.260440510. [DOI] [PubMed] [Google Scholar]
- De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol. 2001;67:1865–1873. doi: 10.1128/AEM.67.4.1865-1873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente L, Montanes E, Meng Y, Li Y, Burr TJ, Hoch HC, Wu M. Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl Environ Microbiol. 2007;73:2690–2696. doi: 10.1128/AEM.02649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol. 2014;24:50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher K, Shen Y, Bassler BL, Stone HA. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4345–4350. doi: 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll WW, Pepper JW. Theory for the evolution of diffusible external goods. Evolution. 2010;64:2682–2687. doi: 10.1111/j.1558-5646.2010.01002.x. [DOI] [PubMed] [Google Scholar]
- Dunne WM., Jr. Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Fritton SP, Weinbaum S. Fluid and Solute Transport in Bone: Flow-Induced Mechanotransduction. Annual review of fluid mechanics. 2009;41:347–374. doi: 10.1146/annurev.fluid.010908.165136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TR, Bhakoo M, Zhang Z. Bacterial adhesion and biofilms on surfaces. Progress in Natural Science. 2008;18:1049–1056. [Google Scholar]
- Gibiansky ML, Conrad JC, Jin F, Gordon VD, Motto DA, Mathewson MA, Stopka WG, Zelasko DC, Shrout JD, Wong GC. Bacteria use type IV pili to walk upright and detach from surfaces. Science. 2010;330:197. doi: 10.1126/science.1194238. [DOI] [PubMed] [Google Scholar]
- Goldman AJ, Cox RG, Brenner H. Slow Viscous Motion of a Sphere Parallel to a Plane Wall .I. Motion through a Quiescent Fluid. Chem Eng Sci. 1967;22:637. [Google Scholar]
- Guasto JS, Rusconi R, Stocker R. Fluid mechanics of planktonic microorganisms. Annual Review of Fluid Mechanics. 2012;44:373–400. Vol 44. [Google Scholar]
- Guttenplan SB, Blair KM, Kearns DB. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 2010;6:e1001243. doi: 10.1371/journal.pgen.1001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempes CP, Okegbe C, Mears-Clarke Z, Follows MJ, Dietrich LE. Morphological optimization for access to dual oxidants in biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:208–213. doi: 10.1073/pnas.1315521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Drescher K, Pak OS, Bassler BL, Stone HA. Filaments in curved streamlines: rapid formation of Staphylococcus aureus biofilm streamers. New Journal of Physics. 2014;065024;16 doi: 10.1088/1367-2630/16/6/065024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostenko V, Salek MM, Sattari P, Martinuzzi RJ. Staphylococcus aureus biofilm formation and tolerance to antibiotics in response to oscillatory shear stresses of physiological levels. FEMS Immunol Med Microbiol. 2010;59:421–431. doi: 10.1111/j.1574-695X.2010.00694.x. [DOI] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Trong I, Aprikian P, Kidd BA, Forero-Shelton M, Tchesnokova V, Rajagopal P, Rodriguez V, Interlandi G, Klevit R, Vogel V, et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer S, Rusconi R, Shen Y, Forsyth A, Vlamakis H, Kolter R, Stone HA. Shear stress increases the residence time of adhesion of Pseudomonas aeruginosa. Biophys J. 2011;100:341–350. doi: 10.1016/j.bpj.2010.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter JA, Thompson MT, Delgadillo M, Nishikawa T, Rubner MF, Van Vliet KJ. Substrata mechanical stiffness can regulate adhesion of viable bacteria. Biomacromolecules. 2008;9:1571–1578. doi: 10.1021/bm701430y. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tay J-H. The essential role of hydrodynamic shear force in the formation of biofilm and granular sludge. Water Res. 2002;36:1653–1665. doi: 10.1016/s0043-1354(01)00379-7. [DOI] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 2012;10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- Meng Y, Li Y, Galvani CD, Hao G, Turner JN, Burr TJ, Hoch HC. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J Bacteriol. 2005;187:5560–5567. doi: 10.1128/JB.187.16.5560-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Millet YA, Alvarez D, Ringgaard S, von Andrian UH, Davis BM, Waldor MK. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 2014;10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitri S, Foster KR. The genotypic view of social interactions in microbial communities. Annu Rev Genet. 2013;47:247–273. doi: 10.1146/annurev-genet-111212-133307. [DOI] [PubMed] [Google Scholar]
- Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadell CD, Drescher K, Wingreen NS, Bassler BL. Extracellular matrix structure governs invasion resistance in bacterial biofilms. The ISME journal. 2015 doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson LM, Thomas WE, Trintchina E, Vogel V, Sokurenko EV. Catch bond-mediated adhesion without a shear threshold: trimannose versus monomannose interactions with the FimH adhesin of Escherichia coli. J Biol Chem. 2006;281:16656–16663. doi: 10.1074/jbc.M511496200. [DOI] [PubMed] [Google Scholar]
- O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Odling-Smee FJ, Laland KN, Feldman MW. Niche construction. Am Nat. 1996:641–648. [Google Scholar]
- Pappelbaum KI, Gorzelanny C, Grassle S, Suckau J, Laschke MW, Bischoff M, Bauer C, Schorpp-Kistner M, Weidenmaier C, Schneppenheim R, et al. Ultralarge von Willebrand factor fibers mediate luminal Staphylococcus aureus adhesion to an intact endothelial cell layer under shear stress. Circulation. 2013;128:50–59. doi: 10.1161/CIRCULATIONAHA.113.002008. [DOI] [PubMed] [Google Scholar]
- Parry JD. Protozoan grazing of freshwater biofilms. Adv Appl Microbiol. 2004;54:167–196. doi: 10.1016/S0065-2164(04)54007-8. [DOI] [PubMed] [Google Scholar]
- Persat A, Stone HA, Gitai Z. The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat Commun. 2014;5:3824. doi: 10.1038/ncomms4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt BL, Dunn AR, Weis WI, Nelson WJ. Mechano-transduction: from molecules to tissues. PLoS Biol. 2014;12:e1001996. doi: 10.1371/journal.pbio.1001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EM. Life at low Reynolds-number. American Journal of Physics. 1977;45:3–11. [Google Scholar]
- Purevdorj B, Costerton JW, Stoodley P. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2002;68:4457–4464. doi: 10.1128/AEM.68.9.4457-4464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel DE, Marin-Medina N, Castro JE, Gonzalez-Mancera A, Forero-Shelton M. Observation of bacterial type I pili extension and contraction under fluid flow. PLoS One. 2013;8:e65563. doi: 10.1371/journal.pone.0065563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SS, van der Walle C, Waigh TA. Microrheology of bacterial biofilms in vitro: Staphylococcus aureus and Pseudomonas aeruginosa. Langmuir. 2008;24:13549–13555. doi: 10.1021/la802442d. [DOI] [PubMed] [Google Scholar]
- Rusconi R, Garren M, Stocker R. Microfluidics expanding the frontiers of microbial ecology. Annu Rev Biophys. 2014;43:65–91. doi: 10.1146/annurev-biophys-051013-022916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi R, Lecuyer S, Autrusson N, Guglielmini L, Stone HA. Secondary flow as a mechanism for the formation of biofilm streamers. Biophys J. 2011;100:3054–3054. doi: 10.1016/j.bpj.2011.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi R, Lecuyer S, Guglielmini L, Stone HA. Laminar flow around corners triggers the formation of biofilm streamers. Journal of the Royal Society Interface. 2010;7:1293–1299. doi: 10.1098/rsif.2010.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Siryaporn A, Lecuyer S, Gitai Z, Stone HA. Flow directs surface-attached bacteria to twitch upstream. Biophys J. 2012;103:146–151. doi: 10.1016/j.bpj.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siryaporn A, Kim MK, Shen Y, Stone HA, Gitai Z. Colonization, Competition, and Dispersal of Pathogens in Fluid Flow Networks. Curr Biol. 2015 doi: 10.1016/j.cub.2015.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siryaporn A, Kuchma SL, O'Toole GA, Gitai Z. Surface attachment induces Pseudomonas aeruginosa virulence. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:16860–16865. doi: 10.1073/pnas.1415712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires TM, Messinger RJ, Manalis SR. Making it stick: convection, reaction and diffusion in surface-based biosensors. Nat Biotechnol. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- Stewart PS. Diffusion in biofilms. J Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS. Mini-review: convection around biofilms. Biofouling. 2012;28:187–198. doi: 10.1080/08927014.2012.662641. [DOI] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Cargo R, Rupp CJ, Wilson S, Klapper I. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J Ind Microbiol Biotechnol. 2002;29:361–367. doi: 10.1038/sj.jim.7000282. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Lewandowski Z, Boyle JD, Lappin-Scott HM. Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid shear: an in situ investigation of biofilm rheology. Biotechnol Bioeng. 1999;65:83–92. [PubMed] [Google Scholar]
- Sycuro LK, Wyckoff TJ, Biboy J, Born P, Pincus Z, Vollmer W, Salama NR. Multiple peptidoglycan modification networks modulate Helicobacter pylori's cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokova V, McVeigh AL, Kidd B, Yakovenko O, Thomas WE, Sokurenko EV, Savarino SJ. Shear-enhanced binding of intestinal colonization factor antigen I of enterotoxigenic Escherichia coli. Mol Microbiol. 2010;76:489–502. doi: 10.1111/j.1365-2958.2010.07116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. Catch bonds in adhesion. Annu Rev Biomed Eng. 2008;10:39–57. doi: 10.1146/annurev.bioeng.10.061807.160427. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Nilsson LM, Forero M, Sokurenko EV, Vogel V. Shear-dependent 'stick-and-roll' adhesion of type 1 fimbriated Escherichia coli. Mol Microbiol. 2004;53:1545–1557. doi: 10.1111/j.1365-2958.2004.04226.x. [DOI] [PubMed] [Google Scholar]
- Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Annu Rev Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- Trejo M, Douarche C, Bailleux V, Poulard C, Mariot S, Regeard C, Raspaud E. Elasticity and wrinkled morphology of Bacillus subtilis pellicles. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2011–2016. doi: 10.1073/pnas.1217178110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang PH, Li G, Brun YV, Freund LB, Tang JX. Adhesion of single bacterial cells in the micronewton range. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nature Reviews Microbiology. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gestel J, Weissing FJ, Kuipers OP, Kovacs AT. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. The ISME journal. 2014;8:2069–2079. doi: 10.1038/ismej.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Weaver WM, Dharmaraja S, Milisavljevic V, Di Carlo D. The effects of shear stress on isolated receptor-ligand interactions of Staphylococcus epidermidis and human plasma fibrinogen using molecularly patterned microfluidics. Lab on a chip. 2011;11:883–889. doi: 10.1039/c0lc00414f. [DOI] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- Whitfield MJ, Luo JP, Thomas WE. Yielding elastic tethers stabilize robust cell adhesion. PLoS Comput Biol. 2014;10:e1003971. doi: 10.1371/journal.pcbi.1003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilking JN, Angelini TE, Seminara A, Brenner MP, Weitz DA. Biofilms as complex fluids. Mrs Bull. 2011;36:385–391. [Google Scholar]
- Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka M, Rehage H, Flemming HC, Wingender J. Rheological properties of viscoelastic biofilm extracellular polymeric substances and comparison to the behavior of calcium alginate gels. Colloid Polym Sci. 2004;282:1067–1076. [Google Scholar]
- Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, Luijten E, Parsek MR, Wong GC. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497:388–391. doi: 10.1038/nature12155. [DOI] [PMC free article] [PubMed] [Google Scholar]