Abstract
The gut microbiota plays a critical role in maintaining physiological homeostasis. This study was designed to evaluate whether gut microbial composition affects hypertension. 16S rRNA genes obtained from cecal samples of Dahl salt-sensitive (S) and Dahl salt-resistant (R) rats were sequenced. Bacteria of the phylum Bacteroidetes were higher in the S rats compared with the R rats. Furthermore, the family S24-7 of the phylum Bacteroidetes and the family Veillonellaceae of the phylum Firmicutes were higher in the S rats compared with the R rats. Analyses of the various phylogenetic groups of cecal microbiota revealed significant differences between S and R rats. Both strains were maintained on a high-salt diet, administered antibiotics for ablation of microbiota, transplanted with S or R rat cecal contents, and monitored for blood pressure (BP). Systolic BP of the R rats remained unaltered irrespective of S or R rat cecal transplantation. Surprisingly, compared with the S rats given S rat cecal content, systolic BP of the S rats given a single bolus of cecal content from R rats was consistently and significantly elevated during the rest of their life, and they had a shorter lifespan. A lower level of fecal bacteria of the family Veillonellaceae and increased plasma acetate and heptanoate were features associated with the increased BP observed in the S rats given R rat microbiota compared with the S rats given S rat microbiota. These data demonstrate a link between microbial content and BP regulation and, because the S and R rats differ in their genomic composition, provide the necessary basis to further examine the relationship between the host genome and microbiome in the context of BP regulation in the Dahl rats.
Keywords: gut, microbial, SCFA, metabolic, metabolomics
the maintenance of blood pressure (BP) homeostasis is a complex process that is carefully orchestrated by a variety of genetic and environmental factors and studied extensively in rat models (2, 5, 11, 15–17, 19, 21–23, 26, 42–44, 50). Dietary salt is one of the prominent environmental factors influencing the development and progression of salt-sensitive hypertension (13, 14, 24, 57). As the consumed salt is transported through the gastrointestinal tract, one of the anatomical sites through which it is absorbed, in addition to the small intestine and the colon, is the cecum. The functions of the cecum are to absorb fluids and salts that remain after completion of intestinal digestion and absorption and to mix its contents with a lubricating substance, mucus. The cecum is also an “anerobic fermentor” as it houses a large number of bacteria that aid in digestion of undigested material in the stomach and small intestine. This is accomplished by a fermentative process that helps in breaking down fibers for their survival (33).
In recent years, there is ample evidence in the literature to suggest that the micro-organisms residing in the gut, collectively termed as gut microbiota, contribute importantly to the pathophysiology of a variety of disorders such as obesity, colitis, inflammatory diseases, metabolic syndrome, and kidney disease (1, 7–10, 28, 30, 31, 35, 45, 46, 52, 53, 55, 56). Recent studies focused on olfactory receptors and G protein-coupled receptors suggest that short chain fatty acids (SCFAs) produced by the gut microbiota likely influence BP regulation that is facilitated by renal sensory receptors (34, 38–41). Besides this, to our knowledge, there are no studies reported that test the influence, if any, of the gut microbiota on hypertension.
Due to the facts that salt is a major environmental factor influencing salt-sensitive forms of hypertension and that salt is absorbed in the cecum, which is also one of the sites where a large number of bacteria reside (18, 47, 48), we hypothesized that interactions between the host and the gut microbiota influence the development of salt-sensitive hypertension. To test this hypothesis, we collected the cecal content from the Dahl salt-sensitive (S) and Dahl salt-resistant (R) rats and administered them orally to R and S rats, respectively. Data obtained from monitoring the BP of these rats are presented.
METHODS
Animals
All animal research protocols were approved by the University of Toledo's Institutional Animal Care and Use Committee. Experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. S and R rats were inbred in our colony.
Physiological Studies
Breeding stocks, dams, and pups (prior to weaning) were maintained on a low salt (0.3% NaCl)-containing diet (Harlan Teklad, 7034). Pups were weaned at 4 wk of age and continued to be maintained on a low-salt diet for experiments conducted under low-salt conditions or fed a high salt (2% NaCl)-containing diet (Harlan Teklad, TD94217) for experiments conducted under high-salt conditions. The animals were implanted with C-10 transmitters (Data Sciences International, St. Paul, MN) as described previously (37) to record systolic BP via radiotelemetry. After the animals recovered from surgery, a baseline systolic BP reading was recorded for 1 h (T = 0 h). To deplete the resident microbiota, we followed the procedure described by Manichanh et al. (32). The animals were treated with 50 mg/kg/day of Vancomycin (Hospira) and 50 mg/kg/day of Meropenem (Novaplus) in their drinking water along with 50 mg/kg/day of Omeprazole (SIGMA) by oral gavage for 3 days. This was followed by transplantation of cecal contents on day 4.
Isolation of cecal content and DNA.
On day 4, cecal contents were collected from S and R rats (n = 6/group). Samples within a group were pooled and immediately transplanted by gavage as quickly as possible (within ∼10–20 min). Before the cecal content was pooled, an aliquot from the cecal content of each animal was snap-frozen on dry ice and processed overnight for DNA isolation with the Promega Wizard SV 96 Genomic DNA Purification System (Promega, Madison, WI). DNA isolated was stored at −80°C to be used at a later time for pyrosequencing.
Transplantation study.
To ensure that there were no pre-existing differences in systolic BP between the experimental groups, we grouped animals so that the average systolic BP for S rat groups were similar and that of the R rat groups were similar (n = 6–10/group). During transplantation, one group of S rats (n = 11) received the pooled cecal content from R rats. This group was designated the R to S group. The second group of S rats (n = 10) received the pooled cecal content from S rats. This group was designated as the S to S group. Similarly, a group of R rats (n = 6) received pooled cecal content from S rats. This group was designated the S to R group. The second group of R rats (n = 6) received pooled cecal content from R rats. This group was designated the R to R group. Each rat received 2 ml of cecal content by oral gavage. Telemetry recording began immediately after transplantation. Rats were maintained from weaning on a high-salt (2% NaCl) diet unless otherwise mentioned as low-salt diet when they were maintained on a 0.3% NaCl diet. Rats in the survival experiment were continued to be fed with 2% NaCl-containing diet until their natural death.
Antibiotic treatment study.
A separate group of S rats was raised, and at 4 wk of age, while on a high-salt diet, they were surgically implanted with C-10 radiotelemetry transmitters. Postrecovery for 3 days, their BP was recorded. The animals were subsequently divided into two groups of n = 10/group and continued to be given a high-salt diet. Only one of the groups was administered 50 mg/kg/day of Vancomycin (Hospira) and 50 mg/kg/day of Meropenem (Novaplus, same class as Imipenem) in their drinking water for the entire experiment. BP of these two groups of S rats was periodically recorded.
Determination of serum and urinary sodium content.
Urine and serum were collected from both pre- and posttransplantation groups of rats on a high-salt diet. Presamples were collected from rats maintained on a 2% NaCl diet from weaning, but before any antibiotic or cecal content delivery. For urine collection, animals in the pretransplant group and 27 days posttransplantation group were transferred into metabolic cages with access to water, but not food, for 24 h urine collection as described previously (20). At the end of the urine collection period, the urine volume was recorded, and before the rats were moved back to their home cages with access to food ad libitum, blood was collected by retro-orbital puncture. Serum was separated by centrifugation at 13,000 g. Both serum and urine were stored at −80°C. Sodium and creatinine contents were assessed with a Beckman Coulter AU680 analyzer by the ion selective electrode diluted (indirect) method.
Genomic DNA Isolation, 16S rRNA Gene Sequencing, and Analysis of Microbiotal Composition
Genomic DNA from both cecal and fecal samples of the high-salt study group were isolated by the Promega Wizard SV 96 Genomic DNA Purification System (Promega) method. The v1–v3 regions of 16s rRNA gene was amplified using 27f (AGAGTTTGATCCTGGCTCAG) and 534r (ATTACCGCGGCTGCTGG). The primers were anchored with adaptor (adapter A: 5′ CCATCTCATCCCTGCGTGTCTCCGACTCAG 3′ and adapter B: 5′ CCTATCCCCTGTGTGCCTTGGCAGTCTCAG 3′) and multiplex identifiers (MIDs; 10 bp long). The multiplexed amplicon was sequenced using the 454 Jr. platform, and the unique MIDs were used to separate the samples from the sequencing run. Microbial data analysis was performed using QIIME platform scripts (http://www.qiime.org) (6). The sequences were rarified at randomly selected 1,000 sequences, and further downstream analysis was performed. Microbial classification was performed with the GreenGenes reference database (gg_otus-4feb2011) using QIIME tools (6). The sequences obtained were picked into operational taxonomic units by clustering 97% sequence similarity (uclust) and classified at various taxonomic ranks (phylum, order, class, family, genus, and species). The beta diversity principle co-ordinate plots were generated using phylogenetic metrics of UniFrac distances. The UniFrac metric distances were used to calculate the significance between two groups using ANOSIM with 999 permutations. Weighted and unweighted UniFrac distances were computed using jackknife beta diversity to estimate the uncertainty principal coordinate analysis (PCoA) plots and hierarchical clustering of microbial communities. In this method, we computed beta diversity distance matrix and built the unweighted pair group method with arithmetic mean, which is a type of hierarchical clustering method using average linkage to interpret the distance matrix.
Targeted Metabolomic Analysis of SCFAs in Plasma
SCFAs were quantified in plasma as described previously (49). In brief, 100 μl of plasma from the high salt-fed rats posttransplantation and BP studies was utilized. Samples were diluted in acidified water spiked with stable isotope-labeled SCFA standards and then extracted with diethyl ether. The ether layer was immediately analyzed by gas chromatography/mass spectrometry using a Phenomenex ZB-WAX column on an Agilent 6890 GC with a 5973MS detector. Quantitation was performed by calibration to internal standards and standard curves on the Mass Hunter quantitative suite version B.06.00 (Santa Clara). All levels are expressed in mM (49).
RESULTS
S and R rats Are Not Identical in Their Cecal Microbiotal Compositions
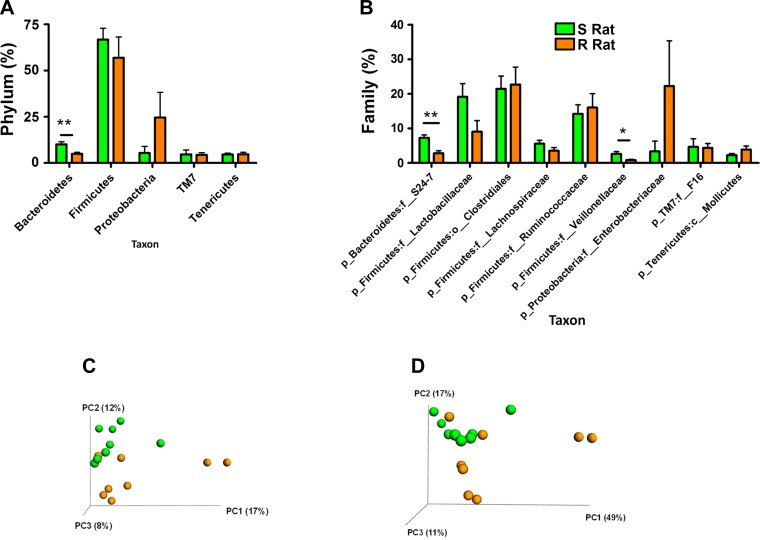
To determine the microbiotal compositions of S and R rats, cecal DNA was isolated from both S and R rats, and the variable regions v1–v3 regions of bacterial 16S rRNA genes were sequenced. We obtained an average of 5,893 quality amplicon sequences/sample. Individual sample unique “quality” amplicon data are provided in Table 1. The 16S rRNA sequences were analyzed using QIIME scripts as described in methods. The microbial composition of S and R rats differed at both the levels of phylum and family (Fig. 1, A and B). Bacteria belonging to the phylum Bacteroidetes was higher in the S rats compared with R rats (Fig. 1A). All other phyla, including Firmicutes, were not altered between S and R rats, although a trend was observed with proteobacteria being lower in the S compared with R rats (Fig. 1A). Further downstream taxonomic analysis at the level of family suggested a specific increase of the family of bacteria under the phylum Bacteroidetes, called S24-7 in the S rats compared with the R rats. In addition, although there were no overall alterations in Firmicutes as a phylum, a family within the phylum Firmicutes, Veillonellaceae, was significantly higher in the S rats compared with R rats (Fig. 1B). Under the phyla proteobacteria, the bacteria belonging to the family Enterobacteriaceae were lower in the S, but not statistically significantly different from that in the R rats (Fig. 1B).
Table 1.
Microbiotal sequence reads data summary
| Rat ID | Source | Quality Unique Amplicons/Sample |
|---|---|---|
| S5842 | Srat_Cecum | 6,713 |
| S5843 | Srat_Cecum | 2,300 |
| S5844 | Srat_Cecum | 6,530 |
| S5845 | Srat_Cecum | 7,077 |
| S5891 | Srat_Cecum | 6,601 |
| S5892 | Srat_Cecum | 5,741 |
| S5893 | Srat_Cecum | 7,142 |
| S5894 | Srat_Cecum | 6,339 |
| R30405 | Rrat_Cecum | 5,860 |
| R30406 | Rrat_Cecum | 3,083 |
| R30419 | Rrat_Cecum | 7,948 |
| R30420 | Rrat_Cecum | 8,059 |
| R30421 | Rrat_Cecum | 2,951 |
| R30427 | Rrat_Cecum | 3,457 |
| R30428 | Rrat_Cecum | 6,245 |
| R30429 | Rrat_Cecum | 8,250 |
| S5354 | Fecal Transfer_StoS | 3,764 |
| S5355 | Fecal Transfer_StoS | 6,310 |
| S5363 | Fecal Transfer_StoS | 7,305 |
| S5365 | Fecal Transfer_StoS | 7,242 |
| S5366 | Fecal Transfer_StoS | 7,721 |
| S5376 | Fecal Transfer_StoS | 4,620 |
| S5334 | Fecal Transfer_RtoS | 4,064 |
| S5336 | Fecal Transfer_RtoS | 3,814 |
| S5342 | Fecal Transfer_RtoS | 6,879 |
| S5345 | Fecal Transfer_RtoS | 8,011 |
| S5346 | Fecal Transfer_RtoS | 1,703 |
| S5352 | Fecal Transfer_RtoS | 5,488 |
| TOTAL sequences | 161,217 |
Total raw number of sequences analyzed: 189,518. Parameters for sequences to be considered: 1) between 200 and 1,000 bp length; 2) quality score > 25. Sequences with homopolymer run exceeding 6 were removed. Amplicon sequences that matched 100% with the barcodes were used for further analysis.
Fig. 1.
Pretransfer profiling of Dahl salt-sensitive (S) and Dahl salt-resistant (R) rat microbiotal compositions. Genomic DNA from S (n = 8) and R (n = 8) rat cecal contents were collected as described in methods and subjected to 16S rRNA sequencing. Bacterial taxonomic classification of S and R cecal content into phyla (A) and families (B). Green bars, bacteria in S; orange bars, bacteria in R. *P < 0.05; **P < 0.001. Bottom: principal coordinate analysis (PCoA) plots: Unweighted (C) and weighted (D) UniFrac distances. Green spheres represent S gut microbiota; orange spheres represent R gut microbiota.
Next, the 16s rRNA gene sequence datasets obtained were analyzed with UniFrac, a tool that computes phylogenetic similarity between microbial communities based on the degree to which their taxa share branch length. PCoA plots indicate that microbiotal composition of S and R rats are grouped within distinct clusters (Fig. 1, C and D). Weighted analysis based on abundance of sequences grouped into operational taxonomic units placed in the context of the bacterial phylogenetic tree indicates that cecal microbiotal composition of S and R rats was distinctly different from each other (Fig. 1C). Similarly, unweighted analysis, which does not consider the abundance of operational taxonomic units but assesses uniqueness of distribution, also indicates that the S and R rat cecal microbiotal compositions were different (Fig. 1D). The statistical analysis of these distance matrices was performed with the ANOSIM test on 999 permutations. The results demonstrate that both weighted and unweighted UniFrac distances were significantly different (P = 0.001) compared with each other, indicating definitive differential microbiota composition in these rats (Fig. 1, C and D).
Host Genomic Sequence Variation of S and R Rats
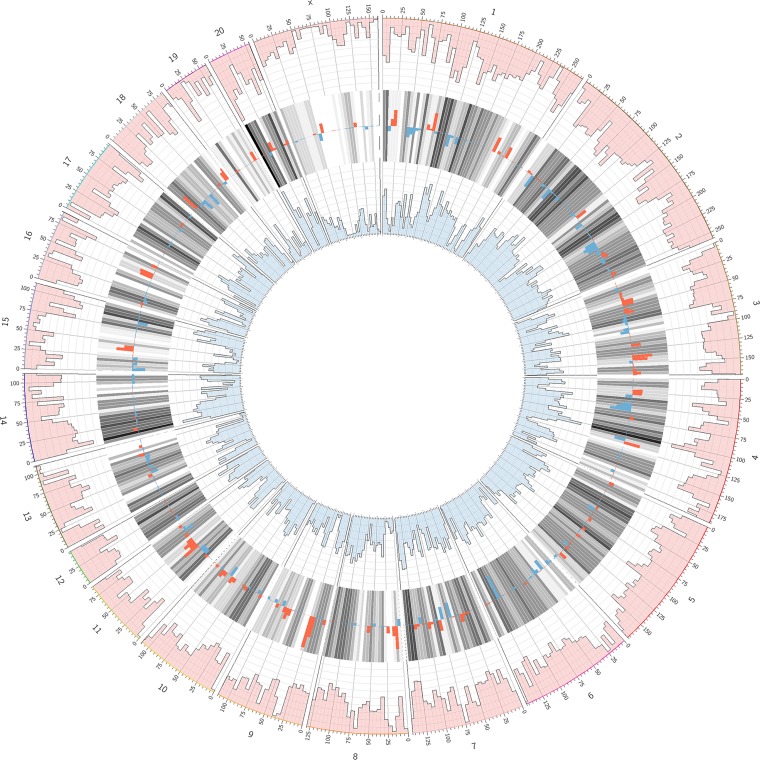
Gut microbial populations are dependent on the host for their survival, and hence differences in the gut microbial populations could be, at least in part, attributed to variations between the host genomes. To assess the variations between the S and R rat genomes, data collected from our previous study through whole genome analyses of these two strains were used to analyze the extent of variations between S and R rats. The densities of variations between S and R rats are shown in Fig. 2. There are a total of 2,058,300 variants estimated between the S and R rat genomes.
Fig. 2.
Circos plot depicting comparison of the genomic variant densities between SR/Jr and SS/Jr genomes. The outermost ring of numbers 1–20 and X indicate rat chromosomes. The numbers below the outer ring that are labels of ticks represent locations on each chromosome in megabases. The pink outer circumference of manhattan plots represents the histogram of variants of the SR/Jr strain compared with the rat reference sequence from Brown Norway rats. The blue innermost circumference of manhattan plots is the histogram of variants of the SS/Jr strain compared with the rat reference sequence from Brown Norway rats. The histograms consist of 10 levels wherein each level represents 2,500 variants. The gray bars in between the 2 histograms (pink and blue) show the average of the 2 densities with darker bars for higher densities. The density of variants is overlapped on the average bars with red bars for higher SR/Jr density and blue bars for higher SS/Jr density. The range of density difference from the bottom to the top of a gray bar is −12,500 to 12,500 variants.
Exacerbated Hypertension in High Salt-fed Dahl S Rats Administered with Microbiota from Dahl R Rats
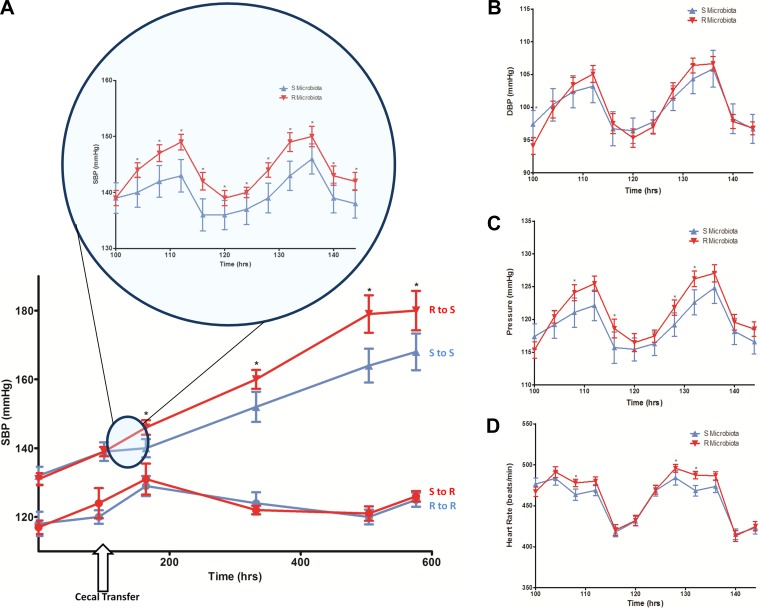
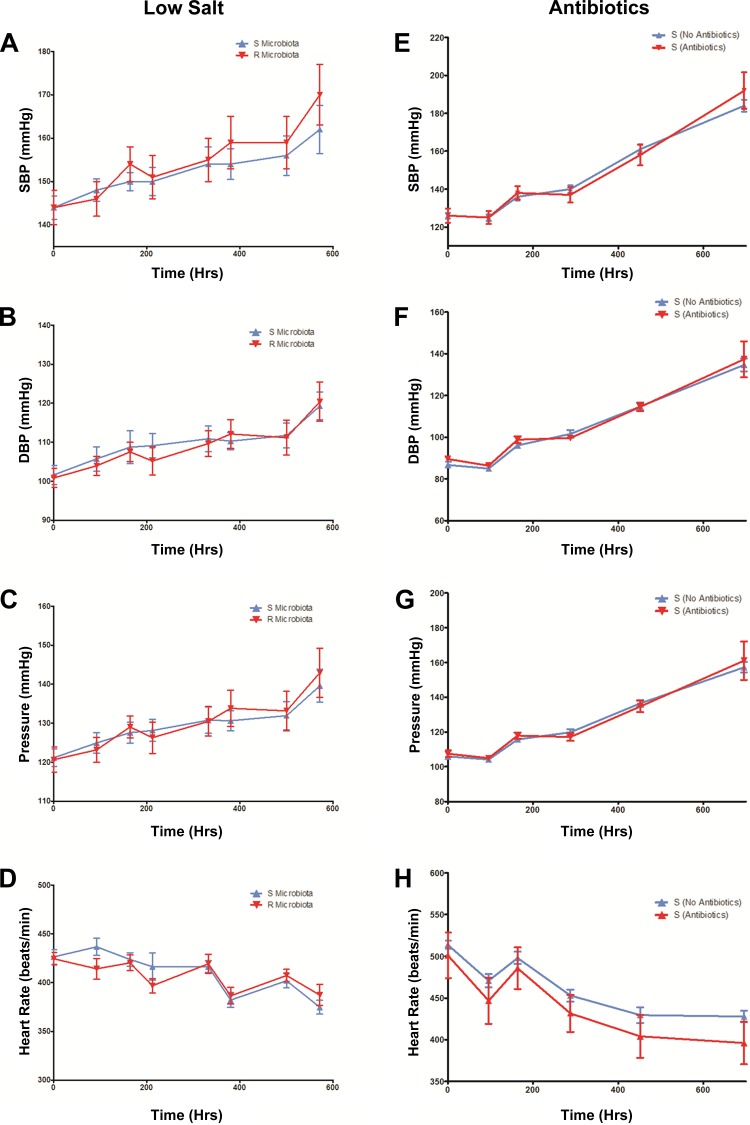
The hypothesis for the study was that microbial content from the hypertensive S rats contributes to the development of hypertension and that the depletion of the same with antibiotics, along with gastric suppression followed by the introduction of the cecal content from the normotensive R rats, should be protective, i.e., attenuate development of hypertension in the S rat. Surprisingly, compared with the S rats administered with S rat cecal content (S to S group in Fig. 3A), the systolic BP of the S rats given a single bolus of cecal content from R rats (R to S group in Fig. 3A) was consistently and significantly elevated throughout their lifetime (Fig. 3A, Table 2). Mean BP, but not diastolic BP, was also similarly elevated in the R to S group compared with the S to S group (Fig. 3, B and C). Heart rates were not different between the groups (Fig. 3D).
Fig. 3.
Blood pressure (BP) in high salt-fed animals posttransplantation of cecal content. Rats were surgically implanted with radiotransmitters, transplanted with cecal contents, and monitored by radiotelemetry. A: average systolic BP ± SE is plotted against time. Red line connecting red triangles: S rats transplanted with R microbiota (R to S, n = 11); blue line connecting blue triangles: S rats transplanted with S microbiota (S to S, n = 10); red line connecting red circles: R rats transplanted with S microbiota (S to R, n = 6); blue line connecting blue circles: R rats transplanted with R microbiota (R to R, n = 6). Inset is the systolic BP recording of 2 days (demonstrating diurnal rhythms), with 4 h moving averages plotted for each day. Statistical analysis was done by 1-way ANOVA. *P < 0.05. B: diastolic BP (DBP), C: mean BP (Pressure), D: heart rate recordings of the R to S (red) and S to S (blue) groups. Recordings are of 2 days (demonstrating diurnal rhythms), with 4 h moving averages plotted for each day. Statistical analysis was done by 1-way ANOVA. *P < 0.05.
Table 2.
Systolic BP of rats receiving cecal transplants
| Days Posttransplant | SBP of S to S Transfer, mmHg | SBP of R to S Transfer, mmHg | SBP of R to R Transfer, mmHg | SBP of S to R Transfer, mmHg |
|---|---|---|---|---|
| 0 | 139 ± 2.7 | 139 ± 1.4 | 120 ± 2.0 | 124 ± 4.4 |
| 3 | 140 ± 2.7* | 146 ± 2.1* | 129 ± 2.9 | 131 ± 4.5 |
| 10 | 152 ± 4.4* | 160 ± 2.7* | 124 ± 3.2 | 122 ± 1.3 |
| 17 | 164 ± 4.9* | 179 ± 5.5* | 120 ± 2.2 | 121 ± 2.1 |
| 20 | 168 ± 5.3* | 180 ± 5.7* | 125 ± 2.0 | 126 ± 1.5 |
SBP, systolic blood pressure recorded by radiotelemetry; day 0, Day of cecal transplantation;
P < 0.05 between S to S and R to S transfers. The data from this experiment are plotted in Fig. 3A.
Similar to the above studies in S rats, groups of R rats were also maintained on a high-salt diet and their blood pressures were monitored following oral administration of either S or R rat cecal contents. R rats did not increase their BP in response to the high-salt diet (Fig. 3A). Systolic, diastolic, mean BP, and heart rates of R rats transplanted with S rat cecal content (S to R group) were not significantly different from that of the R rats that received microbiotal cecal content from R rats (R to R group) (Fig. 3, A–D).
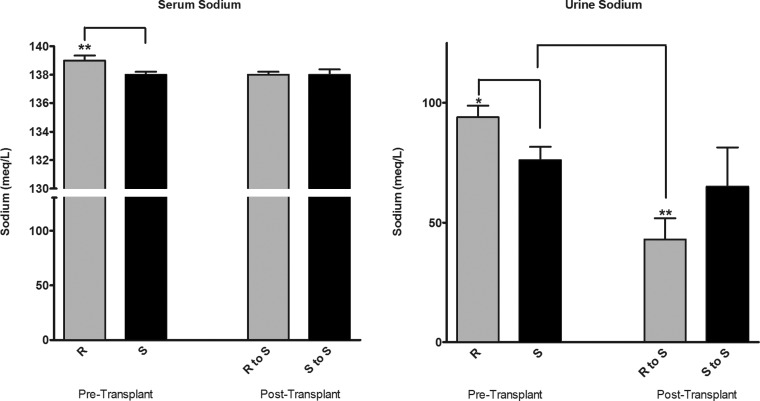
No Change in Fractional Sodium Excretion in the R to S Transplanted Group
To test whether the transplantation of microbes was associated with any alterations in reabsorption of salt, urinary sodium content was examined. Pretransplantation, S and R rats differed in their serum and urine sodium excretions. S rats had lower serum and urinary sodium concentrations compared with R rats (Fig. 4). Posttransplant of cecal content, the S rats in the R to S group and the S to S groups were similar in their serum sodium concentrations. However, the urinary sodium content of S rats transplanted with the R rat microbiota was significantly lower than that of S rats without any transplantation and, although not statistically significant, had a similar trend compared with the S rat group transplanted with S rat microbiota (Fig. 4). However, the fractional excretion of sodium (sodium clearance/glomerular filtration rate) of the R to S transplanted animals was not different from the S to S transplanted group (data not shown).
Fig. 4.
Urine and serum sodium levels. Pretransplantation samples were collected from rats 4 wk of age. Posttransplantation, urine and serum was collected on days 27 and 28, respectively, as described in methods. R to S: S rats transplanted with R rat cecal content; S to S: S rats transplanted with S rat cecal content. Statistical analysis by 1-way ANOVA, *P < 0.05, **P < 0.01.
Comparisons of Lifespan of S Rats Transplanted with S or R rat Microbiota
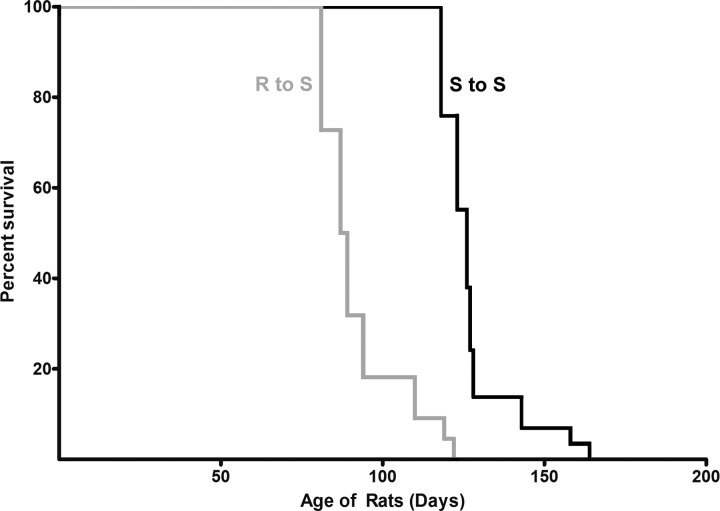
To assess whether the differential increase in hypertension of the S rats transplanted with S or R cecal content, we continued to feed all the S rats in the BP study reported in Fig. 3A (n = 6–10/group) a high-salt diet until their natural death (Fig. 5). The S rats given cecal content from the R rats had a shorter lifespan compared with that of S rats transplanted with S rat cecal content (Fig. 5).
Fig. 5.
Kaplan-Meier plot of rats transplanted with cecal contents. Groups of rats in the high-salt study (shown in Fig. 1) were continued on the high-salt diet until their natural death. On the day of death of each rat, the age of the rat was recorded and imputed in the plot. S rats transplanted with R microbiota are represented by a gray line. S rats transplanted with S microbiota are represented by a black line. t-Test, P < 0.05.
S Rats on a Low-salt Diet Do Not Demonstrate an Increase in BP in Response to Cecal Transplantation from R Rats
The BP monitoring experiments in the high salt-fed groups of S rats given cecal contents from either S or R rats were repeated and corroborated with three different groups of S rats of different age groups, both by telemetry and tail-cuff methods (data not shown). To examine whether the increase in BP of the R to S group of cecal content transplanted rats was a dietary salt-sensitive response, the transplantation studies were conducted in rats maintained on a low salt (0.3% NaCl)-containing diet. As shown in Fig. 6, A–D, systolic, diastolic, mean BP, and heart rates of the S rats given R rat cecal content were comparable to the S rats given S rat cecal content. These data demonstrate that the increase in systolic BP of the R to S group of rats compared with the systolic BP of the S to S group of rats is a salt-sensitive response.
Fig. 6.
A–D: BP in low salt-fed animals posttransplantation of cecal content; E–H: effect of antibiotic treatment on systolic BP of S rats. A–D: systolic BP (SBP), diastolic BP (DBP), mean BP (pressure), and heart rate of rats maintained on a low-salt (0.3% NaCl) diet. We surgically implanted 4 wk old rats BP transmitters. Postrecovery from surgery, rats were transplanted with cecal contents and monitored by radiotelemetry. Average values ± SE are plotted against time. Red line connecting red triangles: S rats transplanted with R microbiota (n = 11); blue line connecting blue triangles: S rats transplanted with S microbiota (n = 10). E–H: SBP, DBP, mean BP (pressure), and heart rate of groups of 4 wk old S rats maintained on a high-salt (2% NaCl) diet. Rats were surgically implanted with BP transmitters and postrecovery, they were continuously administered with (n = 10) or without (n = 10) antibiotic in their drinking water (50 mg/kg/day of Vancomycin and 50 mg/kg/day of Meropenem) and monitored for BP by radiotelemetry. Day 0 = day when antibiotic administration began. Statistical analysis was done by t-test. t-Test, *P < 0.05.
Antibiotic Treatment and BP of the S Rats
To examine the possibility that the observed BP effect in the high salt-fed group of rats is due to the disturbances of the microbial composition as a result of the antibiotics used in the study, we eliminated the cecal transplantation procedure and studied the BP of groups of S rats, with or without antibiotic treatment. Systolic, diastolic, mean BP, and heart rates of S rats given the antibiotics were not significantly different from the group of S rats without antibiotic treatment (Fig. 6, E–H). These data indicate that transplantation of the cecal content from the R rats is specifically responsible for the observed increase in BP of the S rats on a high-salt diet transplanted with R rat cecal content.
Posttransplantation Microbial Profiling
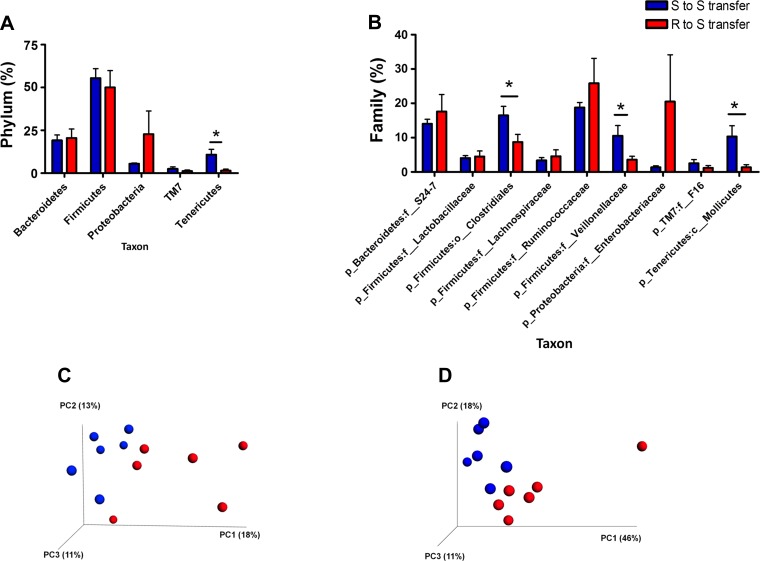
To assess whether cecal transplantation had consequences on the overall microbiotal composition of the rats, posttransplantation of cecal content, and BP monitoring, fecal DNA samples were sequenced with primers for the v1–v3 regions of bacterial 16S rRNA genes. Fecal and not cecal samples were collected because animals were not terminated, but observed for survival. As shown Fig. 7A, there was a significant decrease in bacteria belonging to the phylum Tenericutes in the R to S transfer group compared with the S to S group. We did not observe any significant change in other groups at the phylum level. Interestingly, in the R to S transfer group compared with the S to S group, there was a significant decrease in the bacteria of the order Clostridiales belonging to the phylum Firmicutes as well as bacteria of the class Mollicutes belonging to the phylum Tenericutes. Most importantly, bacteria belonging to Veillonellaceae (phylum Firmicutes) were significantly reduced in the R to S transfer group compared with the S to S group (Fig. 7B), which corroborates very well with differences between the untreated S and R rats (Fig. 1B). The weighted and unweighted PCoA plots also clearly suggest the distinguishable clusters in these transplantation experiments (Fig. 7, C and D), which was statistically significant (P = 0.001) based on the ANOSIM statistical test with 999 permutations.
Fig. 7.
Posttransfer profiling of S and R microbiotal compositions. Posttransfer bacterial taxonomic classification of S and R fecal content into phyla (A) and families (B). Blue bars: bacteria in S rats (n = 6) receiving S rat cecal content; red bars: bacteria in S rats (n = 6) receiving R rat cecal content. *P < 0.05. Bottom: PCoA plots: unweighted (C) and weighted (D) UniFrac distances. Blue spheres represent S to S gut microbiota. Red spheres represent R to S gut microbiota.
Plasma SCFAs of Cecal Transplanted S Rats
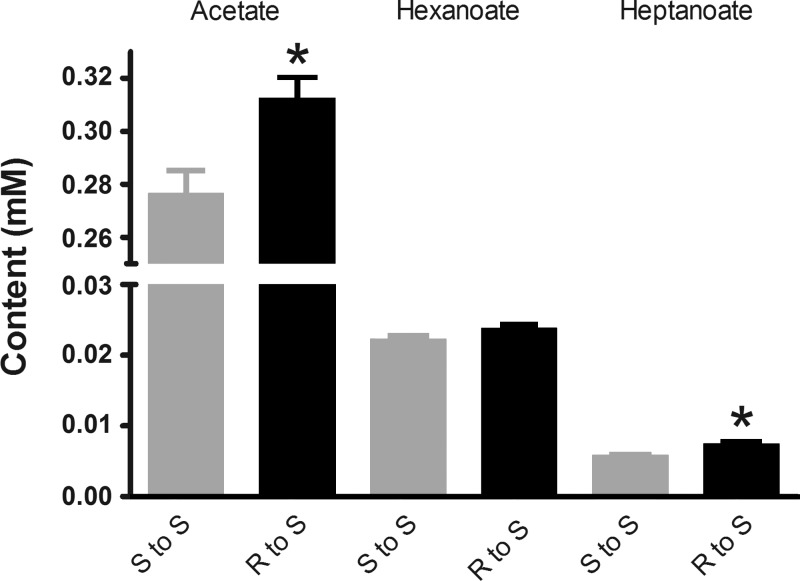
SCFAs are end products of microbial fermentation, reported to be linked to BP regulation (34, 38–41). To examine whether the composition of the S rat SCFA was altered as a result of the differential cecal transplantation experiment, plasma SCFAs were quantitated by mass spectrometry. Compared with the S rats transplanted with cecal content of S rats, plasma levels of acetate and heptanoate were significantly elevated in the S rats transplanted with cecal content of R rats (Fig. 8), suggesting that differential microbial composition affects plasma SCFA levels.
Fig. 8.
Targeted plasma metabolomic profiling for short chain fatty acids. On day 42 posttransplantation plasma samples were collected from S rats transferred with S (n = 5) or R (n = 6) cecal contents. These plasma samples were subjected to a targeted short chain fatty acid analysis by gas chromatography/mass spectrometry as described in methods. Data are expressed as means ± SE; *P < 0.05.
DISCUSSION
The involvement of gut microbiota in regulating metabolic syndrome is unequivocally established in the literature (12, 36, 51, 55, 58). Hypertension is one of the hallmarks of metabolic syndrome, which may or may not be, per se, influenced by gut microbiotal compositions. Evidence for the gut microbiota to influence BP is limited. To date, there are two studies reported (38, 41), which were specifically focused on data obtained from an olfactory G protein-coupled receptor (Olfr78) and G protein-coupled receptor (Gpr41) knockout mice. To our knowledge, there are no gut microbiotal studies reported in genetic models of hypertension.
The S rat and the R rats are genetic models of hypertension and normotension, respectively, that were developed from Sprague-Dawley rats by Lewis K. Dahl (25, 29). While hypertension in the S rats is exacerbated by a high-salt diet, BP of the R rat is unaltered by a high-salt diet (29). Since salt is absorbed by the gut, we were intrigued to construct the hypothesis that genetic factors influencing the gut milieu may contribute to the extent of hypertension in the S rat, but not in the R rat. If this were to be the case, we predicted that one such factor would be microbiotal composition/metabolites, whereby we expected differences in cecal microbiotal compositions between the S and R rats. The data collected from the S and R rats, especially the PCoA plots presented in Fig. 1 indeed confirm that there are significant differences between the cecal microbiota in S and R rats. Because the inbred S and R rats have been maintained in our colony under comparable conditions since they were inbred in the 1970s, we can rule out environmental factors contributing to the observed differences in their cecal microbiota. The evidence therefore points to genomic variation between S and R rats as a likely reason for the observed alterations in their cecal microbial content. According to the NextGen sequencing data obtained from whole genome sequencing of S and R rats (3), the genomic variation between these strains is estimated to be 2,058,300 bp (Fig. 2, estimated from data analyzed by the Rat Genome Database; http://www.rgd.mcw.edu). Since there are no variants in the homologous rat genes for Olfr78 and Gpr41, the two mouse genes reported for gut microbial links to hypertension (38), it is reasonable to conclude that variation within host genomic factors, which are not within Olfr78 and Gpr41, are likely influencing microbiotal compositions of S and R rats.
To test whether these differences in microbiota contribute to the extent of BP in these rats, microbiotal transplantation studies were conducted. R rats are insensitive to a high-salt diet. Their genome is relatively very resistant to alterations in BP (29). Accordingly, it was not surprising to observe that a combination of high-salt diet and microbiotal transfer from the hypertensive S rat did not alter the BP of the R rat (Fig. 3A). Note in passing that the underlying assumption for administering cecal content was that cecal content from the S rat would promote hypertension and that from the R rat would contribute to normotension. We therefore expected that transfer of R rat cecal content (which contributes to normotension) would attenuate hypertension in the S rat. Contradictory to our expectation, S rats given R rat microbiota further exacerbated their hypertension that was sustained throughout their life and adversely affected their lifespan (Figs. 3A and 5). This suggested one of two possibilities: 1) The microbiota resident within the cecum of the S rat, but not the R rat, are in a symbiotic relationship with the host. For instance, they could be halophiles (salt-loving microbes), whereby depletion of such microbiota from the S would increase the bioavailability of salt to the S. Increased salt absorption could then explain the observed exacerbation of the BP in the S rat; or 2) there is no symbiotic relationship between the S rat and the S rat cecal microbiota. The R rat microbiota is responsible for the observed increase in BP of the S rats that received cecal content from the R rats. To explore which of these possibilities is correct, we tested the BP of S rats that were treated exclusively with antibiotics but were not given cecal transplants. Systolic BP of the antibiotic treated S rats did not differ from the BP of the S rats that did not receive the antibiotic treatment (Fig. 6E). This experiment provided evidence to suggest that depletion of S rat microbiota was not sufficient to explain the observed increase in BP of the S rats with cecal transplants from R rats. Thus, we ruled out the first possibility that the S rat microbiota has a symbiotic relationship with the host. The data are therefore interpreted to conclude that the introduction of the R rat microbiota into the S rat is required for the observed increase in the BP of the S rats.
The question of why cecal content from the R rats exacerbated hypertension of the S rat is an important one that is largely beyond the scope of the current study. Nevertheless, microbiotal screening before and after transplantation studies point to a variety of alterations in the overall microbiotal composition, which are occurring along with one consistent factor between the pre- and posttransfer analyses, which is a lower level of the family Veillonellaceae under the phylum Firmicutes. Additional studies will be required to determine whether a reduction in Veillonellaceae is solely responsible for the observed alteration in BP or a combination of the reduction in Veillonellaceae, Clostridiales, and Mollicutes (along with any other bacteria that were not identified in our study) contributes to the increase in BP of the S rats given R rat cecal content.
The observation that R rat cecal microbiota elevates BP of the S rat only when the S rats are on a high-salt diet combined with the observation that urinary sodium is significantly reduced in the R to S group compared with all other experimental groups in the current study was suggestive of an increase in salt reabsorption that may have caused BP to be higher in the R to S group. However, there was no change in fractional excretion of in the R to S group compared with the S to S group, suggesting that the observed differences in BP between these groups is perhaps not due to differential salt reabsorption.
To explore potential mechanisms by which differential microbial populations affect BP in the Dahl rats, we next focused on SCFAs, which are end products of fermentation by the gut microbiota and are absorbed into the circulation. Interestingly, both acetate and heptanoate were higher in the R to S transfer group compared with the S to S transfer group. Both of these SCFAs were not different between the pretransfer S and R groups (data not shown), which lends support to the view that the genome of the S rat is unable to compensate for the excess of plasma acetate to the extent similar to that of R rats. In fact, allelic variations exist between the genomes of the S and R rats for several olfactory receptor genes, Olr129, Olr1392, and Olr1876 and G protein-coupled receptor genes, Gpr37l1, Gpr89b, Gpr110, Gpr155, and Gpr161 (Variant visualizer at http://www.rgd.mcw.edu). Of note, at least one of the G protein-coupled receptors, Gpr43, is a known receptor for acetate (http://cordis.europa.eu/result/rcn/33423_en.html) and is implicated in host-microbial cross talk (4, 27, 54). Thus, the data presented in our study on the increased plasma SCFA in the R to S group prompt further exploration into allelic variants within novel SCFA receptors as candidate genes for host-microbiotal interactions contributing to salt-sensitive hypertension.
GRANTS
This work was supported by National Heart Lung and Blood Institute Grants HL-076709, HL-112641, and HL-020176 to B. Joe and DK-89503, DK-097153, and DK-08194 to S. Pennathur. Authors acknowledge support from the Rat Genome Database (http://www.rgd.mcw.edu) for representation of the S and R genome sequence data.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: B.M., V.R.J., A.V.M., J.B., H.W., and Y.Z. performed experiments; B.M., V.R.J., A.V.M., J.B., Y.Z., and B.J. analyzed data; B.M., V.R.J., A.V.M., J.B., Y.Z., B.H., M.V.-K., S.P., and B.J. interpreted results of experiments; B.M., V.R.J., and A.V.M. prepared figures; B.M., V.R.J., A.V.M., B.H., M.V.-K., S.P., and B.J. edited and revised manuscript; B.M., V.R.J., A.V.M., J.B., H.W., Y.Z., B.H., M.V.-K., S.P., and B.J. approved final version of manuscript; M.V.-K. and B.J. conception and design of research; B.J. drafted manuscript.
REFERENCES
- 1.Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 143: 1006–1016, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atanur SS, Birol I, Guryev V, Hirst M, Hummel O, Morrissey C, Behmoaras J, Fernandez-Suarez XM, Johnson MD, McLaren WM, Patone G, Petretto E, Plessy C, Rockland KS, Rockland C, Saar K, Zhao Y, Carninci P, Flicek P, Kurtz T, Cuppen E, Pravenec M, Hubner N, Jones SJ, Birney E, Aitman TJ. The genome sequence of the spontaneously hypertensive rat: analysis and functional significance. Genome Res 20: 791–803, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atanur SS, Diaz AG, Maratou K, Sarkis A, Rotival M, Game L, Tschannen MR, Kaisaki PJ, Otto GW, Ma MC, Keane TM, Hummel O, Saar K, Chen W, Guryev V, Gopalakrishnan K, Garrett MR, Joe B, Citterio L, Bianchi G, McBride M, Dominiczak A, Adams DJ, Serikawa T, Flicek P, Cuppen E, Hubner N, Petretto E, Gauguier D, Kwitek A, Jacob H, Aitman TJ. Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell 154: 691–703, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bindels LB, Dewulf EM, Delzenne NM. GPR43/FFA2: physiopathological relevance and therapeutic prospects. Trends Pharmacol Sci 34: 226–232, 2013. [DOI] [PubMed] [Google Scholar]
- 5.Braun MC, Herring SM, Gokul N, Monita M, Bell R, Zhu Y, Gonzalez-Garay ML, Wenderfer SE, Doris PA. Hypertensive renal injury is associated with gene variation affecting immune signaling. Circ Cardiovasc Genet 7: 903–910, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Meth 7: 335–336, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho FA, Aitken JD, Vijay-Kumar M, Gewirtz AT. Toll-like receptor-gut microbiota interactions: perturb at your own risk! Ann Rev Physiol 74: 177–198, 2012. [DOI] [PubMed] [Google Scholar]
- 8.Chassaing B, Aitken JD, Gewirtz AT, Vijay-Kumar M. Gut microbiota drives metabolic disease in immunologically altered mice. Adv Immunol 116: 93–112, 2012. [DOI] [PubMed] [Google Scholar]
- 9.Chassaing B, Kumar M, Baker MT, Singh V, Vijay-Kumar M. Mammalian gut immunity. Biomed J 37: 246–258, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr Opin Immunol 22: 455–460, 2010. [DOI] [PubMed] [Google Scholar]
- 11.Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet 7: 829–840, 2006. [DOI] [PubMed] [Google Scholar]
- 12.D'Aversa F, Tortora A, Ianiro G, Ponziani FR, Annicchiarico BE, Gasbarrini A. Gut microbiota and metabolic syndrome. Intern Emerg Med 8, Suppl 1: S11–15, 2013. [DOI] [PubMed] [Google Scholar]
- 13.Dahl LK. Possible role of salt intake in the development of essential hypertension. 1960. Int J Epidemiol 34: 967–972, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Dahl LK. Salt and hypertension. Am J Clin Nutr 25: 231–244, 1972. [DOI] [PubMed] [Google Scholar]
- 15.Delles C, McBride MW, Graham D, Padmanabhan S, Dominiczak AF. Genetics of hypertension: from experimental animals to humans. Biochim Biophys Acta 1802: 1299–1308, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doris PA. The genetics of blood pressure and hypertension: the role of rare variation. Cardiovasc Therapeut 29: 37–45, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, Grzybowski M, Lombard JH, Staruschenko A, Moreno C, Jacob HJ, Geurts AM. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc Natl Acad Sci USA 111: 12817–12822, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escobar E, Ibarra C, Todisco E, Parisi M. Water and ion handling in the rat cecum. Am J Physiol Gastrointest Liver Physiol 259: G786–G791, 1990. [DOI] [PubMed] [Google Scholar]
- 19.Feng D, Yang C, Geurts AM, Kurth T, Liang M, Lazar J, Mattson DL, O'Connor PM, Cowley AW Jr. Increased expression of NAD(P)H oxidase subunit p67(phox) in the renal medulla contributes to excess oxidative stress and salt-sensitive hypertension. Cell Metab 15: 201–208, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrett MR, Joe B, Yerga-Woolwine S. Genetic linkage of urinary albumin excretion in Dahl salt-sensitive rats: influence of dietary salt and confirmation using congenic strains. Physiol Genomics 25: 39–49, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Garrett MR, Meng H, Rapp JP, Joe B. Locating a blood pressure quantitative trait locus within 117 kb on the rat genome: substitution mapping and renal expression analysis. Hypertension 45: 451–459, 2005. [DOI] [PubMed] [Google Scholar]
- 22.Gopalakrishnan K, Kumarasamy S, Abdul-Majeed S, Kalinoski AL, Morgan EE, Gohara AF, Nauli SM, Filipiak WE, Saunders TL, Joe B. Targeted disruption of Adamts16 gene in a rat genetic model of hypertension. Proc Natl Acad Sci USA 109: 20555–20559, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gopalakrishnan K, Kumarasamy S, Mell B, Joe B. Genome-wide identification of long noncoding RNAs in rat models of cardiovascular and renal disease. Hypertension 65: 200–210, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ha SK. Dietary salt intake and hypertension. Electrolyte Blood Press 12: 7–18, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joe B. Dr Lewis Kitchener Dahl, the Dahl rats, and the “inconvenient truth” about the genetics of hypertension. Hypertension 65: 963–969, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joe B, Shapiro JI. Molecular mechanisms of experimental salt-sensitive hypertension. J Am Heart Assoc 1: e002121, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura I, Inoue D, Hirano K, Tsujimoto G. The SCFA receptor GPR43 and energy metabolism. Front Endocrinol 5: 85, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med 3: 14, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knudsen KD, Dahl LK, Thompson K, Iwai J, Heine M, Leitl G. Effects of chronic excess salt ingestion. Inheritance of hypertension in the rat. J Exp Med 132: 976–1000, 1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuczynski J, Costello EK, Nemergut DR, Zaneveld J, Lauber CL, Knights D, Koren O, Fierer N, Kelley ST, Ley RE, Gordon JI, Knight R. Direct sequencing of the human microbiome readily reveals community differences. Genome Biol 11: 210, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science 320: 1647–1651, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res 20: 1411–1419, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mcfarlane GT, CJ. The colonic flora, fermentation, and large bowel digestive function. In: The Large Intestine: Physiology, Pathophysiology, and Disease, edited by Phillips SF, Pemberton JH, Shorter RG. New York: Raven, 1991, p. 51–92. [Google Scholar]
- 34.Natarajan N, Pluznick JL. From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. Am J Physiol Cell Physiol 307: C979–C985, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noel S, Martina-Lingua MN, Bandapalle S, Pluznick J, Hamad AR, Peterson DA, Rabb H. Intestinal microbiota-kidney cross talk in acute kidney injury and chronic kidney disease. Nephron Clin Pract 127: 139–143, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parekh PJ, Arusi E, Vinik AI, Johnson DA. The role and influence of gut microbiota in pathogenesis and management of obesity and metabolic syndrome. Front Endocrinol 5: 47, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pillai R, Waghulde H, Nie Y, Gopalakrishnan K, Kumarasamy S, Farms P, Garrett MR, Atanur SS, Maratou K, Aitman TJ, Joe B. Isolation and high-throughput sequencing of two closely linked epistatic hypertension susceptibility loci with a panel of bicongenic strains. Physiol Genomics 45: 729–736, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes 5: 202–207, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pluznick JL. Gut microbes and host physiology: what happens when you host billions of guests? Front Endocrinol 5: 91, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pluznick JL. Renal and cardiovascular sensory receptors and blood pressure regulation. Am J Physiol Renal Physiol 305: F439–F444, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA 110: 4410–4415, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pravenec M, Churchill PC, Churchill MC, Viklicky O, Kazdova L, Aitman TJ, Petretto E, Hubner N, Wallace CA, Zimdahl H, Zidek V, Landa V, Dunbar J, Bidani A, Griffin K, Qi N, Maxova M, Kren V, Mlejnek P, Wang J, Kurtz TW. Identification of renal Cd36 as a determinant of blood pressure and risk for hypertension. Nat Genet 40: 952–954, 2008. [DOI] [PubMed] [Google Scholar]
- 43.Rapp JP. Genetic analysis of inherited hypertension in the rat. Physiol Rev 80: 135–172, 2000. [DOI] [PubMed] [Google Scholar]
- 44.Rapp JP, Joe B. Do epistatic modules exist in the genetic control of blood pressure in Dahl rats? A critical perspective. Physiol Genomics 45: 1193–1195, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341: 1241214, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogers CJ, Prabhu KS, Vijay-Kumar M. The microbiome and obesity-an established risk for certain types of cancer. Cancer J 20: 176–180, 2014. [DOI] [PubMed] [Google Scholar]
- 47.Rosel E, von Engelhardt W. Sodium transport across the caecal and colonic epithelium of germfree and specific-pathogen free rats. Reprod Nutr Dev 36: 289–299, 1996. [DOI] [PubMed] [Google Scholar]
- 48.Schreiner J, Weber M, Loeschke K. Sodium chloride transport of normal and dietary enlarged rat cecum in vitro. Digestion 59: 676–682, 1998. [DOI] [PubMed] [Google Scholar]
- 49.Theriot CM, Koenigsknecht MJ, Carlson PE Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, Jun L, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5: 3114, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian Z, Liu Y, Usa K, Mladinov D, Fang Y, Ding X, Greene AS, Cowley AW Jr, Liang M. Novel role of fumarate metabolism in dahl-salt sensitive hypertension. Hypertension 54: 255–260, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tilg H. Obesity, metabolic syndrome, microbiota: multiple interactions. J Clin Gastroenterol 44, Suppl 1: S16–S18, 2010. [DOI] [PubMed] [Google Scholar]
- 52.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol 587: 4153–4158, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature 457: 480–484, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulven T. Short-chain free fatty acid receptors FFA2/GPR43 and FFA3/GPR41 as new potential therapeutic targets. Front Endocrinol 3: 111, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328: 228–231, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vijay-Kumar M, Gewirtz AT. Is predisposition to NAFLD and obesity communicable? Cell Metab 15: 419–420, 2012. [DOI] [PubMed] [Google Scholar]
- 57.Zanchetti A. Salt, inflammation, pathophysiological mechanisms, and organ damage in hypertension. J Hypertens 32: 205–206, 2014. [DOI] [PubMed] [Google Scholar]
- 58.Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich S, Jones C, Knight R, Walters WA, Knights D, Mongodin EF, Horenstein RB, Mitchell BD, Steinle N, Snitker S, Shuldiner AR, Fraser CM. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PLoS One 7: e43052, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]