Abstract
Pre-mRNA cleavage and polyadenylation is an essential step for 3′ end formation of almost all protein-coding transcripts in eukaryotes. The reaction, involving cleavage of nascent mRNA followed by addition of a polyadenylate or poly(A) tail, is controlled by cis-acting elements in the pre-mRNA surrounding the cleavage site. Experimental and bioinformatic studies in the past three decades have elucidated conserved and divergent elements across eukaryotes, from yeast to human. Here we review histories and current models of these elements in a broad range of species.
INTRODUCTION
Genes in eukaryotes are transcribed by three RNA polymerases. RNA polymerase II (RNAPII) is responsible for the synthesis of all protein-coding RNAs, most long non-coding RNAs (lncRNAs), and many small non-coding RNAs, such as nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), microRNAs (miRNAs), etc. However, 3′ end formation can vary significantly among the genes transcribed by RNAPII.1,2 Nearly all eukaryotic protein-coding transcripts are subject to 3′ end processing by two tightly coupled reactions: an endonucleolytic cleavage of pre-mRNA and subsequent synthesis of a polyadenylate or poly(A) tail.3 A notable exception in metazoans and some protozoans is a specific set of histone genes that are expressed primarily in S-phase of cell cycle,4 which generate non-polyadenylated transcripts. In this review, we focus on the sequence elements that govern cleavage and polyadenylation of pre-mRNA in diverse eukaryotic species. The process is commonly referred to as 3′ processing, cleavage and polyadenylation, or simply mRNA polyadenylation, while the processing site itself is generally called polyA site.
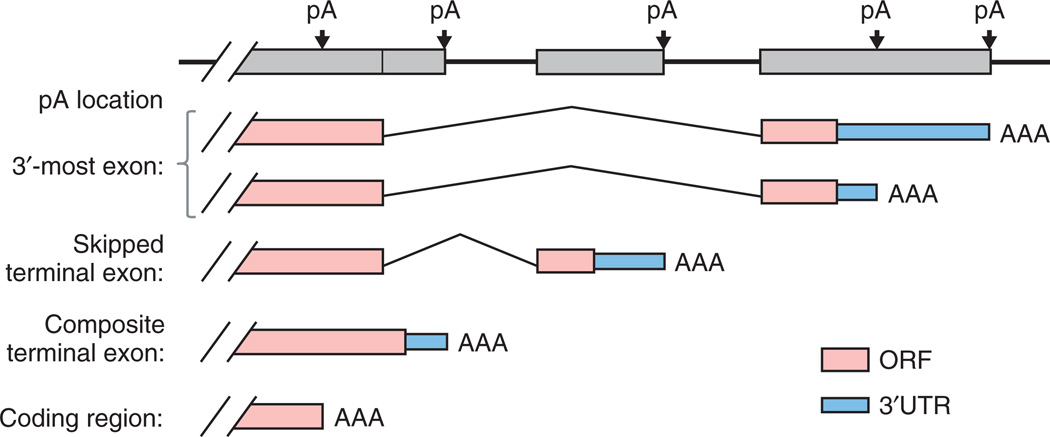
Early molecular biology experiments indicated that one gene can have multiple polyA sites, resulting in mRNA isoforms.5 Recent genomic studies and bioinformatic analyses have established that alternative usage of polyA site, or alternative polyadenylation (APA), is the rule rather than exception in all eukaryotes.6–11 APA can involve selection between sites in a common 3′ terminal exon (tandem polyA sites), or coincide with alternative splicing, involving selection of different 3′ terminal exons (Figure 1). For the latter, there are two scenarios: (1) A terminal exon is either used or skipped in its entirety through splicing. Such exons are referred to as skipped terminal exons (Figure 1). (2) An internal exon either splices to a downstream exon or extends into the adjacent intron and becomes the 3′ terminal exon. The exon therefore contains both internal and terminal exonic regions and is referred to as a composite terminal exon (Figure 1). In a relatively rare occurrence, APA can occur within the coding region, resulting in a truncated transcript without an in-frame stop codon (Figure 1). It is worth noting that nearly all organisms studied to date have demonstrated short-distance variability in cleavage, where the precise polyA site can vary by up to 20–30 nucleotides (nt). This microheterogeneity in alternative cleavage is not generally defined as APA; and its functional consequence is not yet clear.
FIGURE 1.
Alternative polyA sites in a gene. PolyA sites can be located in different parts of a gene, as indicated in the graph. Boxes are exons. pA, polyA site; AAA, poly(A) tail; ORF, open reading frame.
CIS ELEMENTS THAT CONTROL mRNA POLYADENYLATION
Early indications that specific signals are used to guide synthesis of the poly(A) tail of mRNA came from the fact that poly(A) polymerases (PAPs), the enzymes that synthesize the poly(A) sequence, lack substrate specificity.12 Biochemical studies over the last three decades have identified a set of cis elements embedded in the pre-mRNA that control mRNA polyadenylation. Emergence of complete genomic sequences and transcriptome data, such as expressed sequence tags (ESTs) and deep sequencing reads, coupled with advances in bioinformatic analysis, has provided opportunities to systematically establish common and divergent elements across species. Here we provide brief histories of finding polyA signals and present current understandings in the genomic context. We review signals in metazoans, yeast, and plants separately, because these three kingdoms of life have distinct cis element configurations around the polyA site. It is noteworthy that molecular biology and genomics studies of other domains of eukaryotes, such as protists,13 are beginning to shed light on polyA signals in these species. However, they are not discussed here due to limited available information.
Metazoans
Mammals
Experimental Studies
Studies of signals governing mRNA polyadenylation started in the mid-1970s. In the seminal work based on sequence analysis of six mRNAs from human, mouse, rat, and chicken, Proudfoot and Brownlee identified a conserved consensus hexamer AAUAAA, just upstream of the respective polyA sites.14 The critical role of this element in mRNA polyadenylation was later established by Fitzgerald and Shenk using deletion mutants of the simian virus 40 (SV40) late polyA site.15 Subsequent point mutation analyses using adenovirus E1A gene,16 SV40 late gene,17 and the α-globin gene18 further confirmed the specificity of this element. Additional analyses conducted several years later found that a common natural variant AUUAAA has ~80% of the processing efficiency as compared to AAUAAA,19 while most other close variants have significantly reduced efficiency, except for AGUAAA (~30%).20 AAUAAA and AUUAAA are generally considered as canonical hexamers, and are often referred to as the polyadenylation signal, or PAS. For clarity, in this review, all cis elements that can control or modulate polyadenylation are called polyA signals, while the specific acronym PAS is reserved for AAUAAA and its variants.
Shortly after the discovery of AAUAAA, it was reported that sequences downstream of the cleavage site (CS) were also important for polyadenylation. Mutational analyses defined degenerate U-rich and GU-rich sequences in downstream regions.21,22 The multipartite structure of the mammalian polyA site was more definitively established when Levitt et al. engineered a functional synthetic polyA site which contained only the AAUAAA element and a U/GU-rich sequence separated by ~22 random nt.23
Beyond the core structure of polyA site, subsequent reports using viral and cellular genes have detailed a number of auxiliary elements that enhance polyadenylation.24 They are generally located upstream of the PAS and are U-rich in content. In addition, a G-rich element downstream of SV40 late polyA site was reported to regulate polyadenylation.25 More recently, a study of the polyA site of human PAP gene PAPOLG, which does not have a canonical AAUAAA, revealed upstream UGUA element as a general polyA signal.26 Two UGUA elements may be required for the full function, because the protein that binds UGUA, the 25 kDa subunit of cleavage factors I (CF Im), exists as a dimer.27
Bioinformatic Analyses
The emergence of EST data has enabled large-scale analysis of polyA sites and signals since late 1990s. Many EST libraries, particularly early ones, were generated with oligo-dT reverse transcription primers that hybridize to the poly(A) tail, resulting in a significant 3′ end bias of EST sequences, and corresponding utility in the study of polyA sites. On the basis of sequence similarity, Beaudoing and Gautheret clustered ESTs and identified putative alternative polyA sites in human genes.28 The data led to the finding that many single nucleotide variants of the PAS are statistically overrepresented, including AGUAAA, UAUAAA, CAUAAA, GAUAAA, AAUAUA, AAUACA, AAUAGA, AACUAAA, AAG AAA, and AAUGAA, suggesting the PAS is more divergent than previously thought.29 Another genomic study by Graber et al. revealed a common pattern of nucleotide frequencies in the 3′UTR immediately upstream of the polyA sites in several eukaryotes.30
The availability of complete genomic sequences further provided opportunities to extend bioinformatic analysis of polyA sites and signals. Inspection of downstream genomic sequence can significantly mitigate false identification of polyA sites due to internal priming at A-rich sequences in the transcript.30–32 In addition, genomic placement of polyA sites enabled analysis of polyA sites and signals in the context of complete gene structure with splicing information. Using ESTs and genome sequences, Tian et al. and Yan and Marr reported that more than half of the human genes have APA,6,8 much higher than previously thought. In addition, a large number of polyA sites are located in introns, leading to alternative selection of 3′ terminal exons and involving interplay between polyadenylation and splicing.6,33
A Current Model of Mammalian PolyA Sites
Bioinformatic analyses indicate that the nucleotide composition of genomic regions surrounding mammalian polyA sites is generally AU-rich.6,34 This nucleotide bias can be observed within 100 nt up-and downstream of the polyA site.6 No significant elements at the CS have been identified despite that a bias toward CA was found in vitro.35
Consistent with early experimental studies, genomic analyses indicate that the PAS is a dominant signal located within ~40 nt upstream of mammalian polyA sites (Figure 2). The two most common variants, AAUAAA and AUUAAA, are found in ~53–58% and ~15–17% of human polyA sites, respectively.6,29 About 10–20% of human polyA sites are associated with single nucleotide variants of AAUAAA/AUUAAA,6,29 while 5–10% have no easily recognizable PAS in the 40 nt upstream region. The distribution of PAS variants differs among polyA sites, depending upon (1) location of the polyA site and (2) evolutionary conservation.33 Typically, AAUAAA is more frequent for 3′-most polyA sites in terminal exons, as compared to upstream sites in the same exon. However, 3′-most polyA sites in composite terminal exons (Figure 1) typically have a lower frequency of AAUAAA, which may be attributed to the fact that their usage is coupled with alternative splicing.33 In addition, conserved polyA sites are more likely to be associated with AAUAAA, suggesting selection for AAUAAA as PAS. By contrast, AUUAAA shows no such variation, with similar frequencies in different groups of polyA sites.33,36
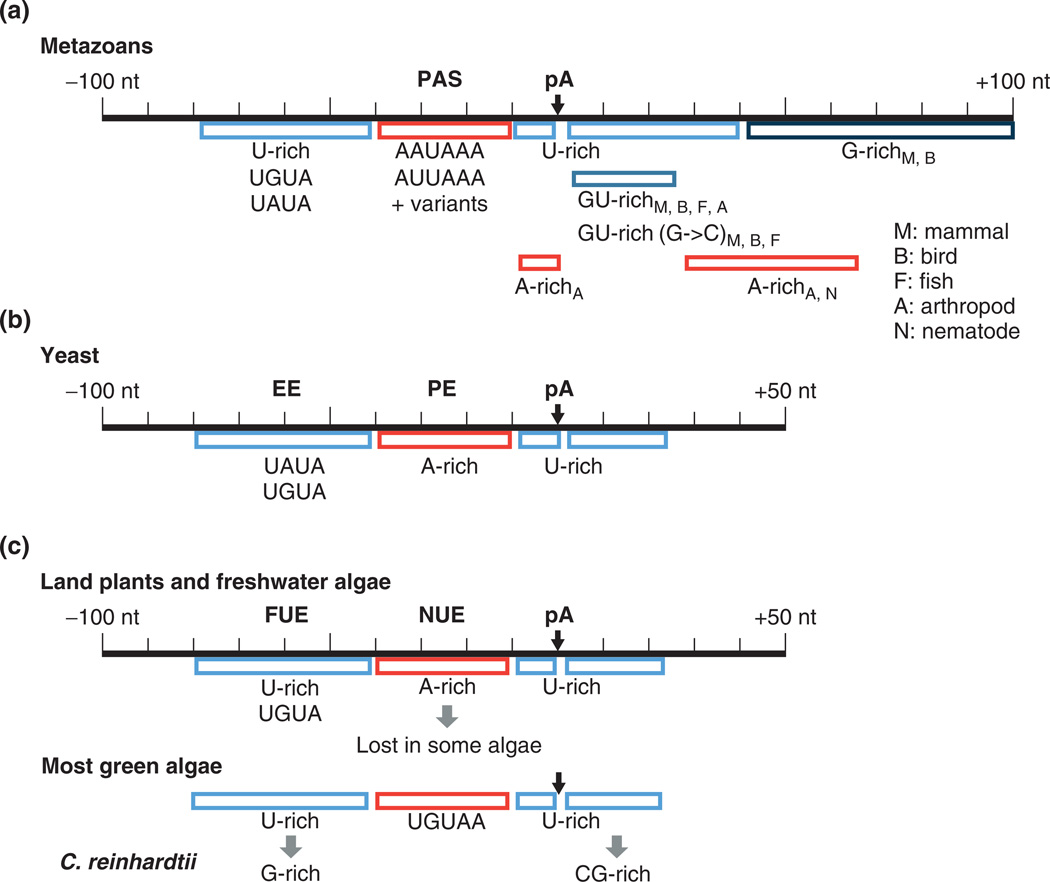
FIGURE 2.
PolyA signals in different species. (a) Metazoans: elements that are restricted to certain species are indicated. M, mammal; B, bird; F, fish; A, arthropod; N, nematode; pA, polyA site. (b) Yeast. (c) Plants, including land plants and freshwater algae (Streptophyta), most green algae (Chlorophyta), and the special case of Chlamydomonas reinhardtii.
Two distinct types of upstream U-rich elements have been reported by bioinformatic analysis,36,37 and can be distinguished by their positioning, either upstream of the PAS or between the PAS and the CS. The former also includes UGUA and UAUA. The placement of U-rich elements between the PAS and the CS is similar to that found in yeast and plants (see below). In addition, an upstream G-rich element was reported to be significantly associated with frequently used polyA sites, albeit with a lower frequency of occurrence than the U-rich elements.36
The region immediately downstream (typically <40 nt) of the polyA site contains distinct U-rich and GU-rich elements, with GU-rich elements generally closer to the polyA site than U-rich elements.36–38 The U-rich elements typically contain a run of ≥3 uracils, and GU-rich elements generally entail GUGU or UGUG. Notably, variants of the GU-rich element, such as GUCU, CUGU, UCUG, and UGUC, also have significant occurrences in the downstream region with positioning profiles similar to the GU-rich element.36,38 While the GU-rich and U-rich elements are considered as ‘core’ elements, functional sites can be found without one or both of these elements.
Mammalian polyA sites also contain distal downstream G-rich elements, typically >30 nt from the polyA site, consistent with experimental findings. The G-rich elements contain runs of guanines that are interrupted by A or U, for example, GGUGGG and GGGAGG. Finally, a putative C-rich element in the distal downstream region (40 to 100 nt from the polyA site)was reported by a bioinformatic analysis,36 although its occurrence is much lower than that of G-rich elements.
Non-Mammalian Vertebrates
To date, chicken (Gallus gallus), fugu fish (Takifugu rubripres), and zebrafish (Danio rerio) are the only non-mammalian vertebrates whose polyA sites have been studied on a genomic scale.37–40 Studies have revealed that avian polyA signals are highly similar to those in mammals, including upstream U-rich, UGUA, and PAS elements, as well as downstream GU-rich, Urich, and G-rich elements.37,38 Both fugu fish and zebrafish have evidence of similar sequence content and positioning for all the mammalian elements, with the exception of the G-rich downstream element,38 which is apparently restricted to amniotes (Figure 2).
Invertebrates
Arthropods
Large-scale bioinformatic analyses of polyA signals in fruit fly (Drosophila melanogaster) and mosquito (Anopheles giambiae)37–39,41 indicate that the polyA signal patterns in these two species are highly similar to each other, and are also similar to those in vertebrates but with some exceptions: As in vertebrates, the PAS is a prominent feature of polyA site. However, its positioning appears to be broadened.38 Like vertebrates, arthropods have a UGUA-like element, but with reduced sequence specificity.37 Interestingly, both arthropods were found to have an increased frequency of A-rich sequences immediately upstream (<5 nt) of the polyA site, a feature not seen in any other organisms studied to date.37
Downstream elements include both a near (<10 nt) GU-rich element and a slightly more distal (10–20 nt) U-rich element, as found in vertebrates.37,38,41 However, the G → C transversion variants of the GU-rich element (e.g., UCUG or UGUC) observed in vertebrates have not been found in arthropods.38 Like fishes, arthropods do not have the distal G-rich element found in mammals. Instead, both arthropods have evidence of distal (≥30 nt) downstream A-rich sequences.37,38
Nematodes
Nematode (Caenorhabditis elegans) polyA signals have been the subject of several large-scale analyses.9,42–44 Like other metazoans, nematodes have a prominent PAS. However, while AAUAAA is the most common hexamer, its frequency is significantly reduced to ~40% and the most common variant is AAUGAA, which is associated with ~10% of the polyA sites.9,44 In addition, compared with polyA sites in other metazoans, nematode sites have higher uracil content and lower occurrence of UGUA in the upstream region42,43 and, strikingly, their downstream region contains the U-rich element but not the GU-rich element.38
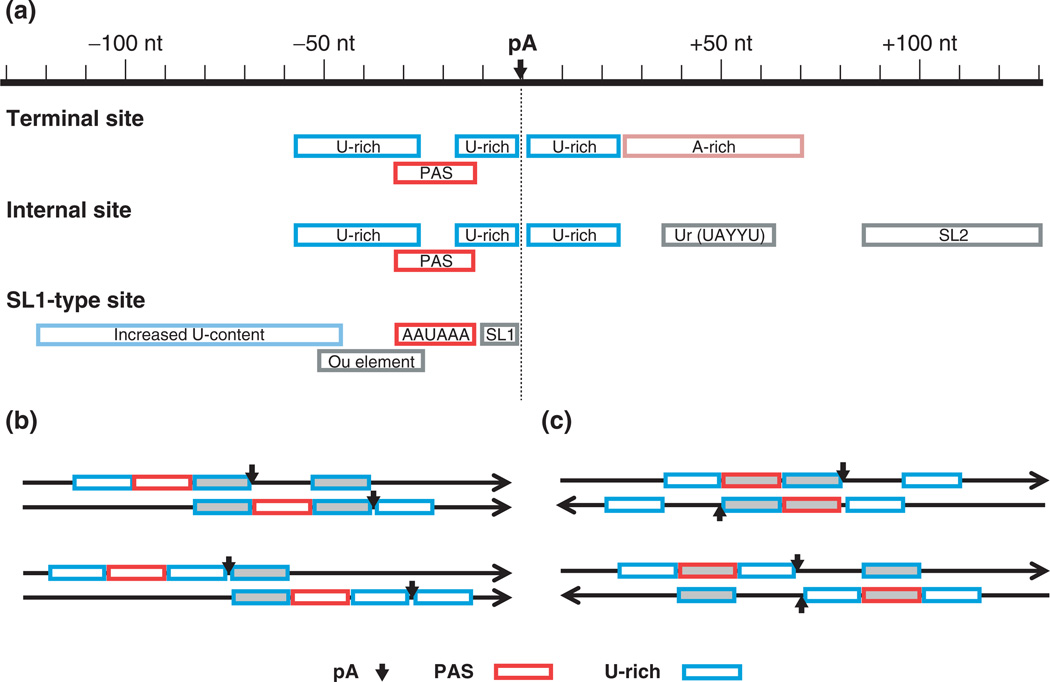
About ~15% of C. elegans genes are embedded in a polycistronic operon structure, in which the pre-mRNA of a downstream gene is trans-spliced to a splice leader (SL) for 5′ end maturation. Like monocistronic genes, each gene in an operon has its own polyA site(s) for 3′ end processing. However, the usage of polyA site of an upstream gene is coupled to trans-splicing of its downstream gene (Figure 3(a)). For internal polyA sites, a ‘Ur’ element (with consensus UAYYU) and the SL2 acceptor are typically located 45–65 nt and ~100 nt downstream, respectively.42 By contrast, terminal polyA sites appear to have Arich sequences ~50 nt downstream. A small group of C. elegans genes (with fewer than 30 validated cases) have a distinct arrangement, in which the polyA site of upstream gene is part of the SL acceptor sequence of downstream gene.45 The PAS of these polyA sites is nearly always the canonical AAUAAA, and the polyA site itself is situated within a consensus UUUUCAG splice acceptor.42,45
FIGURE 3.
PolyA sites in Caenorhabditis elegans. (a) Three types of polyA sites. Terminal sites occur on monocistronic genes, or on 3′ terminal genes in polycistronic operons. Internal sites occur on non-terminal genes in operons. SL-1 type sites are a special class of internal sites characterized by exact overlap of the upstream polyA site and the downstream trans-splicing site. (b) Adjacent polyA sites of the same gene sharing cis elements. Arrowed lines represent pre-mRNAs containing cis elements (shown as boxes) and polyA sites (shown as vertical arrows). Cis elements used by both polyA sites are indicated by shaded boxes. (c) Adjacent polyA sites of genes on opposite strands. As in (b), arrowed lines represent pre-mRNAs with arrows indicating direction of transcription. Shaded boxes indicate that both sense and antisense sequences are polyA signals. pA, polyA site.
Notably, a recent genome-wide polyA site mapping by deep sequencing revealed that a large fraction of polyA sites in C. elegans are adjacent to one other by sharing U-rich elements (Figure 3(b)), for example the U-rich element between the PAS and polyA site of one site is also used as U-rich element upstream of PAS of another site (upper panel, Figure 3(b)); and the downstream U-rich element of one polyA site is used as upstream element of another site (lower panel, Figure 3(b)). Strikingly, about one sixth of all C. elegans genes have polyA sites that are adjacent to polyA sites of the genes on the opposite strand (Figure 3(c)).9 As such, some genomic sequences encode polyA signals on both strands. For example, PAS and U-rich element on one strand are, respectively, U-rich element and PAS on the other. Interestingly, this genomic arrangement of polyA sites appears to be evolutionarily selected, leading to compaction of 3′UTRs in C. elegans.9
Yeast
The discovery that a 38 nt deletion mutation in the 3′UTR of the gene CYC1 resulted in abnormal 3′ end formation in Saccharomyces cerevisiae is arguably the first report concerning polyA signal in yeast.46 Subsequent experiments with genetic constructs revealed both the content and positioning of several putative signals, such as the UAG…UAUGUA, UAUAUA, and UACAUA elements.47 Studies of additional yeast genes, the Ty transposon, and the 2 µm circle plasmid identified UA-rich elements as ‘efficiency’ elements, or EE, for 3′ end processing. Additional studies with the CYC1 system further established the importance of a ‘positioning’ element, or PE, located between the EE and polyA site.48 Mutational analyses showed that the PE tolerates significant degeneracy, while AAUAAA or AAAAAA are the optimal elements. In addition, the CS was shown to follow a preferred but not required consensus pattern of PyA. The tripartite structure (PE-EE-CS) was confirmed with studies of a synthetic polyA site.49
Large-scale bioinformatic analyses of several thousand EST-supported polyA sites largely confirmed the elements identified biochemically, but also revealed two additional U-rich elements flanking the CS,30,50 which were later confirmed by experiments.51
The current model of yeast polyA signals includes four general elements: the EE, with UAUAUA being the optimal sequence, is generally positioned 25–40 nt upstream of the polyA site but can be more distant, possibly up to 75 nt or more. The PE, which is equivalent to the metazoan PAS, is 10–30 nt upstream of the polyA site, with AAUAAA being the optimal sequence (Figure 2). Two U-rich elements are immediately up- and downstream of the CS, respectively. All elements of yeast polyA signals are apparently tolerant of significant variation. For example, the EE can have many single base substitutions, especially those with A → G or U → C transitions, with little or no effect on polyadenylation efficiency.52 Notably, long-and short-lived transcripts appear to have systematic differences in the EE, suggesting additional roles of polyA signals in mRNA stability.53
Plants
Studies of polyA signals in plants in the mid-1980s reported that (1) mammalian polyA signals are not functional in plant cells,54 (2) a plant gene can have multiple closely located polyA signals leading to several CSs,55 and (3) no strong consensus sequence can be found near plant polyA sites.56 Detailed studies of polyA signals in the early 1990s established a general cis element structure of plant polyA site, comprising two groups of ‘upstream elements’, termed near upstream element (NUE, 10–40 nt from the polyA site) and far upstream element (FUE, 30–150 nt from the polyA site).57 NUEs are PAS-like sequences, although highly degenerate, and FUEs are generally U-rich and UG-rich. A general YA (pyrimidine followed by adenine) motif was also reported as the element for cleavage.57
Several species in the two monophyletic lineages of green plants now have been bioinformatically analyzed for polyA signals, including species in the Chlorophyta lineage, such as green algae, and species in the Streptophyta lineage, such as land plants (Arabidopsis and rice) and some freshwater algae. The land plants studied appear to be more similar to yeast than to metazoans (Figure 2), which is consistent with the early biochemical studies implicating the similarity in polyA signals between plants and yeast.58–60 Surprisingly distinct cis element content and positioning have been found in some green algae (Figure 2).
Arabidopsis and Rice
The model plant organism, Arabidopsis (Arabidopsis thaliana), was the first plant to receive large-scale analysis of polyA signals.61,62 Like in yeast, polyA sites in these species are flanked by high uracil content.61,62 An A-rich NUE element, similar to the PE in yeast and to the metazoan PAS, is located 15–25 nt upstream of the polyA site.61–63 An U-rich FUE is located further upstream. While the FUE is similar to the EE in yeast, it has been reported to be functional at greater distances, for example >100 nt from the polyA site.63 In addition, an analysis that takes into account background nucleotide content, specifically compensating for the extreme U-richness in 3′UTRs, revealed distinct UGUA-containing elements in the 30–70 nt region upstream of plant polyA sites.37
PolyA sites in rice (Oryza sativa) have a similar cis element structure to those in Arabidopsis.64–66 Notably, the uracil content of rice 3′UTRs is not as high as that in Arabidopsis, facilitating clear distinction between the UGUA-like element from its surrounding sequences.
Green Algae
Among the algae, the complete polyA signal pattern has been examined only for the Chlorophyte algae Chlamydomonas reinhardtii,66 for which a draft genome sequence is available. In addition, studies focusing on upstream elements only have been performed for seven Chlorophyte and four Streptophyte algae.67 These analyses revealed surprising variations of upstream elements in different algae.66,67
The cis element patterns for Streptophyte algae are highly similar to the closely related land-based plants,67 including a U-rich FUE element, typically manifested as UGUA, an A-rich NUE element, and a U-rich element immediately upstream of the polyA site (Figure 2). However, some Streptophyte algae do not appear to have the NUE element.67
Chlorophyte algae lack standard A-rich NUE in most of the species surveyed,66,67 despite increased adenine content ~20 nt upstream of the polyA site (Figure 2). Instead, a UGUAA element was found to be specifically positioned, consistent with the NUE.67 Strikingly, in the specific case of C. reinhardtii, which has a genome with high GC content, G-rich and CGrich sequences were found upstream of UGUAA and downstream of the polyA site, respectively.66
RECOGNITION OF PolyA SIGNALS BY PROTEIN FACTORS
mRNA polyadenylation is accomplished by the cooperative actions of a number of mutually interacting protein complexes. The components of these complexes are commonly referred to as polyA factors. Starting in the early 1980s, several labs performed extensive and detailed studies to identify and characterize polyA factors, using approaches such as biochemical purification, cDNA cloning, yeast genetics, and, more recently, large-scale proteomics. We briefly describe below the current understanding of the polyadenylation machinery in mammals and yeast, focusing on their relation to polyA signals. Readers are referred to other reviews for more details68 and emerging studies of the polyadenylation machinery in plants.69
Mammalian Factors
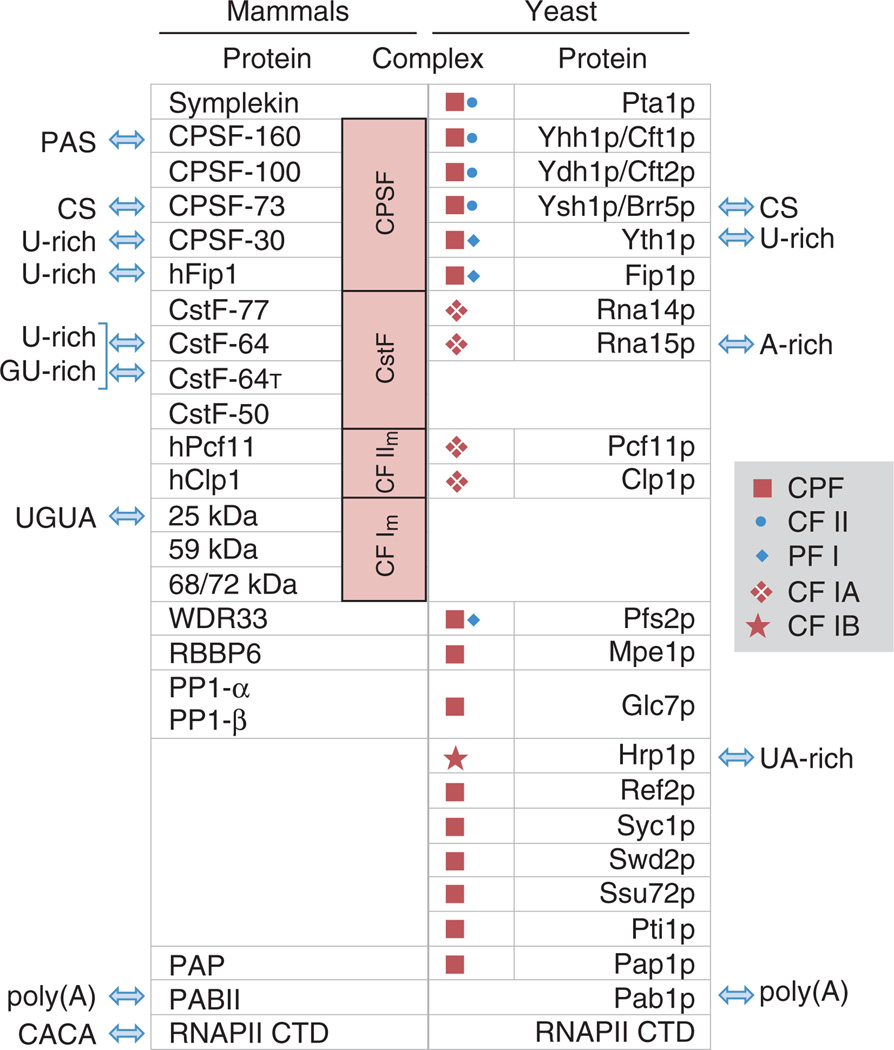
In mammals, the core polyadenylation machinery is composed of four protein complexes, including the cleavage and polyadenylation specificity factor (CPSF), the cleavage stimulation factor (CstF), and CF Im and CF IIm, along with several individual proteins, including Symplekin, PAP, and nuclear poly(A) binding protein (PABP) (Figure 4). In addition, the C-terminal domain (CTD) of RNAPII is directly involved in polyadenylation. A proteomic study of the 3′ processing machinery in human cells identified ~25 core protein factors, along with ~60 additional associated proteins with diverse functions in the cell, including DNA-damage response, transcription, splicing, and translation.70 The discovery of protein factors from distinct cellular processes in complex with the polyadenylation machinery provides new insights into the crosstalk among cellular processes, as well as potential mechanisms controlling APA.
FIGURE 4.
Mammalian and yeast polyA factors and complexes and their interactions with polyA signals. Protein–RNA interactions are indicated by arrows. Homologous proteins are listed in the same row. Both mammalian and yeast complexes and subcomplexes are indicated. CS, cleavage site; poly(A), poly(A) tail.
A large fraction of the core polyA factors have RNA binding activity, some of which are sequence-specific. Within the CPSF complex, CPSF73 is the endonuclease that cleaves pre-mRNA,71 CPSF160 interacts with the PAS,72 and both hFip173 and CPSF3074 bind to U-rich sequences. In addition, the 25 kDa subunit of the CF Im complex binds the UGUA element,75 and CstF-64, a component of CstF, interacts with U-rich and GU-rich elements.76–78 A paralog of CstF-64, CstF-64t, is also encoded in mammalian genomes. CstF-64t has similar but not identical RNA binding specificity compared with CstF-64,79 and was found to be in the human polyadenylation machinery.70 Thus, all cis elements close to the polyA site, including the PAS, UGUA element, upstream and downstream U-rich elements, and downstream GU-rich elements, can be recognized by factors in the core polyadenylation machinery.
Upstream and downstream G-rich elements appear to be recognized by heterogeneous nuclear ribonucleoprotein (hnRNP) H, H′, or F.80–82 However, intrinsic functions of G-rich elements in pausing of transcription83 and forming G-quadruplex structures84 have also been proposed. So far, no factors have been found to bind to the upstream UA-rich element or the downstream C-rich element, both of which were identified by bioinformatic analysis.36
Interestingly, the CTD has been shown to interact with CA-rich RNA sequences.85 As CA-rich sequences can inhibit polyadenylation when placed downstream of a polyA site, this activity of CTD may play a role in inhibition of polyadenylation at undesirable locations in the genome. Consistently, CA-rich elements are significantly depleted in the +1 to +40 nt downstream region of frequently used polyA sites (supplementary table in36).
Yeast Factors
The yeast polyadenylation machinery contains ~20 proteins (Figure 4), which can form several protein complexes and subcomplexes, such as PF I, CPF, CF IA, CF IB. While most of the core polyadenylation factors in mammalian CPSF, CstF, and CF IIm complexes are conserved in yeast, they appear to be organized differently (Figure 4). Some factors appear to have conserved RNA-binding specificities, for example Ysh1p, which, like its mammalian homolog CPSF-73, is the endonuclease that cleaves pre-mRNA; similarly, Yth1p, the homolog of CPSF-30, has conserved binding activity to U-rich sequences.74 In contrast, some other factors have drastic differences between yeast and mammals: for example, Yhh1p/Cft1p, the homolog of CPSF-160, is important for polyA site recognition but does not have binding specificity to AAUAAA/AUUAAA86; similarly, Fip1p is not likely to have similar RNA binding activities to its homolog hFip1 due to lack of the C-terminal arginine-rich domain that is important for binding to U-rich sequences.73 Strikingly, Rna15p, the homolog of CstF-64, was shown to interact with A-rich sequences when it is in the CF IA complex87 despite its in vitro binding activity to U-rich sequences.76 Thus, the A-rich PE sequence in yeast is recognized by Rna15p, and U-rich elements interact with Yth1p. In addition, the yeast protein Hrp1p, which has no known mammalian homolog, binds to sequences containing AUAUAU,88 which corresponds to the EE.
Regulation of PolyA Site Usage
Early studies demonstrated that deletion or absence of some functional elements does not necessarily disable 3′ processing, but instead reduces processing efficiency.52 These results led to the postulation that the complete control sequence acts cooperatively, allowing ‘strong’ elements of one polyA signal to compensate for suboptimal elements of another type.24,77 This may explain why a sizable fraction of EST-supported polyA sites in mammals do not appear to have a strong PAS AAUAAA.6,39 Consistently, A-rich sequences are capable of functioning as PAS in mammalian cells but are often associated with higher uracil content downstream of the polyA site.89
The combinatorial nature of polyA site determination suggests that proteins in the polyadenylation machinery can complement one another in recognition of the polyA site. Conversely, given different polyA sites are surrounded by variable cis elements, it is conceivable that modulation of the relative abundance and/or activity of individual polyA factors can impact alternative polyA site choice. Notably, genomic studies have indicated that most polyA factors appear to be differentially expressed across human tissues, at least at the mRNA level90 and different human tissues have systematic differences in APA.90,91
Recent genomic studies have also uncovered a global trend of 3′UTR shortening via APA in proliferating mammalian cells, as shown in T cell activation,92 embryonic development and cell differentiation,93 and oncogenesis.94,95 Bioinformatic analysis indicated a correlation between expression of polyA factors and 3′UTR length.96 Interestingly, U-rich downstream elements were found to be enriched for polyA sites that are highly regulated during cell proliferation/ differentiation, suggesting potential involvement of the CstF complex in regulation.96 This is consistent with the classic APA model in which the CstF-64 level regulates APA of the IgM heavy chain gene during B cell maturation.97 In a similar vein, knockdown of the 25 kDa submit of CF Im leads to APA of many genes in HeLa cells.98 Therefore, regulation of core polyA factors may be a common mechanism for APA. On this note, mutation of Hrp1 was found to cause changes in APA for many genes in S. cerevisiae.99 Interestingly, this leads to resistance to toxicity of excess copper, a phenotype that is attributed to altered APA of the gene CTR2.
In addition to core factors, a growing number of RNA-binding proteins (RBPs) that interact with specific elements near the polyA site have been shown to positively or negatively influence polyA site usage in mammalian cells, including PTB, Nova, Hu proteins, etc. (reviewed in Ref 100). Interestingly, most of these RBPs have also been shown to play roles in alternative splicing, suggesting diverse roles of nuclear RBPs in pre-mRNA processing. A notable example in yeast is that Npl3, which is homologous to serine/arginine-rich (SR) proteins in higher eukaryotes and has binding preference to G+U sequences, can compete with RNA15p and inhibit mRNA polyadenylation.101,102
CONCLUSION
It has been three decades since the report of the first polyA signal, the AAUAAA hexamer. Experimental studies have identified and characterized many general and site-specific cis elements and their binding factors. Bioinformatic analyses using large-scale genomic data have established general rules and revealed common and divergent signals in a wide range of organisms. Some upstream elements such as A-rich/PAS, UA-rich, UGUA, U-rich appear to have universal presence albeit with different extent of degeneracy and frequency in different species. By contrast, downstream elements vary significantly: GU-rich elements are present only in metazoans other than nematodes; and G-rich elements are restricted to amniotes (chicken and mammals).
While a great deal has been learned about polyA signals, whether some cis elements are still elusive is to be firmly addressed. Notably, a recent study using direct RNA sequencing found that polyA sites of previously unidentified antisense and intergenic transcripts appear to be surrounded by distinct nucleotide contents compared to known sites,11 suggesting novel signals. As sequencing technologies advance, more comprehensive mapping of polyA sites in genomes and deeper understanding of their signals are expected to shed light on this subject. In addition, how different cis elements interact with the polyadenylation machinery and how they co-evolve are yet to be fully examined.
Recent studies have indicated the widespread nature of APA under different cell conditions in mammals. Yet many questions remain. How prevalent is APA in lower species? How does regulation of core polyA factors, including expression and posttranslational modification, impact APA? These questions remain in need of systematic examination. In addition, a growing number of proteins involved in other aspects of cellular functions have been found either to be associated with the polyadenylation machinery and/or to modulate polyadenylation activity. How mRNA polyadenylation affects and is affected by other processes in the cell remains to be defined.
REFERENCES
- 1.Weiner AM. E Pluribus Unum; 3′ end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol Cell. 2005;20:168–170. doi: 10.1016/j.molcel.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Wilusz JE, Spector DL. An unexpected ending: noncanonical 3′ end processing mechanisms. RNA. 2010;16:259–266. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 4.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, Hunt AG. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci USA. 2011;108:12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milossend PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmonds M, Winters MA. Polyadenylate polymerases. Prog Nucleic Acid Res Mol Biol. 1976;17:149–179. doi: 10.1016/s0079-6603(08)60069-0. [DOI] [PubMed] [Google Scholar]
- 13.Clayton C, Michaeli S. 3′ processing in protists. Wiley Interdiscip Rev: RNA. 2011;2:247–255. doi: 10.1002/wrna.49. [DOI] [PubMed] [Google Scholar]
- 14.Proudfoot NJ, Brownlee GG. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald M, Shenk T. The sequence 5′-AAUAAA-3′ forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981;24:251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- 16.Montell C, Fisher EF, Caruthers MH, Berk AJ. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3′ consensus sequence AAUAAA. Nature. 1983;305:600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- 17.Wickens M, Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3′ end formation. Science. 1984;226:1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- 18.Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ. α-Thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- 19.Wilusz J, Pettine SM, Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989;17:3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDevitt MA, Hart RP, Wong WW, Nevins JR. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986;5:2907–2913. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 23.Levitt N, Briggs D, Gil A, Proudfoot NJ. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagga PS, Ford LP, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkataraman K, Brown KM, Gilmartin GM. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q, Coseno M, Gilmartin GM, Doublie S. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 2011;19:368–377. doi: 10.1016/j.str.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gautheret D, Poirot O, Lopez F, Audic S, Claverie JM. Alternate polyadenylation in human mRNAs: a largescale analysis by EST clustering. Genome Res. 1998;8:524–530. doi: 10.1101/gr.8.5.524. [DOI] [PubMed] [Google Scholar]
- 29.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graber JH, Cantor CR, Mohr SC, Smith TF. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JY, Park JY, Tian B. Identification of mRNA polyadenylation sites in genomes using cDNA sequences, expressed sequence tags, and trace. Methods Mol Biol. 2008;419:23–37. doi: 10.1007/978-1-59745-033-1_2. [DOI] [PubMed] [Google Scholar]
- 32.Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci USA. 2002;99:6152–6156. doi: 10.1073/pnas.092140899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legendre M, Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, MacDonald CC, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutchins LN, Murphy SM, Singh P, Graber JH. Position-dependent motif characterization using nonnegative matrix factorization. Bioinformatics. 2008;24:2684–2690. doi: 10.1093/bioinformatics/btn526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salisbury J, Hutchison KW, Graber JH. A multispecies comparison of the metazoan 3′-processing downstream elements and the CstF-64 RNA recognition motif. BMC Genomics. 2006;7:55. doi: 10.1186/1471-2164-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brockman JM, Singh P, Liu D, Quinlan S, Salisbury J, Graber JH. PACdb: polyA cleavage site and 3′-UTR Database. Bioinformatics. 2005;21:3691–3693. doi: 10.1093/bioinformatics/bti589. [DOI] [PubMed] [Google Scholar]
- 40.Lee JY, Yeh I, Park JY, Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007;35:D165–D168. doi: 10.1093/nar/gkl870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics. 2006;7:176. doi: 10.1186/1471-2164-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graber JH, Salisbury J, Hutchins LN, Blumenthal T. C. elegans sequences that control trans-splicing and operon pre-mRNA processing. RNA. 2007;13:1409–1426. doi: 10.1261/rna.596707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hajarnavis A, Korf I, Durbin R. A probabilistic model of 3′ end formation in Caenorhabditis elegans. Nucleic Acids Res. 2004;32:3392–3399. doi: 10.1093/nar/gkh656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams C, Xu L, Blumenthal T. SL1 trans splicing and 3′-end formation in a novel class of Caenorhabditis elegans operon. Mol Cell Biol. 1999;19:376–383. doi: 10.1128/mcb.19.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaret KS, Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982;28:563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- 47.Russo P, Li WZ, Hampsey DM, Zaret KS, Sherman F. Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991;10:563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo P, Li WZ, Guo Z, Sherman F. Signals that produce 3′ termini in CYC1 mRNA of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7836–7849. doi: 10.1128/mcb.13.12.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo Z, Sherman F. Signals sufficient for 3′-end formation of yeast mRNA. Mol Cell Biol. 1996;16:2772–2776. doi: 10.1128/mcb.16.6.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Helden J, del Olmo M, Perez-Ortin JE. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 2000;28:1000–1010. doi: 10.1093/nar/28.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dichtl B, Keller W. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 2001;20:3197–3209. doi: 10.1093/emboj/20.12.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Z, Sherman F. 3′-end-forming signals of yeast mRNA. Trends Biochem Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 53.Graber JH. Variations in yeast 3′-processing cis-elements correlate with transcript stability. Trends Genet. 2003;19:473–476. doi: 10.1016/S0168-9525(03)00196-3. [DOI] [PubMed] [Google Scholar]
- 54.Hunt AG, Chu N, Odell JT, Nagy F, Chua N-H. Plant cells do not properly recognize animal gene polyadenylation signals. Plant Mol Biol. 1987;8:23–35. doi: 10.1007/BF00016431. [DOI] [PubMed] [Google Scholar]
- 55.Dean C, Tamaki S, Dunsmuir P, Favreau M, Katayama C, Dooner H, Bedbrook J. mRNA transcripts of several plant genes are polyadenylated at multiple sites in vivo. Nucleic Acids Res. 1986;14:2229–2240. doi: 10.1093/nar/14.5.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joshi CP. Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucleic Acids Res. 1987;15:9627–9640. doi: 10.1093/nar/15.23.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu L, Ueda T, Messing J. The formation of mRNA 3′-ends in plants. Plant J. 1995;8:323–329. doi: 10.1046/j.1365-313x.1995.08030323.x. [DOI] [PubMed] [Google Scholar]
- 58.Irniger S, Sanfacon H, Egli CM, Braus GH. Different sequence elements are required for function of the cauliflower mosaic virus polyadenylation site in Saccharomyces cerevisiae compared with in plants. Mol Cell Biol. 1992;12:2322–2330. doi: 10.1128/mcb.12.5.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Butler JS, Sadhale PP, Platt T. RNA processing in vitro produces mature 3′ ends of a variety of Saccharomyces cerevisiae mRNAs. Mol Cell Biol. 1990;10:2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irniger S, Egli CM, Braus GH. Messenger RNA 3′-end formation of a DNA fragment from the human c-myc 3′-end region in Saccharomyces cerevisiae. Curr Genet. 1993;23:201–204. doi: 10.1007/BF00351496. [DOI] [PubMed] [Google Scholar]
- 61.Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loke JC, Stahlberg EA, Strenski DG, Haas BJ, Wood PC, Li QQ. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 2005;138:1457–1468. doi: 10.1104/pp.105.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rothnie HM. Plant mRNA 3′-end formation. Plant Mol Biol. 1996;32:43–61. doi: 10.1007/BF00039376. [DOI] [PubMed] [Google Scholar]
- 64.Lu Y, Gao C, Han B. Sequence analysis of mRNA polyadenylation signals of rice genes. Chin Sci Bull. 2006;51:1069–1077. [Google Scholar]
- 65.Dong H, Deng Y, Chen J, Wang S, Peng S, Dai C, Fang Y, Shao J, Lou Y, Li D. An exploration of 3′-end processing signals and their tissue distribution in Oryza sativa. Gene. 2007;389:107–113. doi: 10.1016/j.gene.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 66.Shen Y, Liu Y, Liu L, Liang C, Li QQ. Unique features of nuclear mRNA poly(A) signals and alternative polyadenylation in Chlamydomonas reinhardtii. Genetics. 2008;179:167–176. doi: 10.1534/genetics.108.088971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wodniok S, Simon A, Glockner G, Becker B. Gain and loss of polyadenylation signals during evolution of green algae. BMC Evol Biol. 2007;7:65. doi: 10.1186/1471-2148-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mandel CR, Bai Y, Tong L. Protein factors in premRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hunt AG. Messenger RNA 3′ end formation in plants. Curr Top Microbiol Immunol. 2008;326:151–177. doi: 10.1007/978-3-540-76776-3_9. [DOI] [PubMed] [Google Scholar]
- 70.Shi Y, Di Giammartino DC, Taylor D, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryan K, Calvo O, Manley JL. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murthy KG, Manley JL. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 73.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 75.Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 76.Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J Biol Chem. 1997;272:26769–26779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- 78.Perez Canadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Monarez RR, MacDonald CC, Dass B. Polyadenylation proteins CstF-64 and tauCstF-64 exhibit differential binding affinities for RNA polymers. Biochem J. 2007;401:651–658. doi: 10.1042/BJ20061097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 2002;30:1842–1850. doi: 10.1093/nar/30.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Millevoi S, Decorsière A, Loulergue C, Iacovoni J, Bernat S, Antoniou M, Vagner S. A physical and functional link between splicing factors promotes pre-mRNA 3′ end processing. Nucleic Acids Res. 2009;37:4672–4683. doi: 10.1093/nar/gkp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol Cell Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yonaha M, Proudfoot NJ. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- 84.Zarudnaya MI, Kolomiets IM, Potyahaylo AL, Hovorun DM. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaneko S, Manley JL. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3′ end formation. Mol Cell. 2005;20:91–103. doi: 10.1016/j.molcel.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 86.Dichtl B, Blank D, Sadowski M, Hubner W, Weiser S, Keller W. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 2002;21:4125–4135. doi: 10.1093/emboj/cdf390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gross S, Moore CL. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol Cell Biol. 2001;21:8045–8055. doi: 10.1128/MCB.21.23.8045-8055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3′ UTR recognition by Hrp1. EMBO J. 2006;25:3167–3178. doi: 10.1038/sj.emboj.7601190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nunes NM, Li W, Tian B, Furger A. A minimal functional human Poly(A) site requires only a strong DSE and an A-rich upstream sequence. EMBO J. 2010;29:1523–1536. doi: 10.1038/emboj.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mayr C, Bartel DP. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res. 2009;69:9422–9430. doi: 10.1158/0008-5472.CAN-09-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 98.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 2006;34:6264–6271. doi: 10.1093/nar/gkl794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim Guisbert KS, Li H, Guthrie C. Alternative 3′ pre-mRNA processing in Saccharomyces cerevisiae is modulated by Nab4/Hrp1 in vivo. PLoS Biol. 2007;5:e6. doi: 10.1371/journal.pbio.0050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA. 2007;13:1756–1764. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deka P, Bucheli ME, Moore C, Buratowski S, Varani G. Structure of the yeast SR protein Npl3 and Interaction with mRNA 3′-end processing signals. J Mol Biol. 2008;375:136–150. doi: 10.1016/j.jmb.2007.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]