Abstract
Background:
Hepatitis C virus infection is one of the leading causes of end stage liver diseases. The innate immune response slows down viral replication by activating cytokines such as type I interferon (IFN-α/β), which trigger the synthesis of antiviral proteins and modulate the adaptive immune system. Recently, leucine-rich repeat (in Flightless I) interacting protein-1 (LRRFIP1) was reported contributing to the production of interferon-β in macrophages.
Objectives:
The aim of this study was to assess the role of LRRFIP1 in induction of IFN-β and inhibition of HCV infection in hepatocytes.
Materials and Methods:
Induction of IFN-β by LRRFIP1 in Huh7 and Huh7.5.1 was determined by real-time PCR and western blotting in vitro. Inhibition of HCV replication by LRRFIP1 overexpression in hepatocytes was assessed.
Results:
LRRFIP1 increased the expression of IFN-β in hepatocytes with or without HCV infection. Induction of IFN-β by LRRFIP1 was enhanced with the presence of hepatitis C virus. Overexpression of LRRFIP1 in hepatocytes inhibited HCV replication. However, HCV infection did not regulate intracellular expression of LRRFIP1.
Conclusions:
LRRFIP1 and its mediated production of type I interferon play a role in controlling HCV infection. The findings of this study provide new target for HCV treatment and contribute to development of anti-HCV drugs.
Keywords: LRRFIP1 Protein, Human; Hepacivirus; Interferon Type I
1. Background
Hepatitis C Virus (HCV) is an enveloped and single-stranded RNA virus, which belongs to the Hepacivirus genus. HCV infection mainly affects liver tissues and most infected people have a high risk to develop chronic liver disease, such as liver cirrhosis and hepatocellular carcinoma (HCC) (1). No vaccine is yet available to prevent HCV transmission. Several Direct Acting Antivirals (DAAs) drugs have been approved for HCV treatment (2, 3). However, pegylated interferon with ribavirin (PegIFN/RBV) is still the standard of care against HCV infection in most countries (4).
Studies showed that about 10 - 15% of HCV infected patients recovered without receiving antiviral therapy. Gunduz F et al. reported that early responses of innate immunity such as induction of Type I interferons (IFNs) played a pivotal role in this process (5). Type I IFNs act their antiviral effect by activating transcription of numerous IFN stimulated genes (ISGs), such as ADAR (RNA- specific adenosine deaminase) (6), P56 (7), OAS (oligoadenylate synthetases) (8) and PKR (Protein Kinase R) (9), which have antiviral properties against HCV. Virtually, all human cells are able to synthesize IFN-α/β in response to virus infection, while HCV primarily infects liver cells. Therefore, induction of IFNs in liver cells is especially important for controlling HCV infection.
Recent studies showed that LRRFIP1 might act as a candidate that contributes to the production of IFN-β. LRRFIP1, with a full name of leucine-rich repeat (in Flightless I) interacting protein-1, is originally identified as a protein that plays a role during Drosophila embryogenesis and myogenesis (10, 11). The C-terminus of LRRFIP1 has been described as having nucleic acid-binding activity, including the transactivating response region (TAR) hairpin of HIV (12, 13). Asen Bagashev reported that LRRFIP1 induced type I IFNs in 3T3 cells in the presence of influenza virus (14). Moreover, Yang et al. showed that LRRFIP1 contributed to the production of IFN-β in macrophages induced by VSV (vesicular stomatitis virus) and Listeria monocytogenes (15).
Several studies clarified that HCV RNA is a potent trigger of IFN induction in hepatocytes, leading to establishment of an antiviral status (16). It is not clear whether LRRFIP1 can induce innate immunity in hepatocytes and plays a role in controlling HCV infection.
2. Objectives
This study was performed to investigate the ability of LRRFIP1 to regulate type I IFNs expression in hepatocytes.
3. Materials and Methods
3.1. Plasmids and Cells
LRRFIP1 was cloned from Huh7 cells cDNA by over-lapped polymerase chain reaction (PCR) using primers (Life Technologies, Shanghai, China) as follows (5’ - 3’): GCGAGCTAGCATGACCAGCCCCGCGGCCGCTCAAAG, CAGAACTCCCCTTGAGCCATCCAACTGTTG, CAACAGTTGGATGGCTCAAGGGGAGTT CTG and GGCAGTCTAGATTAGGACATGGTACAGTCTTCTTTG. PCR product was sequenced and submitted to GenBank with an accession number KP455483, which had 100% similarity with Homo sapiens leucine rich repeat (in FLII) interacting protein 1 (LRRFIP1) transcript variant 5 (NM_001137553.1). LRRFIP1 cDNA was cloned into the pcDNA3.1 vector (Promega, San Luis Obispo, CA, USA) using restriction enzyme sites of NheI (upstream) and XbaI (downstream).
Hepatoma cells Huh7 and Huh7.5.1 were cultured in DMEM (Dulbecco’s modified Eagle medium) supplemented with 10% fetal bovine serum, 1% nonessential amino acids and 1% penicillin/streptomycin (Gibco BRL, Grand Island, NY, USA). Transfection was performed using lipofectamin 2000 (Invitrogen, Carlsbad, CA, USA).
3.2. Antibodies
Antibodies used were mouse anti-LRRFIP1 monoclonal antibody (mAb) (Abcam, Cambridge, MA, USA), rabbit anti-IFN beta mAb (Abcam), mouse anti-HCV NS5A mAb (Virogen, Watertown, MA, USA) and mouse anti-GAPDH mAb (Cell Signaling Technology, Danvers, MA, USA). Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG and goat anti-rabbit IgG were purchased from Invitrogen (Carlsbad, CA, USA).
3.3. Immunofluorescence Assay
Huh7 cells were cultured in 96-well plates and fixed for 20 minutes in ice-cold methanol. Cells were then washed with PBS and incubated with PBS containing 3% bovine serum albumin (BSA) for two hours at room temperature. Cells were then incubated with mouse anti-LRRFIP1 mAb (1:1000) at room temperature. Then cells were washed again and incubated with Alexa Fluor 488 conjugated goat anti-mouse IgG (Invitrogen, Carlsbad, CA, USA). At last, 4, 6-diamidino-2-phenylindole (DAPI) was used to stain cell nucleus. Cells were finally viewed using Immunofluorescence microscope (Olympus, Japan).
3.4. Cell Proliferation Assay
Huh7 cells were cultured in 24-well plates and transfected with plasmids pcDNA-LRRFIP1 or mock vector. 48 hours after transfection, cells were allowed to grow in 96-well plates (3000 cells/well) and cultured for 24 hours. Cell proliferation was detected using Cell Counting Kit (Transgen, China) at 12 or 24 hours intervals according to the manufacturer’s instruction.
3.5. Cell-Culture-Derived HCV (HCVcc)
HCVcc was prepared as previously described (17).
3.6. Real-Time PCR
Total cellular RNA was extracted from cultured cells with TRIzol reagent (Invitrogen), which were reversely transcribed to cDNA using cDNA synthesis kit (Promega, San Luis Obispo, CA). SYBR Premix ExTaq2 (Promega, San Luis Obispo, CA) was used for real-time PCR assay in a StepOnePlus real-time PCR system (Applied Biosystems, CA, USA). Specific primers used for real-time PCR were as follows (5’ - 3’): CTTCACGCAGAAAGCGTCTA and CAAGCACCCTATCAGGCA GT for HCV; CCAATGGAGAGACTTCCGACA and GTCATCTTGGTAGGACCTTGGT for LRRFIP1; GCTTGGATTCCTACAAAGAAGCA and ATAGATGGTCAATGCGGCGTC for IFN beta; TGGGCTACACTGAGCACCAG and AAGTGGTCGTTGAGGGCAAT for GAPDH. Data was normalized to GAPDH expression in each sample.
3.7. SDS-PAGE and Western Blotting
Cells were lysed with Radio Immunoprecipitation Assay Reagent (RIPA) (Beyotime, Jiangsu, China) containing protease and phosphatase inhibitors (Sigma-Aldrich, MO, USA). Protein concentrations in the extracts were measured by BCA Protein Assay Kit (Beyotime, China). Equal amounts of cell lysates were separated on SDS-PAGE gels (12.5%) and then transferred onto PVDF membranes (Bio-Rad, CA, USA). Membranes were blocked with PBS containing 5% nonfat dried milk and 0.1% Tween 20 and incubated at 4°C for 16 hours with monoclonal antibodies to LRRFIP1, IFN-β, HCV NS5A and GAPDH, followed by incubation with goat anti-mouse or goat anti-rabbit IgG conjugated with horseradish peroxidase (Invitrogen). Proteins were visualized by the reaction with a chemiluminescent substrate (Thermo Fisher Scientific, Beijing, China).
3.8. Statistical Analyses
The results were analyzed using SPSS Statistics 18.0 program (IBM, New York, USA). Comparisons between groups were performed using One-way ANOVA and Student’s t-test. P values ˂ 0.05 were considered statistically significant.
4. Results
4.1. LRRFIP1 Promotes IFN-β Expression in Hepatocytes
It has been documented that LRRFIP1 contributed to the production of IFN-β in macrophages and fibroblast 3T3 cells (14, 15). Therefore, to elucidate the effect of LRRFIP1 on induction of IFN-β in hepatocytes, LRRFIP1 was cloned as described in methods section. Hepatoma cells Huh7 were transfected with pcDNA-LRRFIP1 and mock vector. LRRFIP1 was over expressed in pcDNA-LRRFIP1 transfected cells (Figure 1 A). We then determined the effect of overexpressed LRRFIP1 on cell proliferation. As shown in Figure 1 B, overexpression of LRRFIP1 had no impact on Huh7 cell proliferation.
Figure 1. Effect of LRRFIP1 on Cell Proliferation.
A, Overexpression of LRRFIP1 in Huh7 cells was detected by immunofluorescence assay. B, LRRFIP1 overexpression had no impact on Huh7 cell proliferation.
Intracellular IFN-β was detected at 48 hours after transfection in Huh7 and Huh7.5.1 cells transfected with pcDNA-LRRFIP1. Results are shown in Figure 2. The background transcriptional level of LRRFIP1 in Huh7 is a little higher than that in Huh7.5.1 cells. The mRNA levels of LRRFIP1 were extremely increased in pcDNA-LRRFIP1 transfected Huh7 or Huh7.5.1 cells compared to that in controls (Figure 2 A). Overexpression of LRRFIP1 upregulated the mRNA levels of IFN-β both in Huh7 and Huh7.5.1 cells (Figure 2 B). Increased induction of IFN-β in LRRFIP1 transfected Huh7 and Huh7.5.1 cells were also observed by western blotting (Figure 2 C). These data indicate that LRRFIP1 can drive a type I IFNs response in hepatocytes independently of virus.
Figure 2. LRRFIP1 Upregulated the Expression of IFN-β in Huh7 and Huh7.5.1 Cells.
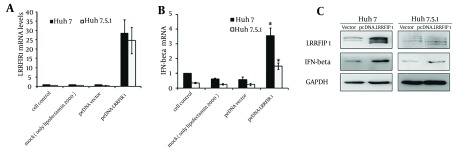
The mRNA levels of LRRFIP1 (A) and IFN-β (B) were determined by real-time PCR at 24 hours after transfection. Data showed means and standard errors of three replicate assays (* P < 0.05, compared with vector). C, Protein levels of LRRFIP1 and IFN-β were quantified by western blotting analyses at 48 hours after transfection.
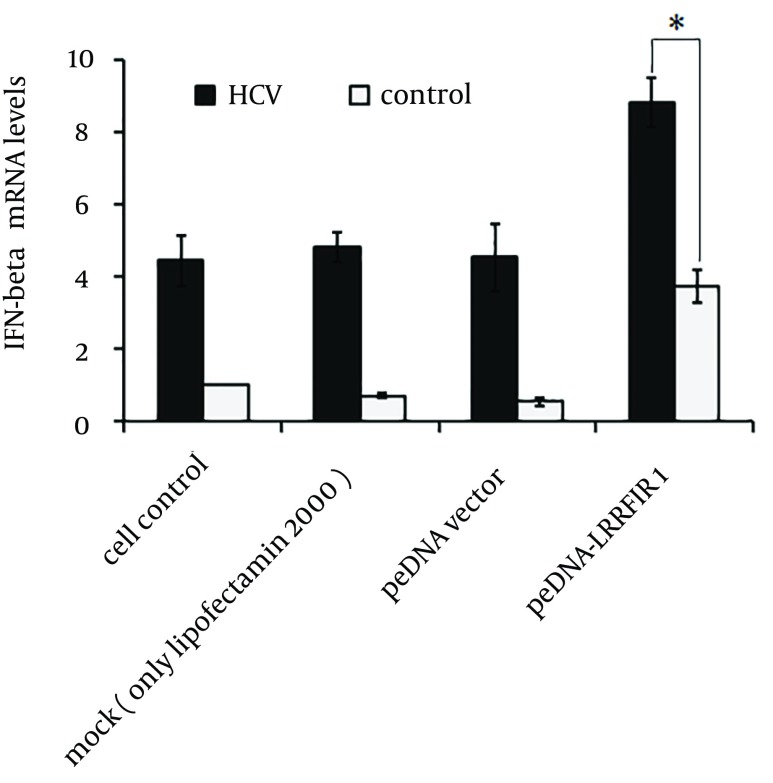
We next analyzed the induction of IFN-β by LRRFIP1 in the presence of HCV. Huh7 cells were infected with HCVcc at 0.5 multiplicity of infection (MOI) for 12 hours and cells were then transfected with pcDNA-LRRFIP1 or mock vector. mRNA levels of IFN-β, shown in Figure 3, were determined by real time PCR at 48 hours after transfection. The expression of IFN-β was remarkably increased in LRRFIP1 over-expressed cells with HCV infection suggesting that the induction of IFN-β by LRRFIP1 was enhanced by HCV infection.
Figure 3. Induction of IFN-β by LRRFIP1 was Enhanced by HCV Infection.
Huh7 cells were infected with 0.5 MOI of HCVcc. Twelve hours later, cells were transfected with pcDNA-LRRFIP1 or mock vector. mRNA levels of IFN-β were determined by real-time PCR at 48 hours after transfection. Data are shown by means and standard errors of three replicate assays (* P < 0.05, compared with control).
4.2. Overexpression of LRRFIP1 in Hepatocytes Inhibits HCV Replication
IFN-β activates the expression of more than 300 IFN-stimulated genes (ISGs) to execute its antiviral functions. We found that LRRFIP1 promoted the induction of IFN-β in hepatocytes. To evaluate the anti-HCV activity of LRRFIP1 in hepatocytes, LRRFIP1 was transfected into Huh7 cells. Twenty-four hours later, the cells were infected with HCVcc at MOI (multiplicity of infection) of 0.5 and 1. IFN-β and HCV non-structural protein NS5A were determined through western blotting at 48 hours after infection. Results are shown in Figure 4 A. IFN-β was upregulated in LRRFIP1 over-expressed cells with or without HCV infection. Notably, NS5A was decreased in LRRFIP1 over-expressed cells both in 0.5 MOI and 1 MOI of HCVcc infected cells, which indicates that overexpression of LRRFIP1 inhibits HCV replication in hepatocytes.
Figure 4. LRRFIP1 Inhibited HCV Infection in Huh7 Cells.
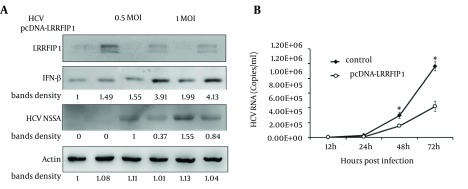
A, Huh7 cells were transfected with LRRFIP1-pcDNA or mock vector. Twenty-four hours after transfection, cells were infected with HCVcc. Protein levels of LRRFIP1, IFN-β and HCV NS5A were determined by western blotting. B, HCV RNA levels were determined by real-time PCR in Huh7 cells transfected with pcDNA-LRRFIP1 or vector control. Data were represented as Means ± SD, n = 3. (* P < 0.05, compared with control).
To further evaluate anti-HCV activity of LRRFIP1, Huh7 cells cultured in 24-well plates were transfected with pcDNA-LRRFIP1. Twenty-four hours after transfection, cells were infected with HCVcc at a MOI of 0.5. HCV RNA levels were determined at 12, 24, 48 and 72 hours post infection. As shown in Figure 4 B, HCV RNA levels were lower in LRRFIP1 overexpressed cells than those in vector control cells at each time points. These data suggest that LRRFIP1 can inhibit HCV replication in hepatocytes.
4.3. HCV Infection Had Limited Effect on LRRFIP1 Expression in Hepatocytes
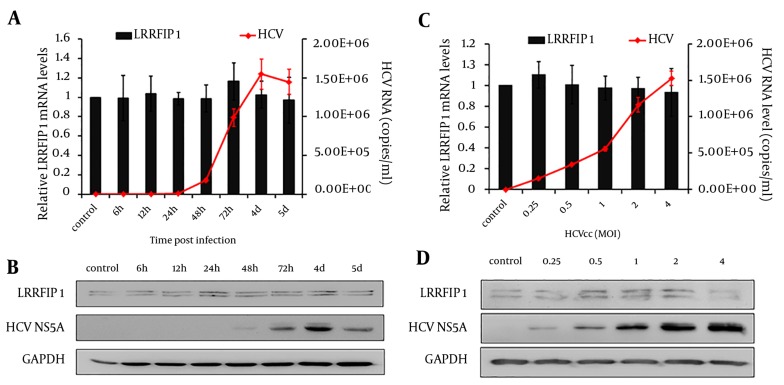
The anti-HCV activity of LRRFIP1 has been identified above. Therefore, we wish to further define whether HCV infection regulates the expression of LRRFIP1 in hepatocytes. Huh7 cells were infected with 0.5 MOI of HCVcc and LRRFIP1 levels were determined at different time points post infection by real-time PCR and western blotting.
As shown in Figure 5A and 5B, HCV replicated in Huh7 cells and had a peak value at 4 days post infection, while the mRNA and protein levels of LRRFIP1 were similar before and after HCV infection. To further explore the effect of HCV infection on LRRFIP1 expression, Huh7 cells were infected with different doses of HCVcc. Seventy-two hours later, the intracellular mRNA and protein levels of LRRFIP1 were determined. Similarly, no change expression of LRFFIP1 was observed in HCV infected Huh7 cells (Figure 5 C and 5D). Similar results were also observed in Huh7.5.1 cells (data not shown). These results suggest that HCV infection had limited effect on LRRFIP1 expression in hepatocytes.
Figure 5. Effect of HCV Infection on Expression of LRRFIP1 in Huh7 Cells.
mRNA levels (A) and protein levels (B) of LRRFIP1 and HCV were detected at different time points post infection. mRNA levels (C) and protein levels (D) of LRRFIP1 and HCV were detected in Huh7 cells infected with different doses of HCVcc. Data were presented by Means ± SD, n = 3.
5. Discussion
Type I interferon (IFN-α/β) is induced in cells immediately after virus infection; therefore, is the first line of defense that inhibits virus replication and infection (18). The signaling pathways of virus-triggered type I IFNs have been clarified by a large number of researches. It is well known that cytoplasmic helicase proteins such as RIG-I and MDA5 sense the presence of viruses and activate adaptor proteins, which mediates the signal to downstream factors and finally inducing the expression of type I IFNs (19, 20).
Despite the advances of findings in signaling pathways of virus-triggered type I IFNs, new mechanisms of virus induction of IFNs have been discovered. LRRFIP1 is recently found as cytosolic nucleic acid-binding protein, which bound exogenous nucleic acids and increased the expression of IFN-β in macrophages and fibroblasts cells (14, 15). LRRFIP1 is primarily found as a protein, which interacts with leucine rich repeat of flightless I. Suriano et al. reported that LRRFIP1 is able to suppress the expression of tumor necrosis factor α (TNF-α) (21). LRRFIP1, localized mainly in the cytoplasm, is able to directly bind to viral dsRNA and induce the expression of type I IFNs (12, 21). Therefore, LRRFIP1 is a potent activator of innate immune responses in mammalian cells that is important for pathogen elimination.
In this study, we showed that LRRFIP1 contributed to HCV-induced production of IFN-β in hepatocytes. Overexpression of LRRFIP1 upregulated the mRNA and protein levels of IFN-β in Huh7 and Huh7.5.1 cells. Moreover, induction of IFN-β by LRRFIP1 is independent of HCV infection.
Previous studies clarified the signaling chain activated by viral RNA in infected cells. Several studies showed that two intracellular RNA helicases, RIG-I and MDA5, act as sentinels for viral RNA (19, 20). Then several adaptor proteins such as Cardif (CARD adaptor inducing IFN-β) and IPS-1 (interferon-β-promoter stimulator 1) were activated and mediated the signal to two IκB kinase (IKK)-related kinases, which then phosphorylate the transcription factor IRF-3 (22). Phosphorylated IRF-3 moves into the nucleus and initiates transcription of genes encoding type I interferons (23).
However, the exact molecular pathway of LRRFIP1 induced IFN-β is still not clear. Studies showed that LRRFIP1 bound and activated β-catenin. The activated β-catenin subsequently bound IRF3, which resulted in the transcription of type I interferons as the regular pathway (15). While Bagashev et al. found that LRRFIP1 sensed early endosomes containing RNA of influenza virus in fibroblasts 3T3 cells. It is possible that LRRFIP1 serves as a bridge for the TLR3 signal pathway (14). The mitogen-activated protein kinases p38 appears to actively regulate LRRFIP1 recruitment to the endosome in this process. However, knockdown of LRRFIP1 did not affect the phosphorylation of p38 (15). Therefore, the exact pathway of LRRFIP1 inducing type I interferons needs further researches.
We identified that LRRFIP1 can induce IFN-β in hepatocytes with or without HCV infection. Induction of IFN-β was enhanced by the presence of HCV. In addition, induction of IFN-β was observed both in Huh7 and Huh7.5.1 cells. The clone Huh7.5.1 of human hepatoma cells contains a mutation in RIG-I, while Huh7 cell has intact RIG-I expression. Therefore, Huh7 cells respond to HCV and induce the transcription of IFNs, while Huh7.5.1 cells induce extremely low levels of IFNs due to containing a mutation in RIG-I. Induction of IFN-β by overexpression of LRRFIP1 was identified both in Huh7 and Huh7.5.1 cells, which indicates that RIG-I mutation does not affect the induction of IFN-β by LRRFIP1. Thus, induction of IFN-β by LRRFIP1 is probably not through RIG-I related pathway.
In addition, Nguyen JB and Modis Y reported a crystal structure of the dimeric coiled-coil domain of LRRFIP1 (13). LRRFIP1 contains an N-terminal domain of unknown function (aa 1 - 162), a conserved 87-amino acid domain predicted to be a coiled coil (aa 163-249) and a nucleic acid binding domain (aa 250 - 808). C-terminal DNA-binding domain (LRRFIP1-DBD, aa 250 - 808) alone is mostly unfolded and has very little secondary structure. In the context of the coiled coil domain, LRRFIP1-CC-DBD (aa 163 - 808) was also unfolded, but was mostly oligomeric. Although LRRFIP1-DBD can bind with DNA, it still lacks secondary structure, while LRRFIP1-CC-DBD can assemble into higher order oligomers in the context of binding with DNA. Moreover, the coiled coil domain is found in all LRRFIP isoform genes and is highly conserved, which indicates its important role. Thus it is interesting to address whether LRRFIP1-DBD or in the context of the coiled coil domain can induce the transcription of Type 1 IFNs. This would be detected in further studies.
In summary, our findings revealed a role for LRRFIP1 in controlling HCV infection. We identified that LRRFIP1 implicated in the innate responses to HCV in liver cells. Although HCV infection did not regulate the expression of LRRFIP1, over-expressed LRRFIP1 in Huh7 and Huh7.5.1 cells inhibit HCV replication. It is an alternative method for antiviral management to mediate the expression of LRRFIP1, which provides new target for HCV treatment and contributes to the development of anti-HCV drugs. Further researches are needed to clarify delicate regulatory mechanisms of LRRFIP1 in inducing type I interferons and its role of antiviral responses. Moreover, factors that mediate the expression of LRRFIP1 should be investigated.
Footnotes
Authors’ Contributions:Yuan Liu: Designed the study and wrote the paper. Ziying Zou, Bing Zhu, Zonghai Hu, Ping Zeng and Lijuan Wu: Contributed in experimental work. Yuan Liu and Lijuan Wu: Analyzed the data.
Funding/Support:This study was supported by the Research Fund of General Hospital of Chengdu Military Region (No. 2013YG-B055) and the National Natural Science Foundation of China (NSFC) (No. 81301445).
References
- 1.Jacobson IM, Davis GL, El-Serag H, Negro F, Trepo C. Prevalence and challenges of liver diseases in patients with chronic hepatitis C virus infection. Clin Gastroenterol Hepatol. 2010;8(11):924–33. doi: 10.1016/j.cgh.2010.06.032. [DOI] [PubMed] [Google Scholar]
- 2.Kayali Z, Schmidt WN. Finally sofosbuvir: an oral anti-HCV drug with wide performance capability. Pharmgenomics Pers Med. 2014;7:387–98. doi: 10.2147/PGPM.S52629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poveda E, Wyles DL, Mena A, Pedreira JD, Castro-Iglesias A, Cachay E. Update on hepatitis C virus resistance to direct-acting antiviral agents. Antiviral Res. 2014;108:181–91. doi: 10.1016/j.antiviral.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Muir AJ. The rapid evolution of treatment strategies for hepatitis C. Am J Gastroenterol. 2014;109(5):628–35. doi: 10.1038/ajg.2014.66. [DOI] [PubMed] [Google Scholar]
- 5.Gunduz F, Mallikarjun C, Balart LA, Dash S. Interferon alpha induced intrahepatic pSTAT1 inversely correlate with serum HCV RNA levels in chronic HCV infection. Exp Mol Pathol. 2014;96(1):36–41. doi: 10.1016/j.yexmp.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol. 2005;79(10):6291–8. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C, Pflugheber J, Sumpter RJ, Sodora DL, Hui D, Sen GC, et al. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77(7):3898–912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JT, Sohn JA, Zhu Q, Seeger C. Mechanism of the interferon alpha response against hepatitis C virus replicons. Virology. 2004;325(1):71–81. doi: 10.1016/j.virol.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 9.Pflugheber J, Fredericksen B, Sumpter RJ, Wang C, Ware F, Sodora DL, et al. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc Natl Acad Sci U S A. 2002;99(7):4650–5. doi: 10.1073/pnas.062055699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu YT, Yin HL. Identification of the binding partners for flightless I, A novel protein bridging the leucine-rich repeat and the gelsolin superfamilies. J Biol Chem. 1998;273(14):7920–7. doi: 10.1074/jbc.273.14.7920. [DOI] [PubMed] [Google Scholar]
- 11.Choe N, Kwon JS, Kim JR, Eom GH, Kim Y, Nam KI, et al. The microRNA miR-132 targets Lrrfip1 to block vascular smooth muscle cell proliferation and neointimal hyperplasia. Atherosclerosis. 2013;229(2):348–55. doi: 10.1016/j.atherosclerosis.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Wilson SA, Brown EC, Kingsman AJ, Kingsman SM. TRIP: a novel double stranded RNA binding protein which interacts with the leucine rich repeat of flightless I. Nucleic Acids Res. 1998;26(15):3460–7. doi: 10.1093/nar/26.15.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen JB, Modis Y. Crystal structure of the dimeric coiled-coil domain of the cytosolic nucleic acid sensor LRRFIP1. J Struct Biol. 2013;181(1):82–8. doi: 10.1016/j.jsb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagashev A, Fitzgerald MC, Larosa DF, Rose PP, Cherry S, Johnson AC, et al. Leucine-rich repeat (in Flightless I) interacting protein-1 regulates a rapid type I interferon response. J Interferon Cytokine Res. 2010;30(11):843–52. doi: 10.1089/jir.2010.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11(6):487–94. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 16.Sumpter RJ, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79(5):2689–99. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Zou Z, Zhu B, Hu Z, Zeng P. CXCL10 decreases GP73 expression in hepatoma cells at the early stage of hepatitis C virus (HCV) infection. Int J Mol Sci. 2013;14(12):24230–41. doi: 10.3390/ijms141224230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perales C, Beach NM, Sheldon J, Domingo E. Molecular basis of interferon resistance in hepatitis C virus. Curr Opin Virol. 2014;8:38–44. doi: 10.1016/j.coviro.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 20.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A. 2004;101(49):17264–9. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suriano AR, Sanford AN, Kim N, Oh M, Kennedy S, Henderson MJ, et al. GCF2/LRRFIP1 represses tumor necrosis factor alpha expression. Mol Cell Biol. 2005;25(20):9073–81. doi: 10.1128/MCB.25.20.9073-9081.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4(5):491–6. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 23.Weber F. Interaction of hepatitis C virus with the type I interferon system. World J Gastroenterol. 2007;13(36):4818–23. doi: 10.3748/wjg.v13.i36.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]