Abstract
Epilepsy is the most common serious neurological disorder affecting 50–60 million persons worldwide. Electrophysiological data recordings, such as electroencephalogram (EEG), are the gold standard for diagnosis and pre-surgical evaluation in epilepsy patients. The increasing trend towards multi-center clinical studies require signal visualization and analysis tools to support real time interaction with signal data in a collaborative environment, which are cannot be supported by traditional desktop-based standalone applications. As part of the Prevention and Risk Identification of SUDEP Mortality (PRISM) project, we have developed a Web-based electrophysiology data visualization and analysis platform called Cloudwave using highly scalable open source cloud computing infrastructure. Cloudwave is integrated with the PRISM patient cohort identification tool called MEDCIS (Multi-modality Epilepsy Data Capture and Integration System). The Epilepsy and Seizure Ontology (EpSO) underpins both Cloudwave and MEDCIS to support query composition and result retrieval. Cloudwave is being used by clinicians and research staff at the University Hospital - Case Medical Center (UH-CMC) Epilepsy Monitoring Unit (EMU) and will be progressively deployed at four EMUs in the United States and the United Kingdom as part of the PRISM project.
Keywords: SUDEP, Electrophysiological signal analysis, EEG monitoring, EEG Signal analysis, Multi-modal seizure data, EDF, Epileptic events, Epilepsy and Seizure ontology
Introduction
Patients with epilepsy suffer from repeated seizures that manifest as physical or behavioral changes, which require intervention using medications or surgery. Multi-modal electrophysiological evaluations, such as electroencephalography (EEG), electrocardiography (EKG), blood oxygen and carbon dioxide levels, and sleep measurements, are essential for diagnosis and treatment of epilepsy. The precision and sensitivity of the EEG, for example Stereotactic EEG (S-EEG), play an important role in pre-surgical evaluation of epilepsy patients to identify the location and extent of epileptogenic network, which is responsible for seizures. Ongoing treatment of patients also requires continuous noninvasive monitoring of brain functions [4].
Monitoring patients over a long period of time involves recording EEG and related signal data from 30–100 channels and requires efficient storage of the resulting large data files. An electrophysiological recording of a patient over a 24 hours period with 100 or more channels results in 5–10GB of data. Hence, a scalable and easy-to-use signal analysis and visualization tool is critical for clinicians and research staff to review these large signal datasets. Effective signal analysis tools require support for multiple data management functionalities including:
Selection of specific sections of the signal data that are likely to feature interesting events, for example seizure-onset or suppression events in EEG data;
Facilitate quantitative measurements for research protocols, for example measurement of multi-modal parameters above or below threshold values. These measurements may be indicative of deteriorating patient condition;
Summarize percentage of seizure occurrences based on events and quantitative measurements for the given patient cohort population; and
Representation of long recordings in a compressed format that allows viewing of several hours of recording in a single “view page”.
The two approaches for developing signal analysis and visualization tools are: (a) Stand-alone tools that need to be downloaded and installed after appropriate configuration of the host computer, and (b) Web-based tools that are accessible through the ubiquitous Web browsers (e.g. Internet Explorer, Google Chrome, and Mozilla Firefox). Stand-alone tools for signal visualization and analysis are usually proprietary software applications or have been developed and extended in an adhoc manner with minimal documentation. These standalone tools have a number of limitations that make it difficult for clinical researchers to use them, including:
Limited re-usability across different institutions and projects due to heterogeneous computing environments, such as operating systems, hardware configuration, and software libraries;
No support for collaborative clinical studies that requires researchers across institutions to synchronously visualize and interact with a single electrophysiological data file; and
Expensive and difficult to maintain over a period of time as each installed version has to be manually configured and updated in response to each change in the original version of the tool.
In contrast, Web-based tools and applications address these challenges by leveraging the widely used Web browser platform, enable consistent user experience across computing environments, and support use of standardized protocols used in signal data analysis. The use of Web-based signal visualization and analysis tool is ideally suited for multi-center clinical studies with multi-user collaborations.
In this paper, we introduce Cloudwave, a Web-based, ontology-driven electrophysiological signal analysis and visualization platform that is integrated with open source cloud computing infrastructure for scalability. Cloudwave has been developed as part of the Prevention and Risk Identification of SUDEP Mortality (PRISM) project with funding support from the U.S. National Institute of Neurological Disorders and Strokes (NINDS). SUDEP or sudden and unexpected death in epilepsy is a poorly understood phenomenon in epilepsy patients and there are no known prevention mechanisms [8].
The aim of PRISM project is to recruit patients for SUDEP study from four participating Epilepsy Monitoring Units at the Case Western Reserve University (CWRU) University Hospitals-Case Medical Center (UH-CMC), Ronald Reagan University of California Los Angeles (UCLA) Medical Center (RRUMC-Los Angeles), the National Hospital for Neurology and Neurosurgery (NHNN, London, UK), and Northwestern Memorial Hospital (NMH Chicago). The aim of Cloudwave is to enable clinical researchers at the participating centers to collaboratively search, query, and visualize clinical events on shared, annotated signal data, such as seizure patterns, onset of generalized tonic phase, and onset of EEG suppression.
Materials
The signal data used in this paper was collected at the UHCMC EMU over a period of five days and consists of EEG, EKG, blood oxygen and carbon dioxide measurements. The signal data files were manually de-identified to remove all protected health information (PHI) and the data was stored in the European Data Format (EDF), which is an eXtensible Markup Language (XML)-based flexible signal data representation format.
European Data Format (EDF) for representing signal data
The European Data Format (EDF) is the de-facto standard for EEG recordings in commercial equipment and multicenter research projects [9]. An EDF data file consists of a header record followed by signal data records. The header record consists of various metadata elements describing the signal data, the version information, patient information, recording, recording date and time, number of data records and the number of signals. The data records contain consecutive fixed-duration “epochs” of polygraphed recordings of the given signal. For example, the duration of an EEG data record. The use of EDF in Cloudwave enhances interoperability and allows the de-identified data files to be shared among the participating EMUs in the PRISM project. Cloudwave stores the EDF data file in a relational database for faster signal access and rendering on a Web browser.
Epilepsy and Seizure Ontology (EpSO)
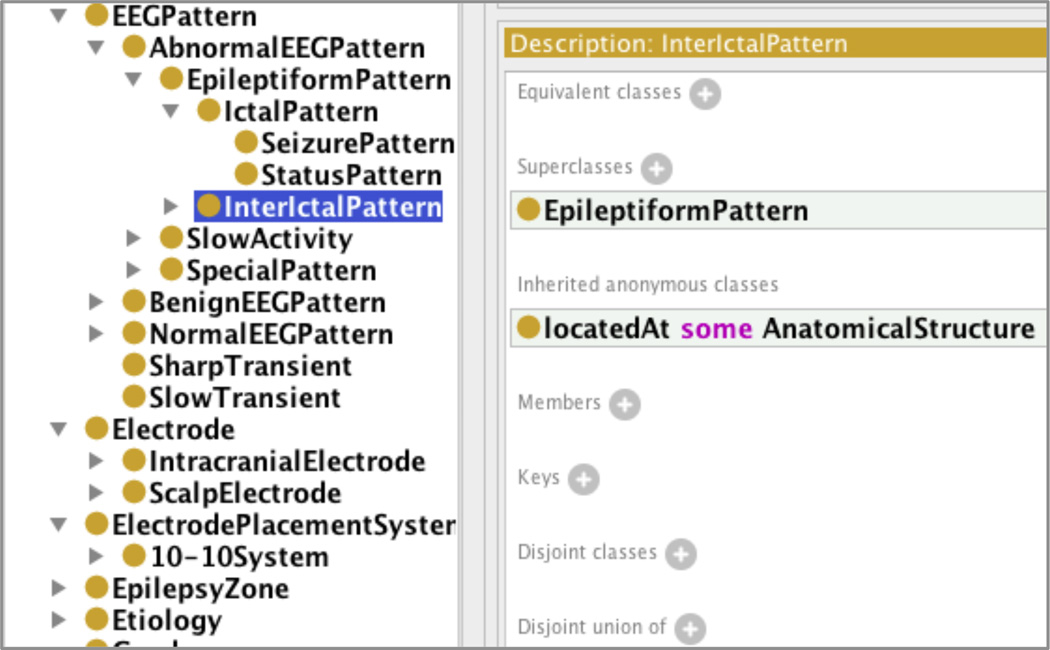
The use of a common terminological system for annotation and query of signal data is essential to reconcile data heterogeneity in multi-center clinical studies. As part of the PRISM project, we developed the Epilepsy and Seizure Ontology (EpSO) as a formal knowledge model representing epilepsy domain knowledge through close collaboration between epileptologists, including members of the International League Against Epilepsy (ILAE), and ontology engineers. EpSO has been modeled using the description logic-based Web Ontology Language (OWL2). EpSO currently models key concepts to effectively annotate signal data, including EEG patterns, both scalp and intracranial electrodes, their placement scheme (e.g. 10–10 system), and detailed brain anatomy to correlate signal events with their location [1][2]. Figure 1 illustrates a snapshot of the EpSO class hierarchy describing the various types of EEG patterns, such as abnormal, benign, and normal EEG patterns. EpSO currently has more than 1000 concepts and their related properties.
Figure 1.
Epilepsy and Seizure Ontology (EpSO)
HighCharts
HighCharts is a charting library that allows creation of timeline charts in the JavaScript language, including sophisticated navigations options like a small navigator series, preset date ranges, date picker, scrolling and panning. Cloudwave uses HighCharts for signal rendering on the web. Some of the key features of HighCharts are compatibility with all modern browsers, free availability for non-commercial applications, open-source, support for JavaScript, and does not require any client-side plug-ins.
HighCharts offers numerous chart types for rendering, simple configuration syntax, support for dynamic update of charts with additional data points or series, range selector for dynamically configuring time ranges, and visual navigation to area of interest on the chart. HighCharts also features event marker flags, allows multiple axes, tooltip labels, configurable date time axis, data grouping, export and print, zooming and panning, and ability to load external data from variety of sources.
Methods
The Cloudwave platform was developed using four design principles:
Fast access to individual signals stored in a multimodal EEG EDF store;
Ability to partition signals into meaningful “segments” based on seizure related events for easier quantitative analysis;
Use of EDF file header record and seizure events as metadata for faster identification of appropriate patient cohorts; and
Polygraph visualization of multiple signals on a single page for visual analysis by human reader.
Use cases for analysis of signal data
A set of use cases were defined and systematically documented to identify appropriate features to be implemented in Cloudwave, such as the ability to:
Search for seizure event information in signal data, including the time of event occurrence or the time duration between start and end of an event. For example, occurrence of ‘Sign-of-Four’ lateralizing sign event, time duration between “onset of jittery phase and end of jittery phase”;
List patients with Cardiac Arrhythmia who also have irregular heartbeat rates. This can be further classified as Bradycardia or Tachycardia, with markings on EKG signal when heartbeat rate is below or above a threshold, such as 60 beats per minute (BPM);
Measure Heart Rate Variability (HRV) for selected patients, which is a physiological phenomenon of variation in the time interval between heartbeats;
List patients with respiratory arrhythmia with markings on the respiration signal where the rate of respiration is above or below a threshold (e.g. 5/minute); and
Provide EEG suppression information for patients, including time duration of “EEG Suppression” in patients after a seizure occurrence and time duration of “EEG suppression to return to baseline” event.
Patients with above set of signal characteristics are of special clinical interest in the PRISM project. These requirements were reviewed to identify the common and distinct set of software functionalities that would need to be implemented in Cloudwave. For example, the functions to identify patients with Bradycardia and patients with abnormal respiration rate can both be implemented using a filtering function based on the value of the signal data. These requirements were identified during iterative collaborative meetings between the informatics team and epileptologists in the PRISM project. Cloudwave was developed using agile software development approach for rapid prototyping and review using the Ruby-on-Rails software technology stack. Figure 2 illustrates the different components of Cloudwave and the flow of data between these components.
Figure 2.
Cloudwave Components and Workflow
Generating EDF files
The signal data generated from patient recordings are stored in a signal data server, which is usually accessed using a commercial tool called Nihon Kohden Neural Workbench [10]. Clinicians and the EMU staff annotate the signal data with various types of seizure events after manual review of the multiple channel signals. The annotations and signal data are de-identified and converted into the EDF format and stored in a relational database. The Cloudwave tool is integrated with the Multi-modality Epilepsy Data Capture and Integration System (MEDCIS) to allow clinicians to query for patients using demography, medication, and diagnosis information. Clinicians can review the signal data of patients in a cohort by navigating from MEDCIS to Cloudwave. Figure 3 illustrates the screenshot of the MEDCIS query interface and the corresponding Cloudwave signal visualization interface [3].
Figure 3.
Cloudwave Access through MEDCIS Web Interface
Parsing the EDF file
The Cloudwave parser module parses an EDF file and extracts the EDF Header and Signal records (we discussed the structure of the EDF file in the Material section). EDF Header consists of two components: patient study header and signal header. Figure 4 illustrates the different components of the EDF file that are extracted and processed by the EDF parser module.
Figure 4.
Cloudwave EDF Parser and Preprocessor modules
Preprocessing and segmentation of signal data
The preprocessor module stores the metadata extracted by the EDF parser module in a relational database and processes the signal data by splitting them into multiple files, one file for each channel. This helps Cloudwave to efficiently store and retrieve the signal segments in response to user queries as compared to the original data files. Each segment corresponds to an individual channel from start to end of recording. Typically, a scalp EEG consists of 30–40 channels, hence splitting a single EDF creates 30–40 signal-segment files. The annotations are marked on the signal during acquisition and the signal-segment file locations are stored in the database for query and subsequent statistical analysis [6]. (Figure 4 illustrates the various processing steps).
The channel-wise segments are well suited for processing and storage on a cloud-computing infrastructure, such as the open source Hadoop platform using the Hadoop Distributed File System (HDFS). In the next version of the Cloudwave (under development), we are implementing parallelized processing of signal data for quantitative signal analysis for all the use cases.
Composing queries using multiple selection criteria
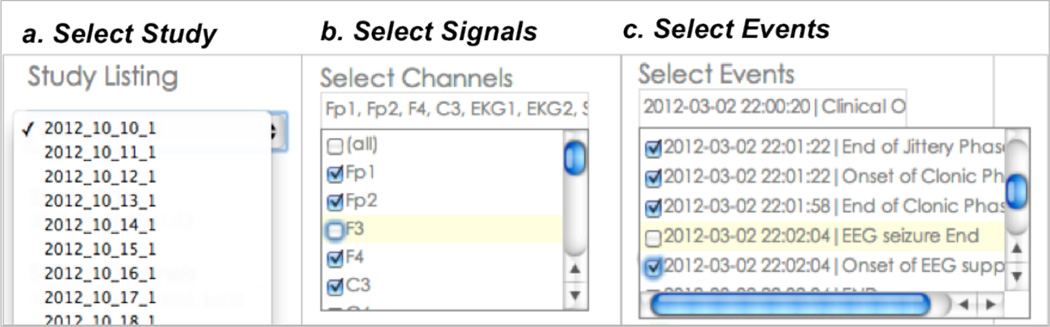
The Cloudwave user interface allows the user to flexibly select specific signal data file, the numbers of channels, and the events of clinical relevance to be rendered on the interface. The selection options are implemented as simple drop down menus with ability to select multiple values in the drop down menu (Figure 5 illustrates the three selection criteria). The query module functionality can be easily extended to support query constraints that involve real time computations over the signal data. For example, users will be able to query for HRV, EEG suppression events in the next version of Cloudwave.
Figure 5.
Cloudwave Query Selection Criteria
The query module uses EpSO-based annotations to reconcile the heterogeneity between terms describing clinical events across different EMUs. The ontology-driven data harmonization plays an important role in ability of Cloudwave to accurately render the correct signal data segment in response to user query on the Web browser (Figure 3 shows a screenshot of the Cloudwave signal rendering section with five annotations).
Rendering the signal data on the Web browser
The Cloudwave rendering module uses the HighCharts software to render the signal data in waveform that can be intuitively reviewed and interpreted by clinicians and EMU staff [5]. The rendering module generates the multimodal polygraph chart for multiple channels that are temporally synchronized in a single screen. This allows the users to review events across different modes of electrophysiology and correlate clinical events, for example an onset of seizure that is detected in the EEG signal channels has direct correlation to the EKG signals showing increase in heart rate and potentially higher blood pressure.
Another key features of the Cloudwave rendering module is dynamic update of events and signals rendered as multiple series in a single chart. The number of data points in a record can be dynamically updated by efficiently accessing the data server without incurring significant delays for very large signal recordings.
While navigating a large EEG signal recording, such as values recorded per minute over several days, it is important to provide a quick way to visualize a specific time range. The rendering module provides a range selector allowing the user to visually zoom in to a particular section of the signal by dragging the scrollbar or the edges of the navigator window to the area of special interest. In addition the user has the option for zooming/panning the time-range of interest using the mouse.
Events related to a seizure can be added as event markers. Different visual styles can be added to the markers to associate them with the related signal. With the exporting option enabled, users can export the chart to PNG, JPG, PDF or SVG format at the click of a button, or print the chart directly from the web page. These features are shown in a screenshot (Figure 6).
Figure 6.
Cloudwave Rendering Module
Downloading signal segments for further processing
Cloudwave allows users to download the specific signal data segments for further processing and analysis in their preferred tools. This feature makes Cloudwave complementary to existing analytical tools, such as MATLAB-based libraries, which can be used to support customized functionality. Cloudwave supports download of files containing the signal data in EDF format and events in plain text format
Cloudwave is an extensible platform that uses the Ruby-on-Rails web development framework based on the Model-View-Controller (MVC) architecture pattern. The MVC pattern enables strict separation of the application logic from the user interface, which allows seamless integration of multicenter EEG signal in Cloudwave. Next, we discuss the current status of Cloudwave and the result of our evaluation in using it for signal visualization and analysis.
Results
Cloudwave has been integrated with the MEDCIS for signal analysis and rendering of patient cohorts at the UH-CMC EMU. At present, Cloudwave database is populated with 1TB of signal data from 85 patients with seizures. A patient examination in the UH-CMC is typically done for five consecutive days after the patient is admitted and signal data are recorded in 6-hour segments, thereby generating 20 studies. Each study generates 1–2 GB of data per study. The evaluation dataset also consists of two patients with surgically implanted intracranial electrodes. Signal data from these patients with intracranial electrodes were recorded a period of one month that also generated about 1 TB of data.
Performance evaluation of Cloudwave
We evaluated the performance of Cloudwave in terms of time taken to:
Render an increasing number of discrete signals per page;
Render signal data with increasing size corresponding to different recording time period;
Time to load each signal (in milliseconds); and
Time to render each signal by the HighCharts charting module (in milliseconds).
The evaluation tests were performed on a standard Web browser (Google Chrome) with both browser cache disabled (to simulate access for each query without caching) and enabled (to evaluate the impact of browser caching). The load time was measured as an average of 3 loads.
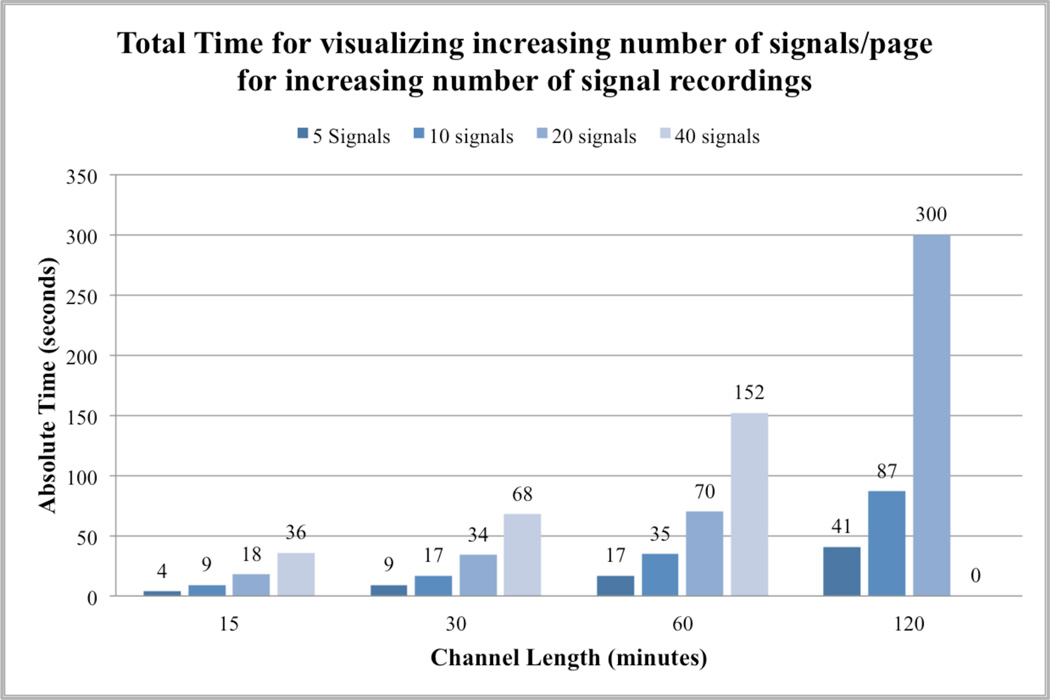
The results of our evaluation are presented in the graphs below. Figure 7 shows that the total time taken to render an increasing number of signals with increasing length of recordings. The total number of signals per page was increased by a factor of 2, starting with 5 channels to a maximum of 40 channels. Similarly, the length of signal recording is doubled starting with 15 minutes to a maximum of 120 minutes. The total time to render the signal recordings is calculated as the sum of average query times for selected signals over 3 runs and the average rendering time on the web interface. It was observed that the total time for rendering 30 or fewer signals per page for 1-hour recording takes less than a minute for query and rendering on the web.
Figure 7.
Total time to render signals with increasing number of signals/page for increasing length of EEG recordings
Figure 8 shows query and rendering time for a 60-minute EEG signal recording for increasing number of signals in increments of five, starting with 5 signals to a total of 50 channels in a single page. The time taken to retrieve all the selected signals increases linearly from 16 seconds (for 5 signals) to 2.5 minutes (for 40 signals). The rendering time is less than 15 seconds for 0–40 signals. It is important to note that the rendering time significantly increases when the number of signals per page is greater than 40. Overall, the results of the Cloudwave performance evaluation demonstrate that Cloudwave is a practical tool and hence it is being deployed at the UH CMC EMU for use by the EMU staff.
Figure 8.
Average Time for loading and rendering 60-min EEG recording for increasing number of signals/page
Discussion
We are in the process of developing a number of quantitative signal analyses (e.g. HRV) using the Map-Reduce paradigm implemented in the Hadoop open source cloud-computing infrastructure [7], which can also support storage of very large signal data. In addition, we also propose to conduct a comprehensive user survey of EMU staff in the four participating centers as part of the PRISM project. This will help us identify additional user requirements that can be added to the Cloudwave user interface.
Conclusion
In this paper we introduced Cloudwave as a Web-based electrophysiological signal visualization and analysis tool that leverages open source cloud computing infrastructure to support very large signal data files. In contrast to existing standalone signal processing tools, Cloudwave does not require installation of a new software, configuration of host computers, and manual maintenance of each installation. Cloudwave features a rich set of functionalities to allow user to select multiple signal channels and event annotations. Cloudwave is in the process of being deployed for use across four participating EMUs in the PRISM project.
Acknowledgements
This research is supported by the PRISM (Prevention and Risk Identification of SUDEP Mortality) Project (1-P20-NS076965-01), and in part by the Case Western Reserve University CTSA Grant NIH/NCATS UL1TR000439.
References
- 1.Cui L, Bozorgi A, Lhatoo SD, Zhang GQ, Sahoo SS. EpiDEA: Extracting Structured Epilepsy and Seizure Information from Patient Discharge Summaries for Cohort Identification; Proceedings of the American Medical Informatics Association (AMIA) Annual Symposium; 2012. [PMC free article] [PubMed] [Google Scholar]
- 2.Sahoo SS, Zhao M, Luo L, Bozorgi A, Gupta D, Lhatoo SD, Zhang GQ. OPIC: Ontology-driven Patient Information Capturing System for Epilepsy. Proceedings of the American Medical Informatics Association (AMIA) Annual Symposium; Nov 2012; Chicago, IL. pp. 799–808. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang GQ, Siegler T, Saxman P, Sandberg N, Mueller R, Johnson N, Hunscher D, Arbandi S. VISAGE: A Query Interface for Clinical Research. Proceedings of the 2010 AMIA Clinical Research Informatics Summit; March 12–13; San Francisco. 2010. pp. 76–80. [PMC free article] [PubMed] [Google Scholar]
- 4.Fisch B. Chapter 7: Epilepsy and Intensive Care Monitoring: Principles and Practice. 2010 [Google Scholar]
- 5.Jain S. MS Thesis. Case Western Reserve University; 2011. Event-Driven Dynamic Query Model for Sleep Study Outcomes Research. [Google Scholar]
- 6.Ng Z. MS Thesis. Case Western Reserve University; 2011. EDF File Preprocessing and the Query Interface PSGQuery. [Google Scholar]
- 7.Hadoop Distributed File System. http://hadoop.apache.org/ [Google Scholar]
- 8.So EL. What is known about the mechanisms underlying SUDEP? Epilepsia. 2008;49(Suppl 9):93–98. doi: 10.1111/j.1528-1167.2008.01932.x. [DOI] [PubMed] [Google Scholar]
- 9.European Data Format (EDF) www.edfplus.info/ [Google Scholar]
- 10.Nihon Kohden. http://www.nihonkohden.com/ [Google Scholar]