Abstract
Protein tyrosine phosphatases (PTPs), which catalyze the dephosphorylation of phosphotyrosine in protein substrates, are critical regulators of metazoan cell signaling and have emerged as potential drug targets for a range of human diseases. Strategies for chemically targeting the function of individual PTPs selectively could serve to elucidate the signaling roles of these enzymes and would potentially expedite validation of the therapeutic promise of PTP inhibitors. Here we report a novel strategy for the design of non-natural allosteric-inhibition sites in PTPs; these sites, which can be introduced into target PTPs through protein engineering, serve to sensitize target PTPs to potent and selective inhibition by a biarsenical small molecule. Building on the recent discovery of a naturally occurring cryptic allosteric site in wild-type Src-homology-2 domain containing PTP (Shp2) that can be targeted by biarsenical compounds, we hypothesized that Shp2’s unusual sensitivity to biarsenicals could be strengthened through rational design and that the Shp2-specific site could serve as a blueprint for the introduction of non-natural inhibitor sensitivity in other PTPs. Indeed, we show here that the strategic introduction of a cysteine residue at a position removed from the Shp2 active site can serve to increase the potency and selectivity of the interaction between Shp2’s allosteric site and the biarsenical inhibitor. Moreover, we find that “Shp2-like” allosteric sites can be installed de novo in PTP enzymes that do not possess naturally occurring sensitivity to biarsenical compounds. Using primary-sequence alignments to guide our enzyme engineering, we have successfully introduced allosteric-inhibition sites in four classical PTPs—PTP1B, PTPH-1, FAP-1, and HePTP—from four different PTP subfamilies, suggesting that our sensitization approach can likely be applied widely across the classical PTP family to generate biarsenical-responsive PTPs.
Keywords: Biarsenicals, FlAsH, Protein tyrosine phosphatases, Allostery, Protein engineering, Inhibitor sensitization
I. Introduction
Protein tyrosine phosphatases (PTPs) catalyze the dephosphorylation of phosphotyrosine residues in protein substrates.1 Regulation of PTP activity is therefore essential to cellular signaling pathways, which hinge on tight control of myriad phosphate-mediated biochemical events. Indeed, a large—and growing—body of work implicates aberrant PTP activity, arising from mutations in PTP genes, in the pathogenesis of human diseases; these diseases range from metabolic disorders to neurodegeneration to cancer.2–3 The emerging therapeutic relevance of PTPs has elicited intensive efforts on PTP-inhibitor-discovery from the pharmaceutical community.4 Despite these and other efforts towards developing tools for deconvoluting PTP signaling, however, the precise biological roles of many individual PTPs have yet to be determined,5 and the potential of PTPs as therapeutic targets has not, to date, been validated in the clinic. Furthermore, chemical approaches for the interrogation of PTP function are hindered by a shortage of selective, cell-permeable PTP inhibitors, as many of the challenges that generally beset inhibitor discovery are exacerbated in PTPs, owing to the high degree of structural similarity between classical-PTP active sites and the low cellular permeability of many active-site-directed PTP inhibitors.4
Small-molecule sensitization through protein engineering presents one putative remedy for the problem of chemically targeting members of a large and homologous protein family.6–7 By introducing a molecular change into a target through protein engineering, novel and specific protein/small-molecule pairs can potentially be generated more efficiently than by the methods of conventional inhibitor design. For PTP inhibitors specifically, a unique inhibitor-sensitizing feature could, in principle, be introduced artificially into the catalytic domain of a PTP. The sensitized mutant would thus represent a distinctive pharmacological target that can be inhibited specifically in the presence of many wild-type PTPs inside a cell. Ideally, the mutant would retain wild-type-like activity in the absence of its small-molecule partner; thus, when expressed in cells in lieu of the wild-type PTP, the mutant would serve as a functional surrogate that can be chemically interrogated in a specific manner. Although the need to engineer a protein target of interest presents a significant limitation for chemical-biology studies (the sensitized mutant must be expressed in place of the wild-type enzyme in an engineered cellular system), inhibitor sensitization through protein engineering potentially presents two key advantages over standard medicinal chemistry. First, high target specificity can be achieved with relatively little small-molecule synthesis and/or screening. Second, if a conserved feature of a large enzyme family (such as the PTPs) is altered in the sensitization strategy, it is likely that the sensitizing mutation(s) can be applied widely across the protein family. In this scenario, the specific sensitizing mutations for different members of the protein family of interest may be easily determined from primary-sequence alignments, and a single inhibitor (or a small number of related compounds) could be used to selectively target many members of the protein family (in separate experiments, in which only a single target in a specifically engineered cell harbors the sensitizing mutation). Successful approaches for the introduction of non-natural inhibitor sensitivity have been described on a number of protein families6, 8–9; particularly noteworthy examples include the “bump-hole” sensitization of the protein kinases10–11 and, more recently, BET bromodomains.12
Our lab has previously generated inhibitor-sensitive mutants of classical PTPs.13–17 Specifically, we have shown that introduction of cysteine-rich motifs into a conserved and catalytically important loop of the PTP domain (the WPD loop) is sufficient to sensitize both non-transmembrane (NT) and receptor-like (R) PTPs to inhibition by the cell-permeable biarsenical compound FlAsH-EDT2 (henceforth “FlAsH” for brevity, Figure 1A).15–18 To sensitize a target PTP to FlAsH, two to four cysteine residues are placed in the WPD loop (either as point mutations or insertions); binding of FlAsH to these cysteines induces inhibition of phosphatase activity, presumably by altering the WPD loop’s conformational switching between “open” and “closed” states.19–20 Targeting engineered cysteine-enriched WPD loops with biarsenicals has proven to be a general approach for PTP inhibition.16, 21–22 However, the WPD loop constitutes part of the PTP active site (in its closed state) and plays a key role in the catalytic mechanism of PTPs, presenting the possibility that mutations on the loop itself could alter the inherent kinetic activity or substrate specificity of the enzyme. Moreover, the presence of catalytically essential residues on the WPD loop significantly constrains the options for its engineering in a manner that is functionally silent (e.g., the “D” of WPD loop is a conserved aspartate residue that functions as a general acid-base in the PTP catalytic mechanism23).
Figure 1.

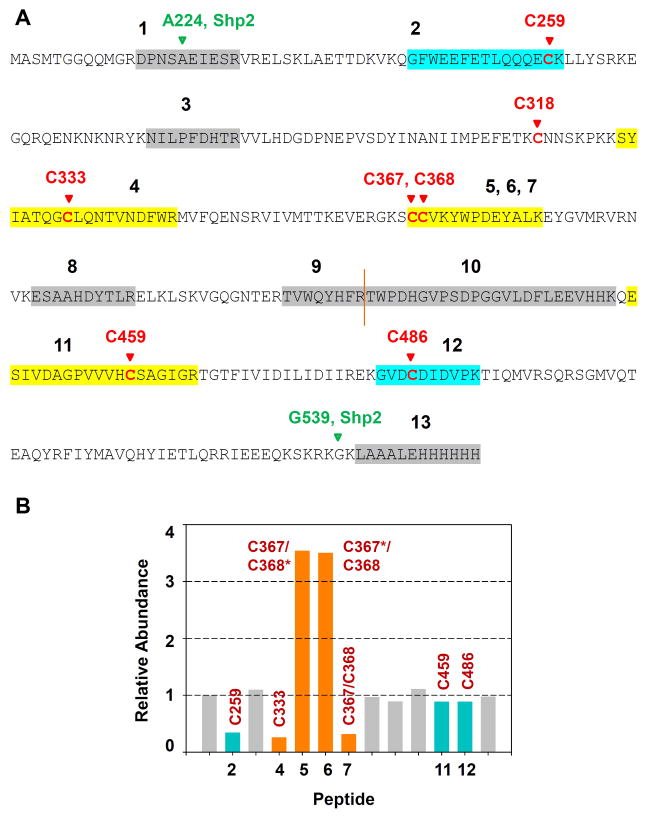
Design of classical PTP mutants that possess unnatural sensitivity to biarsenical compounds. (A) Chemical structure of the biarsenical compound FlAsH. (B) Partial amino-acid sequence alignment of the human PTPs discussed in this study, showing only structural motifs 4 and 7 of the PTP domain (as assigned by Anderson et al1). Highlighted in red are Shp2’s C333 and C367 and their cysteine counterparts in other PTPs. Highlighted in dark yellow are the amino acid residues substituted with cysteine in the present study. (The PTP-domain primary-sequence numbering varies widely due to the diversity in protein size and structure of the PTP family outside of the conserved PTP domain.42) (C) Three-dimensional structure of Shp2’s catalytic domain (PDB ID: 3B7O).30 Shp2 is shown as a blue ribbon, with the conserved active-site motif highlighted in pink and featuring the catalytic cysteine C459. The side chains of C333, C367, V368, and C459 are colored by atom type. The enzyme’s surface is rendered transparently so that the buried residues C333 and C367 can be visualized.
An ideal inhibitor-sensitized enzyme is one in which the sensitized enzyme functions exactly like the wild-type in the absence of ligand; it follows that an ideal PTP-sensitization strategy may be one in which PTP activity is controlled allosterically, at a binding site well removed from the catalytic site. Engineering of allosteric sites, however, represents an exceedingly difficult protein-design challenge, and most inhibitor-sensitization strategies described to date utilize one or more space-creating mutations (“holes”) in the protein target’s active-site. It is difficult to imagine how allosteric-inhibition sites could be designed in PTP domains from first principles.
Recently, however, we serendipitously discovered a rare, naturally occurring PTP allosteric-inhibition site that potentially provides a blueprint for engineering allosteric-inhibitor sensitivity into PTP domains.24 Specifically, we found that the oncogenic PTP Src-homology-2 domain containing PTP (Shp2) is unusual among classical PTPs in that its wild-type catalytic domain is sensitive to potent inhibition by FlAsH (and other biarsenicals), even in the absence of engineering.24 Shp2’s unusual biarsenical sensitivity derives from an allosteric site comprising two cysteine residues—C367, which is situated on PTP Motif 7 and is conserved across the classical PTP subfamily, and C333, which is situated on PTP Motif 4 and lies at a position occupied by proline in almost all other classical PTPs (Figure 1B, C).1 The Shp2-specific cysteine (C333) plays a determinative role in conferring Shp2’s unusual sensitivity to biarsenicals, and when the residue is mutated to proline (C333P), Shp2’s biarsenical sensitivity is abolished.24
The discovery of Shp2’s allosteric site has significant ramifications from both pharmaceutical and inhibitor-sensitization perspectives. Because of the potential importance of Shp2 inhibitors in pharmaceutical development, the enzyme’s unique allosteric site may present a novel target for Shp2-directed drug discovery. More germane to the work presented herein, the “naturally sensitized” Shp2 catalytic domain raises important questions regarding the inhibitor sensitization of PTP domains through protein engineering. First, can the Shp2 allosteric site be engineered to optimize the potency and efficiency of its inhibition? Second, does the existence of Shp2’s allosteric site suggest a strategy for engineering allosteric-inhibitor sensitivity into the catalytic domain of other PTPs? Here we report that the biarsenical sensitivity of the Shp2-specific allosteric site can be optimized through rational design. Moreover, we show that the optimized Shp2 allosteric site presents a template for designing allosteric-inhibition sites in other classical PTPs that are not sensitive to biarsenicals, including PTPs for which no allosteric inhibitors are known. The PTP-engineering strategy we report here could thus provide a general means for the introduction of allosteric-inhibition sites in PTP domains, toward the facilitation of the chemical-genetic analysis of the cell-signaling roles of classical PTPs.
2. Material and methods
2.1. General
FlAsH was synthesized as described.25 All PTP assays were performed in triplicate; error bars and “±” values represent the standard deviations of at least three independent experiments.
2.2. Cloning and mutagenesis of PTP-encoding genes
The plasmids encoding His6-tagged catalytic domains of Shp2 (pJGO0001),24 Shp1 (pACB149),24 PTP1B (pOBD0002),21 PTPH-1 (pERB047),26 HePTP (pBAD-HePTP),16 and FAP-1 (pAML001)16 have been previously described. All site-directed mutations were introduced using the Quikchange mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Desired mutations were confirmed by sequencing. Sequences of all mutagenic primers are provided as Supplementary Material.
2.3. Protein expression and purification
The His6-tagged catalytic domains of Shp2,24 Shp1,24 PTP1B,21 PTPH-1,26 HePTP,16 and FAP-116 were expressed and purified as described previously.
2.4. Phosphatase activity and inhibition assays with purified enzymes
PTP assays using para-nitrophenylphosphate (pNPP) as substrate were carried out in a total volume of 200 μL, containing PTP buffer (50 mM 3,3-dimethylglutarate at pH 7.0, 1 mM EDTA, 50 mM NaCl), enzyme (50 nM), β-mercaptoethanol (0–1 mM), and pNPP (0.625–10 mM). PTP reactions were quenched by the addition of 40 μL of 5 M NaOH, and the absorbances (405 nm) of 200 μL of the resulting solutions were measured on a VersaMax plate reader (Molecular Devices). Kinetic constants were determined by fitting the data to the Michaelis-Menten equation using SigmaPlot 12.3. For inhibition experiments, PTP samples (50 nM) were pre-incubated with FlAsH (varying concentrations), or with DMSO only (1% v/v), for 120 minutes at 37 °C prior to initiation of the PTP assays. The PTP activities of the solutions were then measured by assaying them at 37 °C with pNPP as described above at a pNPP concentration equal to the previously determined KM of the enzyme. 50% inhibitory concentration (IC50) values were estimated by fitting the inhibition data to a Four Parameter Logistic Equation for IC50 determination in SigmaPlot 12.3. For FlAsH-inhibition time-dependence experiments, pNPP (at a final concentration equal to the previously determined KM of the enzyme) was incubated with FlAsH (or DMSO, 1%) in PTP buffer at 37 °C, and PTP reactions were initiated by adding enzyme (50 nM). Changes in absorbance at 405 nm were measured continuously at 37 °C.
2.5. Phosphatase activity with whole-cell lysates
BL21(DE3) cells containing the appropriate PTP-encoding plasmid were grown overnight at 37 °C in LB. Cultures were diluted, grown to mid-log phase (OD600 = 0.5), induced with IPTG (1 mM), and shaken at room temperature overnight. The cells were harvested by centrifugation, resuspended in binding buffer (50 mM Tris at pH 7.8, 500 mM NaCl, 5 mM imidazole), and lysed by French Press at ~2000 psi. Lysates were clarified by centrifugation and incubated with FlAsH (varying concentrations) or DMSO (1%) for 120 minutes at 37 °C, after which PTP activities of the lysates were measured using pNPP as described above.
2.6. SDS/PAGE and fluorescence visualization of protein/FlAsH complexes
Samples of purified enzymes (2 μg) were incubated with FlAsH (6 μM) in SDS/PAGE loading buffer (Life Technologies) at room temperature for 30 minutes. After pre-incubation, the samples were heated at 96°C for 3 minutes for protein denaturation. Samples were loaded into a NuPAGE Bis-Tris precast gel (Life Technologies) and run per the manufacturer’s instructions. The fluorescence of protein/FlAsH conjugates was visualized under UV light.
2.7. Mass-spectrometry-based FlAsH-induced cysteine-protection assay
Samples for liquid chromatography-tandem mass spectrometry (LC/MS/MS) were prepared by incubating V368C Shp2 (100 μg, 2.7 μM) with either DMSO or FlAsH (27 μM) for 30 minutes in Shp2-catalytic domain storage buffer (50 mM 3,3-dimethyl glutarate at pH 7.0, 1 mM EDTA, 1 mM TCEP). Samples were then incubated with iodoacetic acid (IAA, 50 mM) for 30 minutes to modify free cysteine residues. IAA labeling was quenched with TCEP (100 mM), and the samples were precipitated with trifluoroacetic acid and run on an SDS-PAGE gel. Coomassie-stained gel slices were cut into 1×1 mm pieces and treated with trypsin. Digested samples were dissolved in acetonitrile and trifluoroacetic acid and were injected onto a custom packed 2 cm × 100 μm C18 Magic 5 μm particle trap column. Labeled peptides were then eluted and sprayed from a custom packed emitter (75 μm × 25 cm C18 Magic 3 μm particle) with a linear gradient from 95% solvent A (0.1% formic acid in water) to 35% solvent B (0.1% formic acid in acetonitrile) in 60 minutes at a flow rate of 300 nL/min on a Waters Nano Acquity UPLC system. Data dependent acquisitions were performed on a Q Exactive mass spectrometer (Thermo Scientific) according to an experiment where full MS scans from 300–1750 m/z were acquired at a resolution of 70,000 followed by 12 MS/MS scans acquired under HCD fragmentation at a resolution of 35,000 with an isolation width of 1.2 Da. Raw data files were processed with Mascot Distiller (version 2.5) prior to searching with Mascot Server (version 2.4) against a SwissProt Human database containing the construct sequences. Search parameters utilized were fully tryptic with two missed cleavages, parent mass tolerances of 10 ppm and fragment mass tolerances of 0.05 Da. Variable modifications of acetyl (protein N-term), pyro glutamic for N-term glutamine, oxidation of methionine, and carboxymethyl cysteine were considered. Search results were used to create spectral libraries for the Skyline software (University of Washington), which was used to quantitate selected peptides using precursor intensity data from extracted ion chromatograms. Intensities for peptides identified in the FlAsH-treated protein sample were compared to the corresponding peptides identified in the DMSO-treated sample, and their resulting intensity ratios were calculated. For peptides detected multiple times, the mean intensity ratios were calculated (peptide 1: n = 2, standard deviation (SD) = 0.167; peptide 2: n = 4, SD = 0.0404; peptide 3: n = 5, SD = 0.0846; peptide 4: n = 4, SD = 0.0691; peptide 5: n = 1; peptide 6: n = 1; Peptide 7: n = 1; peptide 8: n = 3, SD = 0.122; peptide 9: n = 2, SD = 0.0868; peptide 10: n = 3, SD = 0.0226; peptide 11: n = 4, SD = 0.117; peptide 12: n = 3, SD = 0.0755; peptide 13: n = 2, SD = 0.0663). Intensity ratios were then divided by the average of all non-cysteine-containing peptide ratios to determine normalized relative abundance values.
3. Results and discussion
3.1. Rational design of a Shp2 allosteric-inhibition site with increased sensitivity
Due to the presence of a recently discovered allosteric site, the wild-type Shp2 catalytic domain possesses unusual sensitivity to inhibition by biarsenical compounds.24 The naturally occurring biarsenical-binding site on Shp2 consists of two cysteines (C333 and C367), which are located close to one another in three-dimensional space (5.7 Å from Cβ to Cβ), despite lying 34 residues apart in the protein’s amino-acid sequence. The primary-sequence distance between these biarsenical-reactive cysteines stands in remarkable contrast with the canonical FlAsH-binding motif used to tag proteins for FlAsH-labeling, which has the linear tetracysteine sequence CCXXCC,18, 27 although precedent for biarsenical engagement of non-linear multi-cysteine motifs does exist.28–29 Regardless of the sequential arrangement of the cysteine residues, the fact that only two cysteines exist in Shp2’s biarsenical-binding site suggest that the strength of its binding interaction with FlAsH may be weak compared to the canonical biarsenical/CCXXCC interaction. The sub-optimal nature of the Shp2 biarsenical site is further suggested by the observation that the apparent inhibitory potency of FlAsH on Shp2 is reduced substantially by the presence of competing proteins in a complex proteomic mixture.24
3.1.1. Design of a cysteine-enriched Shp2 allosteric site
To potentially enhance the interactions between Shp2’s allosteric site and biarsenical compounds, we selected a site at which to introduce a third cysteine, in close proximity to C333 and C367 (Figure 1C). We selected valine 368 (human Shp2 numbering) as a promising site for several reasons: (i) V368 is directly adjacent to C367, so a V368C mutation would produce a contiguous CC motif, akin to “half” of a canonical CCXXCC biarsenical-binding sequence. (ii) Valine at position 368 is not conserved among classical PTPs (Figure 1B).1 Thus, we expected that mutation of this position would not inherently disrupt enzymatic function. (iii) V368 contains the most solvent-accessible side chain adjacent to either C333 or C367. In fact, V368’s side chain is significantly more solvent-accessible than either the C333 or C367 side chains, which are buried in Shp2 crystal structures (Figure 1C).30–31 We therefore hypothesized that addition of a third, solvent-exposed cysteine to the otherwise “cryptic” Shp2 allosteric site, would enhance the overall accessibility of the site. Based on this analysis we used site-directed mutagenesis to generate V368C Shp2 as a mutant with putatively enhanced biarsenical sensitivity.
3.1.2. PTP activity of V368C Shp2
To ensure that the V368C mutation is not deleterious to Shp2’s inherent catalytic activity, we performed Michaelis-Menten kinetic analysis on the wild-type and V368C Shp2 catalytic domains in the absence of inhibitor. We found that the V368C mutation has no negative effect on Shp2’s catalytic activity (Table 1). In fact, compared to wild-type, V368C Shp2 possesses a modestly increased catalytic efficiency (kcat/KM) that derives from both a small increase in the enzyme’s rate constant (kcat) and a marginally decreased Michaelis constant (KM) for the small-molecule substrate para-nitrophenyl phosphate (pNPP). More broadly, the kinetic characterization of V368C Shp2 suggests that the 368 position of PTP domains may be generally suitable for functionally silent protein engineering, as mutagenesis of this position can be performed without introducing defects in PTP activity.
Table 1.
Kinetic constants of wild-type and engineered Shp2 assayed with pNPP.
| enzyme | kcat(s−1) | KM (mM) | kcat/KM (mM−1 s−1) |
|---|---|---|---|
| Wild-type Shp2 | 8.7 ± 0.54 | 5.6 ± 0.027 | 1.6 |
| V368C Shp2 | 13 ± 1.7 | 4.1 ± 0.83 | 3.2 |
3.1.3. Enhanced biarsenical sensitivity of V368C Shp2
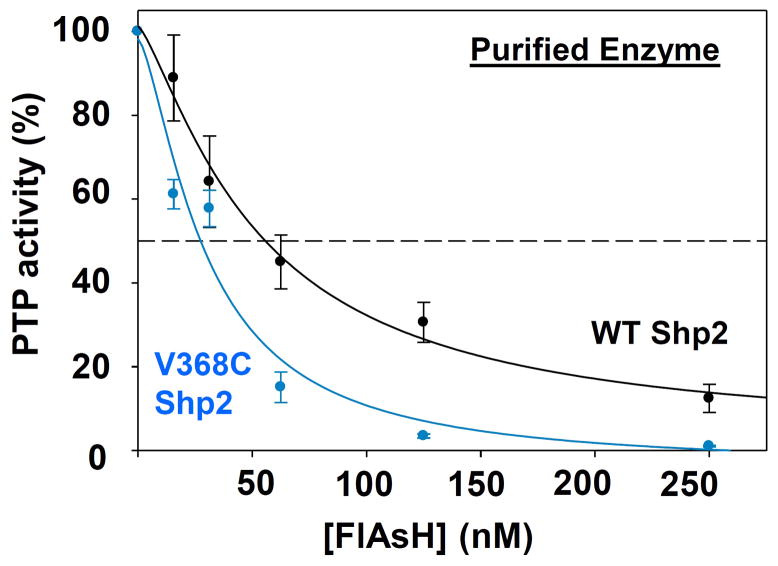
To investigate the effects of the V368C substitution on Shp2’s sensitivity to FlAsH, we compared the potencies of FlAsH-induced inhibition for purified wild-type and V368C Shp2 (Figure 2). Although the biarsenical compound inhibits both Shp2 variants nearly stoichiometrically—both enzyme/FlAsH 50% inhibitory-concentration (IC50) values approach or are lower than the concentration of enzyme in the assay (50 nM)—the V368C mutation does appear to increase the sensitivity of Shp2 to inhibition by FlAsH: a lower IC50 is distinctly observable for V368C Shp2 (IC50 = 30 nM), as compared to wild-type (IC50 = 53 nM). (The inhibition data for wild-type Shp2 are consistent with previously reported results.24) Thus, the V368C mutation does appear to increase the sensitivity of Shp2’s allosteric site, as designed.
Figure 2.
V368C Shp2 is inhibited more potently by FlAsH than wild-type Shp2. The phosphatase activities of purified wild-type and V368C Shp2 (50 nM) were measured with pNPP in the presence of the indicated FlAsH concentrations after 120-minute pre-incubations. The activities were normalized to DMSO-only controls for the corresponding enzymes.
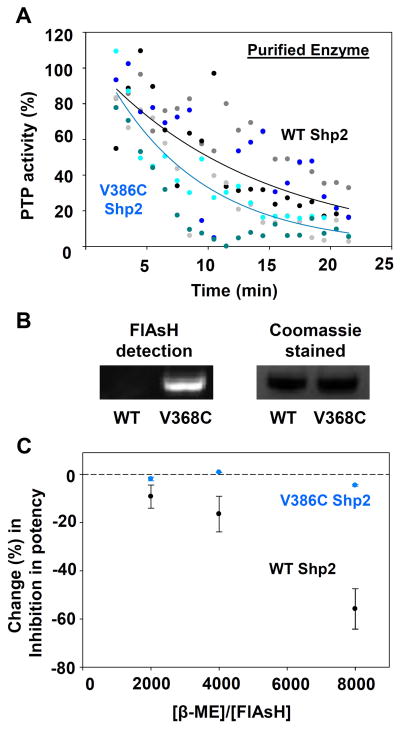
The dose-response experiments described above suggest an enhancement in the potency of FlAsH’s interaction with the expanded, tricysteine site in V368C Shp2 following pre-incubation. We next investigated whether the V368C mutation gives rise to a more kinetically accessible binding site by continuously monitoring the time-dependent change in the PTP activities of wild-type and V368C Shp2 in the presence of a fixed concentration of FlAsH (Figure 3A). Somewhat surprisingly, the rate of inhibition was not dramatically enhanced by the V368C mutation. Although V368C Shp2 appears to undergo a modestly more rapid FlAsH-induced loss of activity than does wild-type Shp2 (wild-type Shp2 time to 50% activity: ~ 11 minutes; V368C Shp2: ~7 minutes, Figure 3A), the difference is not statistically significant.
Figure 3.
V368C Shp2 displays enhanced allosteric-site interactions with FlAsH. (A) Wild-type and V368C Shp2’s rates of dephosphorylating pNPP in the presence of 500 nM FlAsH over 1-minute windows were normalized to a DMSO-only control. The results of three independent experiments (grayscale for wild-type Shp2, bluescale for V368C Shp2) were averaged and the averaged data was fitted as single-variable exponential decays. (B) Purified wild-type and V368C Shp2 were treated with FlAsH, run on an SDS/PAGE gel, and visualized both under UV light and by Coomassie staining. (C) After incubation with 125 nM FlAsH and varying concentrations of β-mercaptoethanol (β-ME), PTP activities of purified wild-type and V368C Shp2 were measured with pNPP and normalized to no-FlAsH controls.
To further probe the strength of FlAsH’s interactions with the wild-type and expanded FlAsH-binding sites, we assessed the ability of the protein/FlAsH conjugates to withstand denaturation and gel electrophoresis. After treating both Shp2 constructs with FlAsH, we performed SDS/PAGE on the samples and used fluorescence to visualize the FlAsH-bound proteins (Figure 3B).27 We found that FlAsH-treated V368C Shp2 gives rise to a strongly fluorescent band at approximately 37 kD, the molecular weight of our Shp2 construct. By contrast, the corresponding band is not present in the lane containing FlAsH-treated wild-type Shp2. The data suggest that the two-cysteine site of wild-type Shp2 does not make a strong enough interaction to persist through SDS/PAGE, whereas the FlAsH/tricysteine site interaction of V368C Shp2 is sufficiently strong to withstand SDS/PAGE conditions, as has been previously observed for canonical tetracysteine motifs.27–28
Finally, we measured the biarsenical sensitivity of the Shp2 constructs in the presence of free thiols in solution, which could serve to compete with cysteine-containing protein motifs for binding with FlAsH. We observed that the thiol-containing reagent β-mercaptoethanol (β-ME) attenuates FlAsH’s inhibitory effects on wild-type Shp2, whereas it induces no significant changes in the potency of FlAsH inhibition on V368C Shp2 (Figure 3C). The latter observation holds true even when the concentration of β-ME present is 8,000 times in excess of that of FlAsH, a clear indication of the enhanced strength of engagement between FlAsH and the engineered tricysteine motif in the V368C mutant.
In sum, the data presented in Figures 2 and 3 are consistent with the rational design of the V368C Shp2 mutant: the experiments suggest that the affinity of the protein/FlAsH interaction is increased in V368C Shp2, and that the biarsenical compound is engaging the expanded, tricysteine site through an additional arsenic-thiol covalent interaction at C368.
3.1.4. Cysteine-protection assay to identify FlAsH/allosteric-site interactions
To potentially gain more direct evidence of an interaction between C368 and FlAsH in the allosteric site of V368C Shp2, we performed a cysteine-protection assay in which Shp2’s cysteine residues were labeled with iodoacetic acid (IAA) in the presence or absence of FlAsH.32 Previously, we used this assay to establish the presence of the unexpected allosteric site in wild-type Shp2 by showing that FlAsH strongly and specifically protects the site’s constituent cysteine residues (C333 and C367) from carboxymethylation by IAA.24 To assess FlAsH’s protective effects on V368C Shp2 in the current study, we incubated the mutant enzyme with either FlAsH or vehicle and then labeled the protein’s free cysteines with IAA. Tryptic liquid chromatography-tandem mass spectrometry (LC/MS/MS) analysis of the samples revealed that FlAsH strongly protects C333, C367, and C368 from modification by IAA (Figure 4). Protection of the allosteric site is evident in the low abundances of two peptides—the peptide containing carboxymethylated C333 (Figure 4, peptide 4) and the peptide containing C367 and C368 in which both cysteines are carboxymethylated (Figure 4, peptide 7). These cysteine-protection data suggest that FlAsH directly associates with all three cysteines, C333, C367, and C368, as rationally designed.
Figure 4.
FlAsH protects cysteines 333, 367, and 368 in V368C Shp2 from modification by iodoacetic acid. V368C Shp2 (2.7 μM) was incubated with DMSO or FlAsH (27 μM), followed by addition of iodoacetic acid (50 mM). The labeled protein was trypsinized and the abundances of the resulting peptides were quantitated by LC/MS/MS. Relative abundances indicate the normalized intensities of the indicated peptides in a FlAsH-treated sample as compared to its no-FlAsH control. Peptides containing positions 333, 367, and 368 are highlighted in orange; peptides containing other carboxymethylated cysteines are highlighted in blue; other non-cysteine-containing peptides appear in gray. The sequences of peptides 1–13 are provided in panel A. Asterisks in panel B indicate non-carboxymethylated (free) cysteine residues detected in the LC/MS/MS experiment.
Our LC/MS/MS experiment yielded an additional and unexpected piece of data. In addition to the doubly modified C367 and C368-containing peptide mentioned above, singly modified peptides—each containing one carboxymethylated cysteine and one unmodified cysteine—were also identified (Figure 4B, peptides 5 and 6). Furthermore, these peptides are present in substantially greater amounts in FlAsH-treated samples as compared to vehicle-treated samples. The FlAsH-induced enrichment of the singly modified peptides suggests that carboxymethylation of either C367 or C368 is made more facile when only one of the two residues is engaged by FlAsH. (The FlAsH/Shp2 interaction does not persist through the mass-spectrometry experiment, so whichever cysteine was engaged with FlAsH during the incubation step would appear as an unmodified cysteine, C367* or C368*, in the LC/MS/MS data.) These intriguing results suggest that FlAsH binding promotes a Shp2 conformational change that increases the IAA accessibility of the C367/368 peptide motif, consistent with a conformational-switching hypothesis we have previously proposed to potentially explain how the buried residues C333 and C367 site could be “revealed” to an inhibitor. We speculated that Shp2 must sample at least two conformational states: an active state in which the allosteric site is buried and an inactive state in which it is exposed.24 If this hypothesis is correct, FlAsH binding (either partial or complete) would promote formation of the latter conformational state, so that structural elements of Shp2’s allosteric site might remain in their more solvent-exposed positions. In line with this model, C367 and C368 could be found to be individually carboxymethylated more readily in the presence of FlAsH, as the biarsenical promotes the formation of V368C Shp2’s “exposed” state. The observed relative abundances of the singly modified peptides might thus constitute the first evidence pointing to the existence of the proposed Shp2 conformational equilibrium.
3.1.5. Inhibition of V368C Shp2 in a complex proteomic mixture
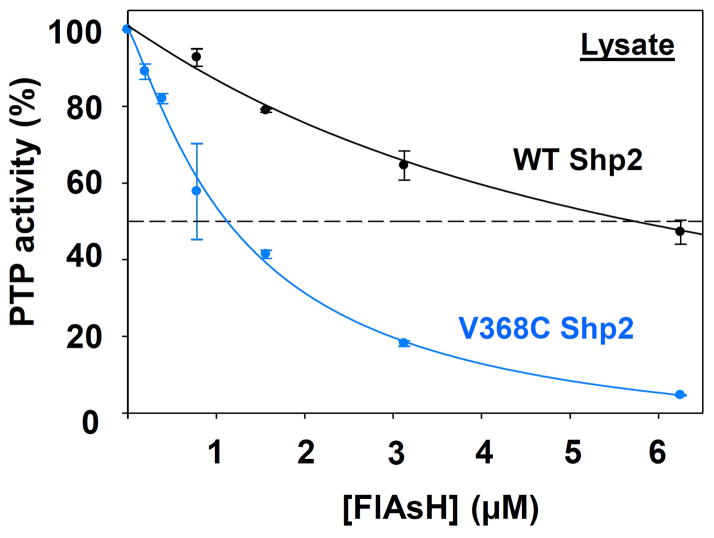
To test whether a stronger interaction between FlAsH and V368C’s expanded allosteric site translates to enhanced potency in a complex proteomic mixture, we measured the effects of FlAsH on total PTP activities of crude lysates from bacteria that express wild-type or V368C Shp2. (The E. coli genome does not encode for PTPs, so total-lysate PTP activity can be used as a proxy for the activity of a bacterially overexpressed mammalian PTP.33) Strikingly, the apparent IC50 values differ by a factor of four between wild-type and V368C Shp2 (Figure 5), with more potent inhibition (IC50 = 1.3 μM) observed for the mutant-expressing lysates than for the wild-type-expressing lysates (IC50 = 6.0 μM). These data suggest that, in a cellular environment, biarsenical compounds might target V368C Shp2 more potently than wild-type Shp2. Expression of V368C Shp2 in lieu of the wild-type enzyme may therefore yield the advantage of facilitating the use of lower biarsenical concentrations for effective cellular perturbation of Shp2 signaling.
Figure 5.
V368C Shp2 is targeted more potently by FlAsH than wild-type Shp2 in a complex proteomic mixture. Crude lysates from bacterial cells expressing wild-type or V368C Shp2 were treated with the indicated FlAsH concentrations. Total PTP activities of the lysates were then measured with pNPP and normalized to DMSO-only controls for the corresponding enzymes.
3.2. Rational design of de novo allosteric sites in PTP domains: PTP1B as prototype
The data presented above establish that Shp2’s unique allosteric site, consisting of C333 and C367, can be further sensitized by the addition of a third cysteine residue at position 368. Only two human PTPs, Shp2 and its closest homolog Shp1, contain cysteine residues at position 333 and no human PTPs naturally have cysteine residues at all three positions (333, 367, and 368). Our findings on Shp2, therefore, suggest the possibility that cysteine mutagenesis at these key positions could be used to introduce novel allosteric-inhibitor sensitivity into PTP domains that do not possess natural sensitivity to biarsenicals. Moreover, the heightened sensitivity of V368C Shp2’s tricysteine allosteric site, which has no equivalent in any wild-type PTP, suggests the possibility that engineered de novo tricysteine sites could surpass the sensitivity of even Shp2’s naturally occurring site.
To test the idea that Shp2 could serve as a blueprint for the introduction of biarsenical-reactive sites in PTP domains that are not sensitive to FlAsH, we selected protein tyrosine phosphatase 1B (PTP1B), a classical PTP that has inspired intensive drug-discovery efforts,4, 34–37 as our initial candidate for sensitization. PTP1B and Shp2 share only a moderate level of homology (39% PTP-domain identity) and are members of different PTP subfamilies (NT1 and NT2, respectively).1 We surmised, therefore, that successful sensitization of PTP1B as a (somewhat arbitrarily) selected prototype might suggest that members of still other PTP subfamilies could also be sensitized through analogous strategies.
Primary-sequence alignment of the PTP1B and Shp2 catalytic domains reveals that PTP1B residues P87 and A122 occupy the equivalent amino-acid positions as Shp2’s C333 and V368 (Figure 1B).1 To test whether PTP1B could be sensitized to FlAsH via the same allosteric mechanism observed in wild-type Shp2, we substituted P87 of PTP1B with cysteine. The resulting cysteine mutant, P87C PTP1B, carries a dicysteine structural motif (C87/C121) analogous to the naturally FlAsH-sensitive site in wild-type Shp2 (C333/C367). To further test our hypothesis that our findings on V368C Shp2 can inform rational design on PTP1B, we also generated a PTP1B mutant in which both P87 and A122 are substituted with cysteines. The resulting dicysteine mutant, P87C/A122C PTP1B, carries a tricysteine structural motif (C87/C121/C122) analogous to the expanded FlAsH-binding site in V368C Shp2 (C333/C367/C368).
3.2.1. Enzymatic activity of PTP1B mutants
We first asked whether the catalytic activity of PTP1B is retained in the presence of the P87C and A112C substitutions. To do so, we performed Michaelis-Menten kinetics on wild-type, P87C, and P87C/A122C PTP1B in the absence of FlAsH to determine if the mutants’ catalytic competency was affected by the protein engineering (Table 2). Both mutants display moderately reduced catalytic efficiencies: relative to the wild-type enzyme, the catalytic efficiencies of the monocysteine and dicysteine mutants are lowered approximately four- and six-fold, respectively. Specifically, the P87C substitution both diminishes the kcat and raises the KM two-fold, while the additional A122C substitution slightly lowers the kcat value further. The small kcat-lowering effect of the A112C mutation is a bit surprising as the analogous mutation in the Shp2 catalytic domains (V368C) gave rise to a modest increase in kcat (Table 1). While we do not currently understand the molecular basis of these small effects, and it remains to be seen which of the effects will be the more general one, all of the changes in catalytic parameters observed in engineering both Shp2 and PTP1B are quite modest, and the magnitude by which the mutants’ catalytic efficiencies differ from their wild-type activities is within the range observed in other enzyme-sensitization approaches.10, 15
Table 2.
Kinetic constants of wild-type and engineered PTP1B assayed with pNPP.
| enzyme | kcat(s−1) | KM (mM) | kcat/KM (mM−1 s−1) |
|---|---|---|---|
| Wild-type PTP1B | 14 ± 1.9 | 2.5 ± 0.37 | 5.6 |
| P87C PTP1B | 7.1 ± 0.97 | 4.0 ± 0.82 | 1.8 |
| P87C/A122C PTP1B | 4.8 ± 0.17 | 4.8 ± 0.40 | 1.0 |
3.2.2. FlAsH sensitivity of PTP1B mutants
To test whether P87C PTP1B and/or P87C/A122C PTP1B possess non-natural sensitivity to biarsenicals, we subjected these mutants to an array of tests analogous to those described above for V368C Shp2. First, we tested whether the enzymatic activity of the various PTP1B constructs could be inhibited by FlAsH (Figure 6A). In agreement with previous observations, we found that the activity of wild-type PTP1B is unaffected by FlAsH.16, 21 By contrast, a single cysteine substitution renders P87C PTP1B’s phosphatase activity sensitive to strong inhibition by FlAsH. The inhibition of P87C PTP1B is both potent—in the presence of 50 nM enzyme, the IC50 value of FlAsH is approximately 120 nM (Figure 6A)—and rapid—in the presence of 500 nM FlAsH, the total phosphatase activity of P87C PTP1B decreases to 50% of its initial rate in approximately 10 minutes (Figure 6B). These data establish that, with a single cysteine substitution at the amino-acid position equivalent to Shp2’s C333, a dicysteine allosteric-inhibition site can be introduced into PTP1B, and that the sensitivity of P87C’s non-natural inhibitor-binding site is comparable to that of the naturally occurring allosteric-inhibition site in Shp2.
Figure 6.
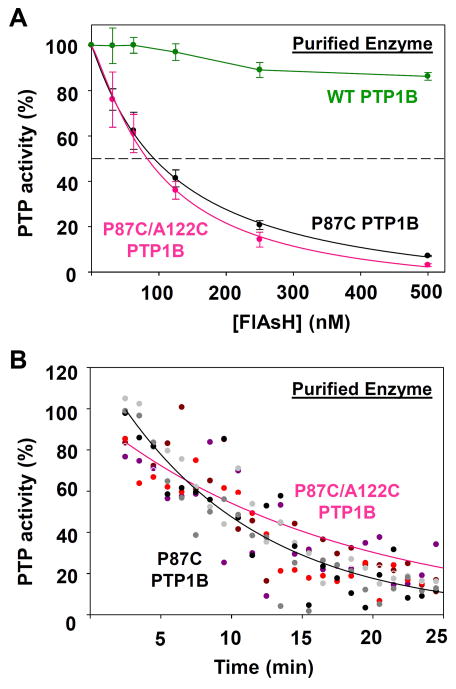
Purified P87C PTP1B and P87C/A122C PTP1B exhibit marginal differences in FlAsH sensitivity. (A) The phosphatase activities of purified wild-type and engineered PTP1B enzymes (50 nM) were measured with pNPP in the presence of the indicated FlAsH concentrations after 120-minute pre-incubations. The activities were normalized to DMSO-only controls for the corresponding enzymes. (B) The PTP1B mutants’ rates of dephosphorylating pNPP in the presence of 500 nM FlAsH over 1-minute windows were normalized to a DMSO-only control. The results of three independent experiments (grayscale for P87C PTP1B, redscale for P87C/A122C PTP1B) were averaged and the averaged data was fitted as single-variable exponential decays.
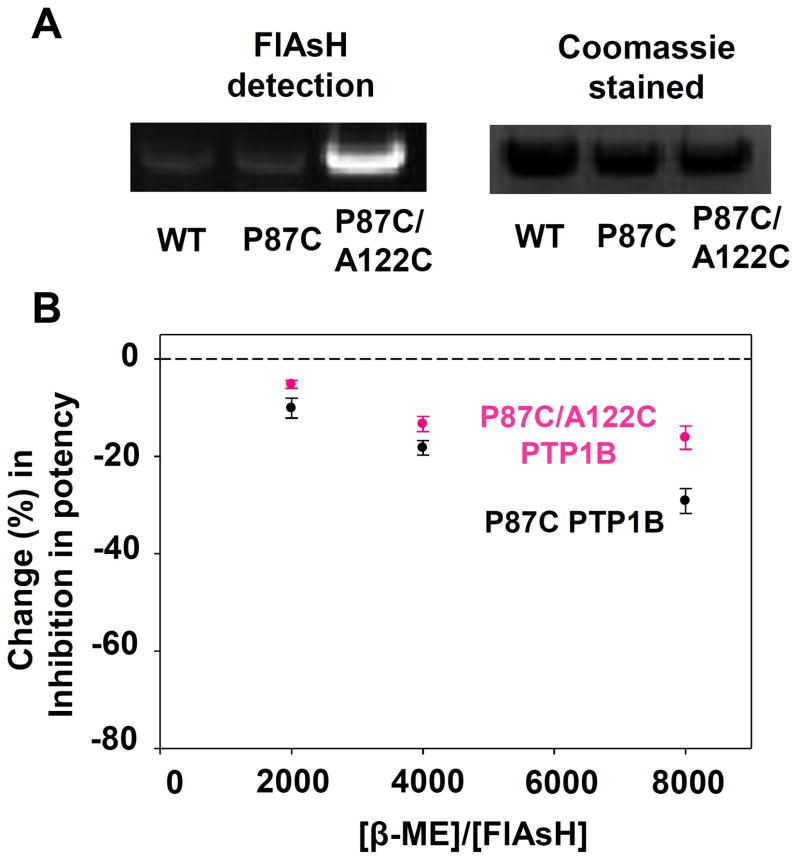
As expected based on the results for P87C PTP1B, the double-cysteine mutant P87C/A122C PTP1B is also potently inhibited by FlAsH in a time-dependent manner. Somewhat surprisingly, however, the A122C mutation appears to confer only marginal differences (compared to P87C alone) in the dose-dependence and time-dependence of FlAsH inhibition as measured with PTP assays on purified protein (Figure 6A, B; compare black lines to pink lines). To more closely investigate potential differences in the strength between the biarsenical/dicysteine (P87C) and biarsenical/tricysteine (P87C/A122C) interactions, we subjected the PTP1B variants to further experimental challenges, analogous to those presented earlier for V368C Shp2 (Figure 7).22 In an SDS-PAGE/fluorescence experiment, we found that only P87C/A122C PTP1B produces a strongly fluorescent band at 45 kD (the molecular weight of our PTP1B construct), whereas wild-type PTP1B and P87C PTP1B both fail to produce strongly fluorescent bands (Figure 7A). Similarly, a β-ME-challenge experiment indicated that the additional A122C mutation permits the target site in PTP1B to compete more effectively with free thiols in solution for binding with FlAsH (Figure 7B). In the presence of an 8000:1 β-ME:FlAsH ratio, the potency of FlAsH’s inhibitory effects on P87C/A122C PTP1B suffers a significantly smaller reduction (16.2%) than the potency of FlAsH’s inhibitory effects on P87C PTP1B (29.1%).
Figure 7.
The tricysteine allosteric site of P87C/A122C PTP1B binds FlAsH more tightly than the dicysteine allosteric site of P87C PTP1B. (A) Purified wild-type and engineered PTP1B variants were treated with FlAsH, run on an SDS/PAGE gel, and visualized both under UV light and by Coomassie staining. (B) After incubation with 125 nM FlAsH and varying concentrations of β-mercaptoethanol (β-ME), PTP activities of purified PTP1B variants were measured with pNPP and normalized to no-FlAsH controls.
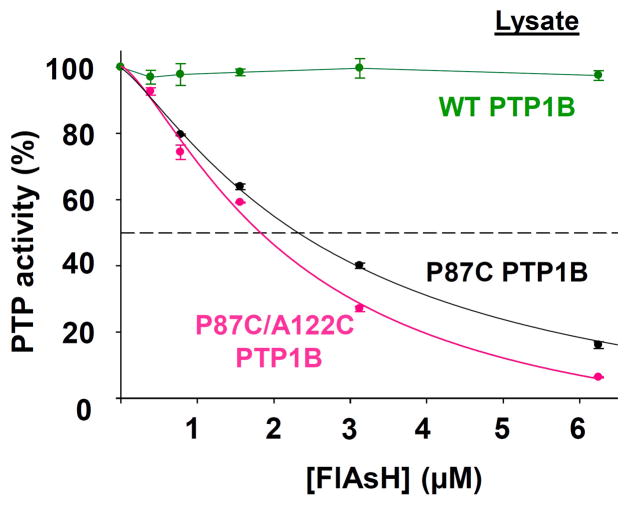
In total, the data for P87C and P87C/A122C PTP1B are generally consistent with the corresponding data for Shp2 and indicate a positive correlation between the number of cysteines at the allosteric site and the strength of FlAsH’s interaction with the site. Although the “third” cysteine of PTP1B’s engineered tricysteine site (C122) does not appear to dramatically change the biarsenical sensitivity as measured on purified enzyme (Figure 6), the SDS-PAGE and β-ME-challenge experiments (Figure 7) suggest that C122 does enhance the protein/biarsenical interaction and could potentially improve the specificity of allosteric-site targeting in the presence of the many competing proteins in a complex proteomic mixture. To investigate the latter question directly, we measured the effects of FlAsH on total PTP activities of crude lysates of E. coli that expressed either P87C or P87C/A122C PTP1B (Figure 8). Consistent with the hypothesis of enhanced potency in the FlAsH/tricysteine-site interaction, P87C/A122C PTP1B-expressing lysate exhibits a modestly but measurably lower IC50 of FlAsH inhibition (2.2 μM) than does P87C PTP1B-expressing lysate (2.8 μM). The combined data from the Shp2 and (engineered) PTP1B allosteric sites imply that cysteine-enrichment of a biarsenical-binding site (in particular, from di- to tri-) could be a general strategy for enhancing the specificity of FlAsH-induced inhibition.
Figure 8.
FlAsH-induced inhibition of P87C PTP1B and P87C/A122C PTP1B in a complex proteomic mixture. Crude lysates from bacterial cells expressing wild-type PTP1B, P87C PTP1B, or P87C/A122C PTP1B were treated with the indicated FlAsH concentrations. Total PTP activities of the lysates were then measured with pNPP and normalized to DMSO-only controls for the corresponding enzymes.
3.3. Rational design of de novo allosteric sites in PTP domains: Testing the scope
Following our success at introducing “Shp2-like” sensitivity in PTP1B, we sought to test the allosteric-engineering strategy more widely in the family of classical PTPs. For our sensitization targets, we selected four additional biologically important classical PTPs, representing both non-transmembrane and receptor-like phosphatases: PTPH1 (subtype NT5), FAP-1 (subtype NT7), HePTP (subtype R7), and Shp1 (subtype NT2).1 Like PTP1B, the PTP domains of PTPH-1, FAP-1, and HePTP have previously been shown to be insensitive to FlAsH,16 whereas the PTP domain of Shp1 exhibits weak biarsenical sensitivity, consistent with its containing a cysteine residue (C328) at the position corresponding to C333 of Shp2 (Figure 1B).24
3.3.1. PTP mutant design
The Shp2- and PTP1B-sensitizing efforts described above showed that a tricysteine allosteric site is generally more potently targetable than a dicysteine site. As such, we elected to test PTP mutants carrying the tricysteine structural motif analogous to the expanded FlAsH-binding site of V368C Shp2 (and P87C/A122C PTP1B). Catalytic-domain-sequence alignments of the above PTPs and Shp2 again permitted facile identification of the PTP residues equivalent to Shp2’s C333 and V368 (Figure 1B).1 To generate PTP mutants that can potentially be regulated by the allosteric-inhibition mechanism of Shp2, we used site-directed mutagenesis to substitute the appropriate residues with cysteines, producing the following panel of mutants: P2285C/Q2320C FAP-1, P160C/V184C HePTP, P720C/H755C PTPH-1, and V364C Shp1, all of which bear the tricysteine motif analogous to that of C333/C367/C368 in V368C Shp2. (Because the FAP-1, HePTP, PTPH-1, and Shp1 mutants all contain analogous biarsenical-binding motifs—and only one mutant was prepared for each PTP—they will often be referred to simply as “mutant.” In Figure 9, the tricysteine-containing constructs of Shp2 (V368C) and PTP1B (P87C/A122C) are likewise presented as “mutant” for easy comparison.)
Figure 9.
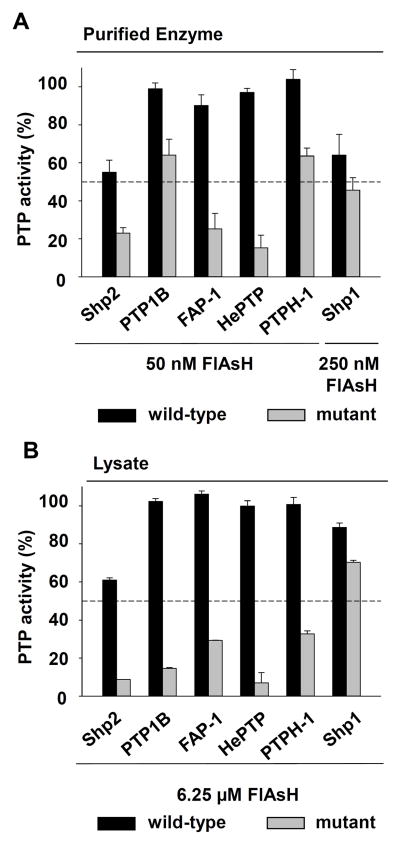
Allosteric-site engineering confers unnatural FlAsH sensitivity on PTPs from multiple subfamilies. (A) The phosphatase activities of purified wild-type and engineered PTPs (50 nM) were measured with pNPP in the presence of the indicated FlAsH concentrations after 120-minute pre-incubations. The activities were normalized to DMSO-only controls for the corresponding enzymes. (B) Crude lysates from bacterial cells expressing wild-type or engineered PTPs were treated with the indicated FlAsH concentrations. Total PTP activities of the lysates were then measured with pNPP and normalized to DMSO-only controls for the corresponding enzymes.
3.3.2. Enzymatic activity of PTP mutants
We first assessed the effects of our protein-engineering on the catalytic activities of the panel of PTP mutants. Table 3 contains Michaelis-Menten kinetic constants for the mutant enzymes, as well as their wild-type counterparts. The magnitude of changes conferred by the cysteine mutations falls broadly within the range observed with V368C Shp2 and P87C/A122C PTP1B—all four of the panel’s mutant PTPs showed either no significant change in catalytic efficiency (HePTP, Shp1) or a modest drop in catalytic efficiency (FAP-1, PTPH-1). Interestingly, the V364C mutation of mutant Shp1 conferred a modest drop in kcat, whereas the directly analogous V368C mutation in the closely related Shp2 catalytic domain gave rise to a small increase in kcat (Table 1), suggesting that it will be difficult to predict the precise catalytic effects of sensitizing cysteine mutations based on sequence homology.
Table 3.
Kinetic constants of wild-type and engineered PTPs assayed with pNPP.
| enzyme | kcat(s−1) | KM (mM) | kcat/KM (mM−1 s−1) |
|---|---|---|---|
| Wild-type FAP-1 | 3.2 ± 0.40 | 0.82 ± 0.082 | 3.9 |
| P2285C/Q2320C FAP-1 | 2.4 ± 0.28 | 1.9 ± 0.40 | 1.3 |
|
| |||
| Wild-type PTPH-1 | 3.5 ± 0.82 | 0.67 ± 0.062 | 5.2 |
| P720C/H755C PTPH-1 | 2.2 ± 0.24 | 1.6 ± 0.17 | 1.4 |
|
| |||
| Wild-type HePTP | 1.3 ± 0.13 | 9.1 ± 1.4 | 0.14 |
| P160C/V184C HePTP | 1.2 ± 0.19 | 11 ± 2.90 | 0.11 |
| Wild-type Shp1 | 4.1 ± 0.41 | 3.5 ± 0.60 | 1.2 |
| V364C Shp1 | 2.4 ± 0.28 | 1.9 ± 0.40 | 1.3 |
3.3.3. FlAsH-sensitivity of PTP mutants
To test whether cysteine mutagenesis was sufficient to sensitize the PTP domains of FAP-1, PTPH-1, and HePTP to inhibition by FlAsH, we measured the phosphatase activities of purified wild-type and mutant enzymes (50 nM, the same concentration used in previous dose-dependence experiments) after incubation with one equivalent of FlAsH. Consistent with precedent, the wild-type enzymes were not significantly inhibited by the biarsenical compound.16, 24 By contrast, the FAP-1 and HePTP mutants exhibited drastically reduced catalytic activity in the presence of FlAsH; the potencies of inhibition of mutant FAP-1 and HePTP are comparable to that observed with V368C Shp2 (Figure 9A). The PTPH-1 mutant also possesses novel sensitivity to FlAsH, albeit a slightly lower level, comparable to the tricysteine-containing mutant of PTP1B (P87C/A122C PTP1B).
Inhibition experiments on whole-cell lysates from E. coli that express the mutant PTPs corroborated the results on purified enzymes. We found that, in the context of a complex proteomic mixtures, the mutant forms of FAP-1, PTPH-1, and HePTP were all strongly inhibited at FlAsH concentrations that had no effects on the activity of the corresponding wild-type PTPs (Figure 9B). It should be noted that in the context of cell lysates, the tricysteine-containing mutants of PTP1B, FAP-1, PTPH-1, and HePTP are all inhibited more potently than the “naturally sensitized” wild-type Shp2 catalytic domain, suggesting that biarsenicals could be used to selectively target suitably sensitized PTPs even in mammalian cells that endogenously express Shp2.
3.3.4. The unique case of Shp1
Puzzlingly, among the classical PTPs investigated, Shp1 proved to be singularly refractory to allosteric-site sensitization. In experiments utilizing both the purified Shp1 catalytic domain and Shp1-expressing lysates, mutant Shp1 demonstrated only a 15–20% greater reduction in activity, as compared to wild-type Shp1 (Figure 9A, B). Given that Shp1’s closest homolog, Shp2 (59% PTP-domain identity), provided the initial precedent for the sensitization strategy described here, the Shp1 results are particularly perplexing. Even if one takes a more nuanced structural view, it is difficult to rationalize Shp1’s recalcitrance to sensitization: a superimposition of PTP crystal structures reveals the highly conserved nature of PTP motifs 4 and 7—the structural elements containing the cysteines that comprise the FlAsH-binding site—among the PTPs investigated in this report,30, 38–41 and, in particular, these motifs in Shp2 and Shp1 are essentially indistinguishable in three-dimensional space (Figure 10, compare red and magenta). Although the weak sensitivity of Shp1 is difficult to explain, it is consistent with the previous observation that wild-type Shp1 is less sensitive to inhibition by FlAsH than wild-type Shp2,24 even though both PTP domains contain naturally occurring dicysteine sites (Figure 1B). For reasons that remain to be elucidated, the Shp1 catalytic domain appears to possess an unusual inherent resistance to inhibition by biarsenicals, a phenomenon that is consistently observed in both its dicysteine (wild-type) and tricysteine (mutant) variants.
Figure 10.
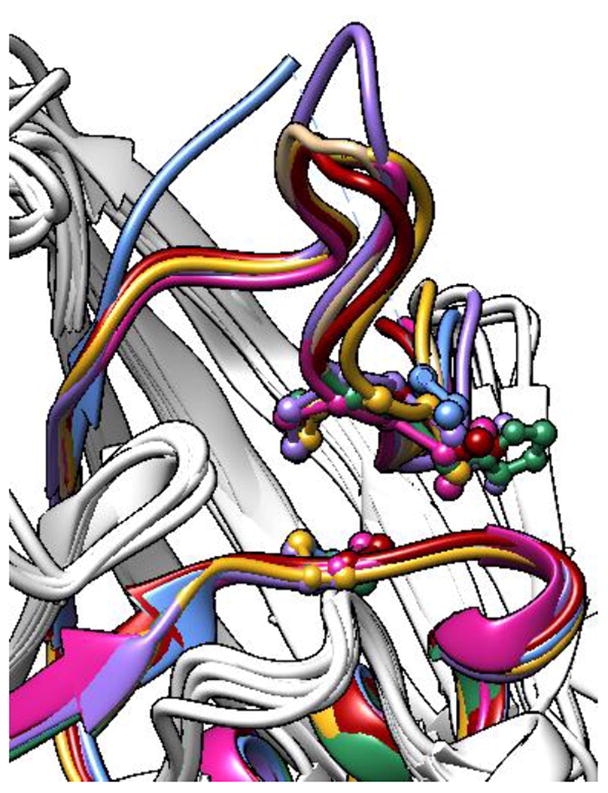
Structural conservation of PTP motifs 4 and 7. The PTP-domain structures of Shp2 (red, PDB ID: 3B7O),30 PTPH-1 (sea green, PDB ID: 2B49),30 FAP-1 (violet, PDB ID: 1WCH),38 Shp1 (magenta, PDB ID: 1GWZ),39 HePTP (blue, PDB ID: 2A3K),40 and PTP1B (gold, PDB ID: 1SUG)41 were aligned and superimposed using the UCSF Chimera package.43 For clarity, only the ribbons corresponding to residues 327–373 (human Shp2 numbering) are colored.
4. Conclusions
Drawing on the unique, naturally occurring allosteric site of Shp2 as an inspiration for an inhibitor-sensitization strategy, we have used rational design to install allosteric-inhibition sites in classical PTP domains. With PTP1B as a prototype, we found that unnatural sensitivity to inhibition by biarsenical compounds can be introduced by a single cysteine substitution at the amino-acid position equivalent to Shp2’s cysteine 333, a non-active-site amino-acid position that is rarely occupied by cysteine in wild-type PTP domains. In a sensitized PTP domain, the engineered cysteine residue at position 333 couples with a highly conserved, naturally occurring cysteine residue to constitute a dicysteine allosteric-inhibition site, akin to that of wild-type Shp2. To further strengthen the inhibitor/engineered allosteric-site interaction, we expanded the biarsenical-binding site by adding a third, highly solvent-accessible cysteine residue to generate a tricysteine allosteric site that does not exist in any wild-type PTP domain and that can be targeted with higher potency than Shp2’s dicysteine motif. The sensitizing positions for generating both dicysteine and tricysteine allosteric sites are readily identifiable from primary-sequence alignments for any classical PTP, and we have demonstrated the scope of the approach by generating four highly sensitized PTPs from four distinct subfamilies. In sum, the present study provides a general method for the installation of allosteric-inhibition sites in classical PTPs, utilizing a minimal amount of protein engineering that avoids alterations to the PTP active site. Our findings therefore provide a novel tool for the specific small-molecule control of PTP activity and may facilitate the chemical-genetic analysis of PTP function in engineered cells and model organisms.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R15GM071388. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors also gratefully acknowledge the Henry Dreyfus Teacher-Scholar Awards Program (A.C.B.) and Amherst College for funding. LC/MS/MS experiments were carried out by the University of Massachusetts Medical School’s Proteomics & Mass Spectrometry Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Mol Cell Biol. 2001;21:7117. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hendriks W, Pulido R. Biochim Biophys Acta. 2013;1832:1673. doi: 10.1016/j.bbadis.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 3.Hardy S, Julien SG, Tremblay ML. Anticancer Agents Med Chem. 2012;12:4. doi: 10.2174/187152012798764741. [DOI] [PubMed] [Google Scholar]
- 4.Blaskovich MA. Curr Med Chem. 2009;16:2095. doi: 10.2174/092986709788612693. [DOI] [PubMed] [Google Scholar]
- 5.Tiganis T, Bennett AM. Biochem J. 2007;402:1. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop A, Buzko O, Heyeck-Dumas S, Jung I, Kraybill B, Liu Y, Shah K, Ulrich S, Witucki L, Yang F, Zhang C, Shokat KM. Annu Rev Biophys Biomol Struct. 2000;29:577. doi: 10.1146/annurev.biophys.29.1.577. [DOI] [PubMed] [Google Scholar]
- 7.Shokat K, Velleca M. Drug Discov Today. 2002;7:872. doi: 10.1016/s1359-6446(02)02391-7. [DOI] [PubMed] [Google Scholar]
- 8.Alaimo P, Knight ZA, Shokat KM. Bioorg Med Chem. 2005;13:2825. doi: 10.1016/j.bmc.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Lin Q, Jiang F, Schultz PG, Gray NS. J Am Chem Soc. 2001;123:11608. doi: 10.1021/ja011423j. [DOI] [PubMed] [Google Scholar]
- 10.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. Nature. 2000;407:395. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 11.Bishop AC, Buzko O, Shokat KM. Trends Cell Biol. 2001;11:167. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 12.Baud MG, Lin-Shiao E, Cardote T, Tallant C, Pschibul A, Chan KH, Zengerle M, Garcia JR, Kwan TT, Ferguson FM, Ciulli A. Science. 2014;346:638. doi: 10.1126/science.1249830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman HE, Blair ER, Johndrow JE, Bishop AC. J Am Chem Soc. 2005;127:2824. doi: 10.1021/ja043378w. [DOI] [PubMed] [Google Scholar]
- 14.Bishop AC, Zhang XY, Lone AM. Methods. 2007;42:278. doi: 10.1016/j.ymeth.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang XY, Bishop AC. J Am Chem Soc. 2007;129:3812. doi: 10.1021/ja069098t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XY, Chen VL, Rosen MS, Blair ER, Lone AM, Bishop AC. Bioorg Med Chem. 2008;16:8090. doi: 10.1016/j.bmc.2008.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XY, Bishop AC. Biochemistry. 2008;47:4491. doi: 10.1021/bi800014c. [DOI] [PubMed] [Google Scholar]
- 18.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 19.Khajehpour M, Wu L, Liu S, Zhadin N, Zhang ZY, Callender R. Biochemistry. 2007;46:4370. doi: 10.1021/bi602335x. [DOI] [PubMed] [Google Scholar]
- 20.Kamerlin SC, Rucker R, Boresch S. Biochem Biophys Res Commun. 2007;356:1011. doi: 10.1016/j.bbrc.2007.03.093. [DOI] [PubMed] [Google Scholar]
- 21.Davis OB, Bishop AC. Bioconj Chem. 2012;23:272. doi: 10.1021/bc200562y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walton ZE, Bishop AC. Bioorg Med Chem. 2010;18:4884. doi: 10.1016/j.bmc.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang ZY, Wang Y, Dixon JE. Proc Natl Acad Sci USA. 1994;91:1624. doi: 10.1073/pnas.91.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chio CM, Lim CS, Bishop AC. Biochemistry. doi: 10.1021/bi5013595. [DOI] [Google Scholar]
- 25.Adams SR, Tsien RY. Nat Protoc. 2008;3:1527. doi: 10.1038/nprot.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blair ER, Hoffman HE, Bishop AC. Bioorg Med Chem. 2006;14:464. doi: 10.1016/j.bmc.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 27.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. J Am Chem Soc. 2002;124:6063. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 28.Krishnan B, Gierasch LM. Chem Biol. 2008;15:1104. doi: 10.1016/j.chembiol.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luedtke NW, Dexter RJ, Fried DB, Schepartz A. Nat Chem Biol. 2007;3:779. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barr AJ, Ugochukwu E, Lee WH, King ONF, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Muller S, Knapp S. Cell. 2009;136:352. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Cell. 1998;92:441. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 32.Chen CY, Willard D, Rudolph J. Biochemistry. 2009;48:1399. doi: 10.1021/bi801973z. [DOI] [PubMed] [Google Scholar]
- 33.Bishop AC, Blair ER. Bioorg Med Chem Lett. 2006;16:4002. doi: 10.1016/j.bmcl.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 34.Wiesmann C, Barr KJ, Kung J, Zhu J, Erlanson DA, Shen W, Fahr BJ, Zhong M, Taylor L, Randal M, McDowell RS, Hansen SK. Nat Struct Mol Biol. 2004;11:730. doi: 10.1038/nsmb803. [DOI] [PubMed] [Google Scholar]
- 35.Hansen SK, Cancilla MT, Shiau TP, Kung J, Chen T, Erlanson DA. Biochemistry. 2005;44:7704. doi: 10.1021/bi047417s. [DOI] [PubMed] [Google Scholar]
- 36.Zhang S, Zhang ZY. Drug Discov Today. 2007;12:373. doi: 10.1016/j.drudis.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 37.Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, Jensen MR, Gauss CM, Page R, Blackledge M, Muthuswamy SK, Peti W, Tonks NK. Nat Chem Biol. 2014;10:558. doi: 10.1038/nchembio.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villa F, Deak M, Bloomberg GB, Alessi DR, van Aalten DM. J Biol Chem. 2005:8180. doi: 10.1074/jbc.M412211200. [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Liang XS, Niu TQ, Meng WY, Zhao ZZ, Zhou GW. J Biol Chem. 1998;273:28199. doi: 10.1074/jbc.273.43.28199. [DOI] [PubMed] [Google Scholar]
- 40.Eswaran J, Debreczeni JE, Longman E, Barr AJ, Knapp S. Protein Science. 2006;15:1500. doi: 10.1110/ps.062128706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen AK, Peters GH, Moller KB, Iversen LF, Kastrup JS. Acta Cryst D. 2004;60:1527. doi: 10.1107/S0907444904015094. [DOI] [PubMed] [Google Scholar]
- 42.Tonks NK, Neel BG. Curr Opin Cell Biol. 2001;13:182. doi: 10.1016/s0955-0674(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.