Abstract
Purpose
Neurological diseases have a devastating impact on millions of individuals and their families. These diseases will continue to constitute a significant research focus for this century. The search for effective treatments and cures requires multiple teams of experts in clinical neurosciences, neuroradiology, engineering and industry. Hence, the need to communicate a large amount of information with accuracy and precision is more necessary than ever for this specialty.
Method
In this paper, we present a distributed system that supports this vision, which we call the CranialVault Cloud (CranialCloud). It consists in a network of nodes, each with the capability to store and process data, that share the same spatial normalization processes, thus guaranteeing a common reference space. We detail and justify design choices, the architecture and functionality of individual nodes, the way these nodes interact, and how the distributed system can be used to support inter-institutional research.
Results
We discuss the current state of the system that gathers data for more than 1,600 patients and how we envision it to grow.
Conclusions
We contend that the fastest way to find and develop promising treatments and cures is to permit teams of researchers to aggregate data, spatially normalize these data, and share them. The Cranialvault system is a system that supports this vision.
Keywords: Neuromodulation database, DBS, Distributed database, data normalization, data sharing, collaborative
Introduction
Because neurodegenerative diseases strike primarily in mid- to late-life, the incidence is expected to soar as the population ages. By 2030, as many as 1 in 5 Americans will be over the age of 65. If left unchecked 15 years from now, more than 12 million Americans will suffer from neurologic diseases. The urgency of finding treatments and cures for neurodegenerative diseases is thus increasing.
Neurodegenerative diseases require solutions that involve a broad range of expertise encompassing such different fields as neurology, genetics, brain imaging, drug discovery, neuro-electrophysiology, stereotactic neurosurgery, and computer science, all of which generate large data sets related to very small targets within the brain. Responding to the need to collect and share data, a number of research tools have been developed. The most successful examples focus on specific data types. In 2005, Marcus et al. [1] offered to the community the XNAT system whose primary objective is to support the sharing of clinical images and associated data. This is a goal shared by other successful initiatives such as NiDB [2], LORIS [3], and COINS [4]. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) unites researchers with study data as they work to define the progression of Alzheimer’s disease [5], Ascoli et al focus on sharing cell data [6] [7], Kotter [8] on cortical connectivity and Van Horn on fMRI [9] to give only a few examples. Another example is the Collaborative Research in Computational Neuroscience (CRCNS) program funded by the US National Institutes of Health (NIH) and National Science Foundation that focuses on supporting data sharing. The CRCNS provides a resource for sharing a wide variety of experimental data that is publically available [10]. In parallel, tools to support structured heterogeneous data acquisition within or across institutions have been developed such as the REDCap [11] system already used for more than 138,000 projects with over 188,000 users spanning numerous research focus areas across the REDCap consortium.
Neurodegenerative diseases such as the Parkinson’s disease have been studied through the prism of neuromodulation. As a result of pioneering work by Drs. Benabid and DeLong, deep brain stimulation is now a primary surgical technique that reduces tremors and restores motor function in patients with advanced Parkinson’s disease, dystonia, and essential tremor. Much of the future potential in precisely focused therapy such as DBS, has to do with understanding the exact location of delivered therapy and its impact on circuitry at a millimetric level of precision; and subsequently, the impact such therapy has on clinical outcomes. The variation in human brain anatomy is large enough that simple aggregation of data without accurate co-representation within a common space leads to vague generalization and comparisons of treatment modalities and diagnostic capabilities. The lack of analysis of data with the level of precision needed is a result of the complexity and heterogeneous nature of the data acquired during neuromodulation procedures. A system that can store and share data efficiently should be able to connect imaging datasets to electrophysiological neuronal signals, patient’s responses to stimulation to disease progression and follow up scores, and drug levels to neuromodulation parameters. The lack of widespread standards for data acquisition and processing further complicates the task. As a result, most neuromodulation databases still are homegrown by laboratories needing to manage their own data. These systems are often highly customized to a site’s particular data and to the preferences of the laboratory that built the systems and do not easily permit inter-institutional collaborations.
Over the last decade we have developed a system designed to handle applications that involve images and information that can be spatially related to these images. It consists of a database and associated processing pipeline system that are fully integrated into the clinical flow. This system permits spatial normalization of the data, data sharing across institutions, and handles the issue of data ownership, privacy, and compliance with the Health Insurance Portability and Accountability Act (HIPAA).
This system called CranialVault was originally developed at Vanderbilt to store, manage and normalize multiple types of data acquired from patients undergoing DBS procedures[12]. The deep brain procedure generates a large amount of data such as microelectrode recordings, patient’s responses to stimulation, and neurological observation. Prior to the CranialVault, only minimal patient related information often not including much more than the final position of the implant was stored in the clinical archive. Therefore, research projects required the manual extraction of data from patient and imaging archive, a process that did not facilitate studies that required a large amount of data.
The first implementation of the CranialVault system consisted of an SQL-based architecture to store any deep brain stimulation related data; a set of connected clinical tools, called CRAVE (CranialVault Explorer) used to collect the data without disturbing the clinical flow; and a processing pipeline used to normalize the data acquired from each patient onto a common reference system, called the atlas.
The second and third iterations of the system improved performance, extended the type of stored data to any data acquired during the neurological stereotactic procedure, and added the functionality required to handle data ownership and data sharing across users and research groups. During the development of CranialVault, we identified several key features that a clinical imaging-based data normalization system needs to possess to become a common data-sharing resource: 1) association of all data with a particular subject, 2) clinical applications and/or API to interface with existing third-party application to import structured heterogeneous data, 3) automated data quality checks, 4) modular normalization system, 5) real time queries, and 6) simple export of data.
The previous iteration of the CranialVault system met these requirements. In its previous iteration, the CranialVault system was a centralized system in which data from any partnering group was stored in virtual private databases. While data is easier to manage and share across groups with this type of architecture, it raises concerns over data ownership, data security, ethics (Institutional Review Board - IRB) issues, as well as, the permanence of a resource supported mainly by investigator-initiated federal grants.
To address these issues, a new version of the system was developed around a cloud-based architecture. This new system called the CranialVault Cloud consists of a set of interconnected CranialVault systems with each institution owning its own CranialVault node called a CranialVault Box. Patient health information (PHI) data, if used, stays within the institution in a separate encrypted part of the system to limit the risks of patient data infringement.
To foster data sharing, the infrastructure was designed to allow the creation of “social networks”-like research networks permitting researchers to share data only with other researchers they want to share data with instead of relying on an open-ended sharing agreement.
Additional effort was made to interact with third party databases such as RedCap, XNAT and PACS systems.
Finally, to connect the system to other third-party data gathering applications, we developed an API and present our data definition as a standard format for neuro-related data sharing and open it to the community as a starting point for future standard.
Method
To the best of our knowledge, the CranialVault is the only system that can be distributed and combines patient’s image data, data that can be localized within them such as micro-electrode recordings, patient’s response to stimulation, implant location and any demographic or disease-related information. The CranialVault system normalizes the patient’s data onto a common reference system using the patient MRI images and registration algorithms. Once normalized, the data can be analyzed through various queries.
The CranialCloud consists of a set of distributed CranialVault nodes on the network, also called CranilVault Boxes.
Each CranialVault node uses a subject-centric model to organize data. The subject is considered the unique root object and all other data are dependent on it. A unique identifier (UID) is assigned to each subject in the database. The UID is a combination of a unique number that identifies a specific node (UNI) and a unique id within the node generated locally to make each patient unique in the entire CranialCloud domain. PHI information is separated from any other information and kept in a highly encrypted section of the CranialVault database. Only accredited medical teams can access the PHI information related to the patient data. Patients are enrolled in studies defined as ethically (IRB) approved protocols. The IRB information and accesses to the de-identified data are managed locally at the level of each node.
The CranialVault system requires at least one reference image for each subject to that is used for normalization purposes (see [12] for details). Therefore, each patient is attached to a primary reference image and multiple additional imaging studies.
Each data point, localized in patient-space, is identified by a unique point ID. During the normalization process, the coordinates from all the points are projected using the transformation computed by the registration algorithms in the pipeline module and the results are stored in specific tables. The ID of the point is associated with its normalized coordinates. Meta-data can therefore be easily analyzed in any reference system.
Subject Uniqueness
Following the DICOM format for generating unique identifiers, each subject UID is composed of two parts, an <org root> and a <suffix> (UID = <org root>.<suffix>). The <org root> portion of the UID uniquely identifies a CranialVault Node, and is composed of a number of numeric components as defined by ISO 8824. The <suffix> portion of the UID is also composed of a number of numeric components, and is unique within the scope of the <org root>. Subjects are unique within a node based on name, medical record number, date of birth, and gender. While there is a risk of creating two entries within the CranialCloud for a patient that would register in two different CranialVault nodes, in most instances, these criteria will ensure data integrity with limited chance for error because of duplication.
CranialVault Node System Architecture
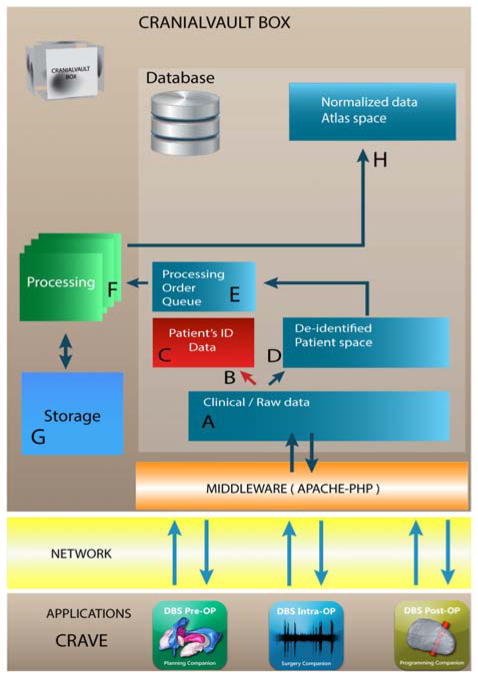
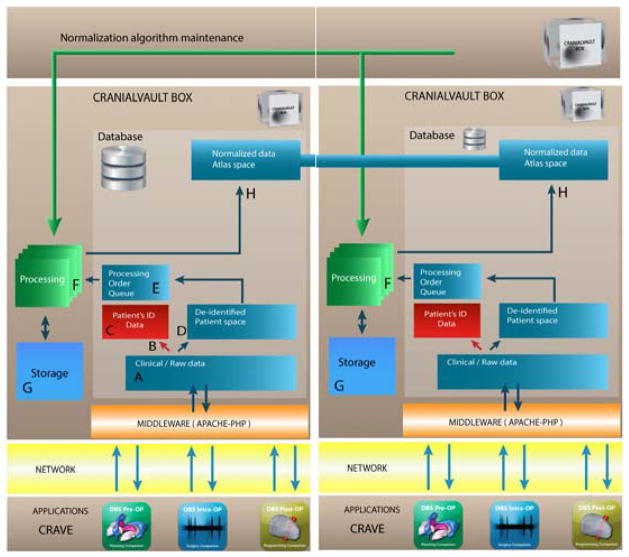
Each CranialVault system node uses a multi-tier architecture. Figure 2 illustrates the flow. The client applications layer such as the CRAVE (CRAnialVault Explorer) modules[12], communicate with the database through the middleware layer that consists of a PHP-Apache interface that prepare the data for and from the database. The data (A-D) is then de-identified and stored within the database. The data are then normalized in the processing tier (F) before being stored and prepared for data sharing (H). While historically, the database system has been an Oracle Enterprise database (currently running Oracle 12c), another version of the system has been developed using MySQL essentially to offer an alternative to expensive licenses costs. With minor changes MySQL could be exchanged for any other RDBMS. While the system has been designed in a way that does not depend on the database system used, Oracle offers additional options and advanced security that MySQL does not. To that extent, we have decided to keep both versions.
Figure 2.
The data is captured from the clinical flow with applications compatible with the CranialVault such as the CRAVE modules. The applications communicate with the CranialVault Box through secure channels to a middleware layer (Apache/PHP) to send data to the database and storage. PHI information is separated from the rest of the clinical data and stored in a secure location, patient de-identified data is then processed through a pipeline module to be normalized onto the CranialVault Atlas.
PHP is used as the client–server based language to handle the communication between the client applications and the database. Using a web-based approach as the front end helps transferring data through very secure institutions without the need to open new ports into firewalls, thus facilitating deployment.
Processing is done through the use of the pipeline module and an asynchronous queuing model. Once patient’s data is added to the database, a processing order is added to the Processing Queue. When first run, a pipeline instance checks for any unprocessed order in the queue and consumes the next order. The system can scale easily by adding processing pipelines that can all subscribe to the processing queue and process orders simultaneously. The processing pipeline runs a sequence of registrations algorithms to normalize patient’s data onto a common reference system as described in details in [12]. In order to maintain the best normalization service, the CranialVault architecture has been designed to permit replacing current algorithms with better, more efficient ones as they are developed. Algorithms are stored with a definition of their parameters in a specific database table and can be called automatically through a generic procedure.
Data organization
As introduce before the Cranialvault handles 1) “non-localized” patient data, including historical or current clinical data, such as demographics, drugs, disease state, imaging, or longitudinal observations through scores of pre-defined rating scales such as the unified Parkinson’s disease rating scale (UPDRS); 2) “localized” data or data that can be associated with a specific location in the brain, such as the recording of the neuronal activity at a specific location or the response of the patient to intra-cranial stimulation; and 3) multiple data normalization algorithms and atlases. Moreover, the Cranialvault should also permit to access identified or de-identified patient data depending on the credentials and applications.
The patient data is organized in a relational model where the patient ID data is isolated from all other information by means of a specific table. A virtual private database (VPD) masks data from that table so that only a subset of the data appears to exist depending on the user’s credentials. This prevents HIPAA sensitive data to be accessed by everyone but also allows more privileged users to access the whole table.
Non-localized data are then stored by type in different tables: the images table will store all information pertaining to data acquisition as well as a pointer to raw image data, the demographics table will contain any static information about the patient (gender, handedness, …), scores from user-defined or global rating scales are stored in a third tables system. Each of these tables is structured in a very scalable way to allow users of the CranialVault to store any similar data without the need to define new columns or tables.
Localized data follows the following scheme. Each data point is kept in a records table that stores x, y, z coordinates in patient-space and assign a unique record id. The associated recorded data is then split through different tables based on the type of information. While designing the database we split records into very general types to avoid being limited to DBS procedures. These are for instance response to stimulation, signals (micro-electrode recordings, SEEG, …) or implants.
Because multiple algorithms exists that can normalize data onto multiple atlases, the database stores algorithms within the database in a specific table and store the output transformations they create in another. Each algorithm can be associated with one or more set of parameters. Today, the Cranialvault has the following algorithms: ABA[13], SyN[14], F3D[15] and ART[16] as they are among the most efficient and robust ones [17].
Once normalized onto one or more atlases, coordinates of the records are stored in an atlas_record table. One benefit of this approach is that patient data can be accessed either in patient or atlas space.
CranialCloud and Data security
Nodes can either be hosted physically within a specific medical or research institution or deployed in a public cloud such as the Amazon or Google Cloud. As opposed to CranialVault boxes that are physically deployed in the IT architecture of a medical center, a CranialVault box deployed in a public cloud such as the Amazon Cloud is exposed to usual cloud risks including hacking, rogue administrators, accidents, complicit service provides, and snooping governments. Client-side cryptography is a good solution to these issues as it allows users to protect their own data with individual, per-file encryption and protect access to that data with user-controlled keys. The system uses an AES-256 encryption mechanism. The encryption, decryption and key management are all done on the end user’s computer or device, meaning the data in the cloud only exists in its encrypted state.
As explained earlier, the CranialVault database stores two kind of data: data that contains patient identifier to give clinician providing care to the patient access to the patient information, and de-identified patient data that could be shared after normalization with specific users. The CranialVault Cloud system uses a mix of shared and unshared keys: a unique key that is generated for each CranialVault node and is kept within the client institution to provide access to the patient data, while another key is shared among all CranialVault nodes to allow the decryption of de-identified data.
Data Sharing
All nodes within the CranialCloud consist of CranialVault Boxes. All boxes are self-sufficient and have the same functionality. In addition, each node has the capability to connect to others to enable data sharing of de-identified and normalized data.
Sharing normalized data is a concept different than sharing raw data: it involves 1) normalization of the data and, 2) a common reference system. Sharing a common and unique normalization method and a common reference system are thus crucial for this project. In order to maintain the capability to compare data generated from different CranialVault nodes, all nodes have to stay in sync, i.e., the same normalization protocol needs to be used by each box.
In order to achieve this, each node is registered to a central node, called the CranialVault Core. Its role is to update the node periodically with database structural changes, new processing algorithms and to update the list of existing nodes with their addresses in order for the node to stay connected with the complete CranialCloud network. While the CranialVault Core is based on the same 251722752architecture as a node, its purpose is not to store any data but only to maintain other nodes centrally.
A decentralized model is used for data sharing. Once connected to the CranialVault Core node, each individual site has access to information about other nodes on the network and can share data without interacting with a central repository.
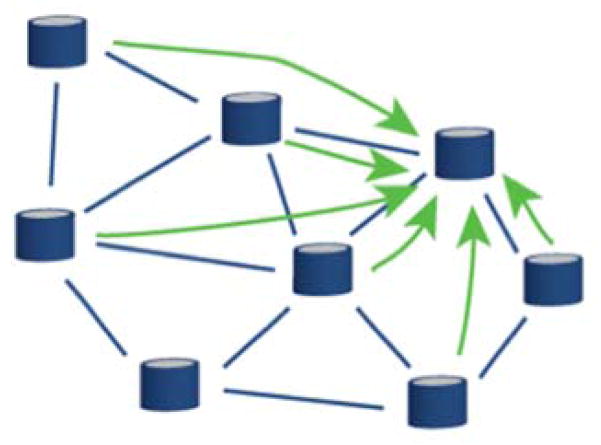
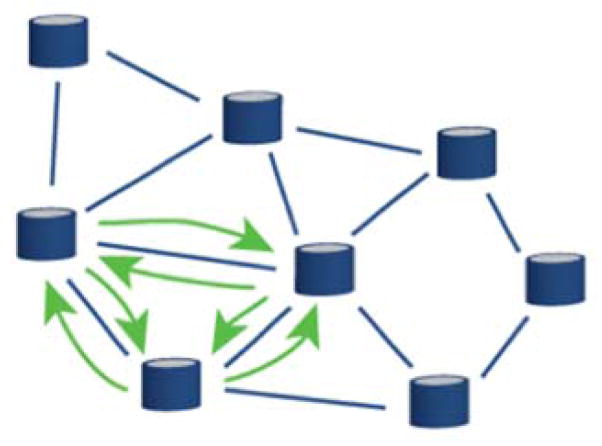
Data is shared between nodes using a three-step process: 1) the sharing site creates a research study associated with the data to be shared, 2) the sharing site selects the node(s) and user(s) with whom to share the data, 3) views are automatically created from queries generated to fetch the shared data from each node and results are only available to the members of the study across institution. Sharing can be done in two ways (Figures 3 & 4): either unidirectionnally, i.e., a group shares its findings with others on a read-only mode; or, bidirectionally, i.e., multiple groups collaborate on a specific study, each bringing data that can be aggregated with data brought in by the other members.
Figure 3.
Unidirectional sharing. A single node, with proper privileges, can query the CranialCloud with a specific request. Data will then be fetched through the Cloud and returned to the instance initiating the query.
Figure 4.
Bidirectional sharing. Multiple instances share data. Each node can query the Cloud with a specific request. Data is fetched across each participating node transparently.
Advantages of a decentralized system are: 1) data sharing is the responsibility of the group sharing the data 2) no PHI or image data is shared, only de-identified patient information normalized in a common reference system 3) connections between groups are only known by the nodes involved.
Querying normalized data onto the Cranialvault atlas
Data can be queried using several criteria regrouping subject, study or any specific patient information. By default, all searches are cross-projects, with the only limitation being access permissions.
Analyzing neurodegenerative disease-related data often consists of comparing regions or locations within the patient’s brain that correspond to an observation. While the normalization of the data allows comparing points located in brains with different anatomies through the use of a common reference atlas, multiple references currently exist without a single one being recognized as the universal reference. Research in the area of data normalization also suggests that the use of multiple atlases improves the results of data normalization. The CranialVault system has been designed to handle multiple atlases that can be updated through the use of the CranialVault Core. At the time of writing, the CranialVault system uses 10 atlases built from the brain of 5 male and 5 female individual subjects ranging form 30 to 55 y/o. Each atlas consists of multiple MRI sequences acquired both on 3T and high-field 7T scanners. To provide anatomical and white matter tracts information, each atlas has been segmented carefully following a similar method as described in [18]. Major fiber tracts such as the corticospinal tract were extracted by experts using DTI studio and following approaches described in [19], [20].
While the results of each query can be exported for statistical analysis in system such as R, excel or MATLAB, the visualization of the results is done using the CRAVE Atlas module. The CRAVE Atlas module is a graphical user interface that allows each user to create his/her own database requests, set up the access and privileges, and visualize the results. An interface lets the user create new queries, save them as Reports and manage access to the data. Depending on access, these reports will fetch data from the local node or the CranialCloud.
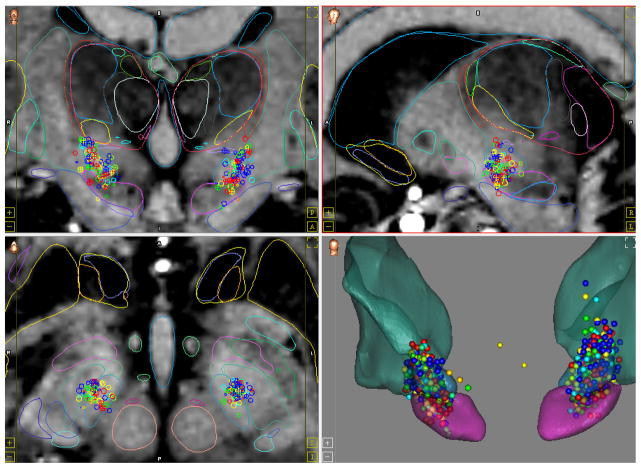
Data returned from the queries are then shown as points as illustrated in Figure 6, or statistical maps on top of the atlas MRI and the atlas anatomical structures. Multiple reports can be shown simultaneously for data comparison. To illustrate this by means of an example, a user could create two reports showing the normalized lead positions for patients suffering from Parkinson’s disease, one for good and the other for bad responders. While this example is trivial, queries can be as complex as the data will support. Once a report is created, it can be automatically updated with new patients added in the CranialCloud.
Figure 6.
CranialVault Atlas module visualization interface. This particular set of data represents normalized intra-operative stimulations. Each sphere is color coded to reflect how well the symptoms of the patient are controlled by the stimulation: from blue: no control on symptoms to red: full control of the symptoms. The data are shown as cluster of points on top of the atlas high field MRI and anatomical structures segmentations.
CranialCloud Governance
It is reasonable that most of the services managed by the Cranialvault Core, including updates of algorithms and database structural changes should be provided by a small core team of theoretical neuroscientists, imaging scientists, and image processors. Thus core services are being developed to implement and administer standardized procedures that can be used across resources that are shared through the CranialCloud. Further, for steering and evaluation of the activities a governance board will be established. The board will include noninvolved experts in the field, engineers, database experts, experimental and theoretical neuroscientists, some active resource contributors, representatives of related activities and other experts. The board’s responsibilities will be to guide important decisions concerning the core services. Conversely, the experience gathered in this data-sharing project will be brought back to the relevant communities in workshops and publications.
Results
Vanderbilt University has been the primary site for the CranialVault development and has been converted into a CranialVault node recently. Ohio State University Medical Center, Wake Forest University Medical Center, the VA in Richmond, which were part of the initial CranialVault consortium now have their individual node. Some of them are still hosted at Vanderbilt University while waiting to be transferred inside the institution. While historical the Cranialvault focused on DBS-related data, we have started to broaden the spectrum of use of the Cranialvault to other disorders such as epilepsy.
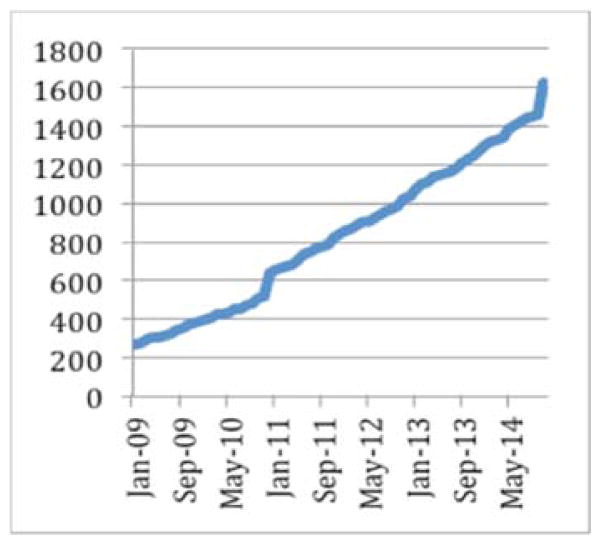
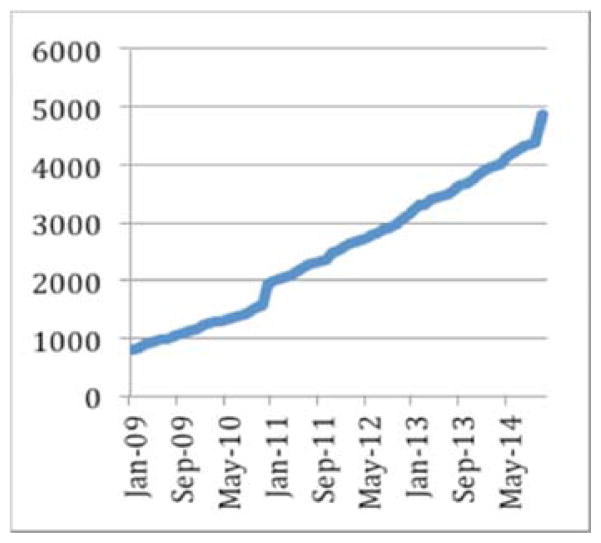
The CranialCloud has normalized data related to more than 1600 patients, generating 32,000 registrations and 5 terabytes of raw data. The CranialCloud manages more than 51,000 intra-operative records related to observations such as patient’s responses to stimulation, micro-electrode recordings that can be correlated to patient demographics, motor and/or cognitive scores to answer specific questions. The DBS application generates on average 32 new subjects each months resulting in 900 Gb of results generated in 16 hours of CPU time.
Conclusion
In recent years, scientists have made great strides in understanding the underlying causes of neurodegenerative diseases. As we uncover more details about these diseases, we hope that, in time, we will be able to better treat and possibly prevent them. As in most major endeavors, the greater the resources we bring to bear, the faster the progress toward our goal. If we want to avoid the looming and dramatic impact of these diseases on our increasingly elderly population, we need as a research community to share data to reach statistical significance within our analyses in the hope of finding solutions. In the process of creating a database that satisfies our local needs, we were able to create a system that we believe will satisfy the needs of many other researchers and research groups, and solve many of the challenges related to data sharing for neurodegenerative diseases. While we hope that the CranialCloud will speed the process of scientific discovery by making data available across institution, this system could also be used by any images or signals processors, to test and compare their algorithms on data of variable quality acquired with different protocols from all around the world. The governance board could then decide to upgrade the processing algorithms of the CranialCloud to the best preforming ones. We are currently working on the design to make this possible without the need to transfer image or signal data from each institution to the testing one. Tests we have done so far allow us to automatically run algorithms on a large set of data using Vanderbilt’s ACCRE (Advanced Computing Center for Research and Education) high performance computing cluster and we are exploring options to run algorithms in the Cloud.
Today, the system has been deployed at a limited number of sites because its installation still requires technical expertise and resources; it is currently done by the Vanderbilt team with the local IT team. Each site manages its own ethics requirements and patients’ consents, through its local ethics board. Each group is also added onto the main IRB managed by Vanderbilt University. Until now, we have worked with teams and institutions that are willing to invested some IT resources and who had or were willing to use the CRAVE tools in their clinical flow for non-disruptively acquire the data. We will continue to work with new group who are willing to do the same as our resources permit. Our goal is to reach a point at which our system can be installed and managed with minimum local technical expertise, thus permitting its wide dissemination.
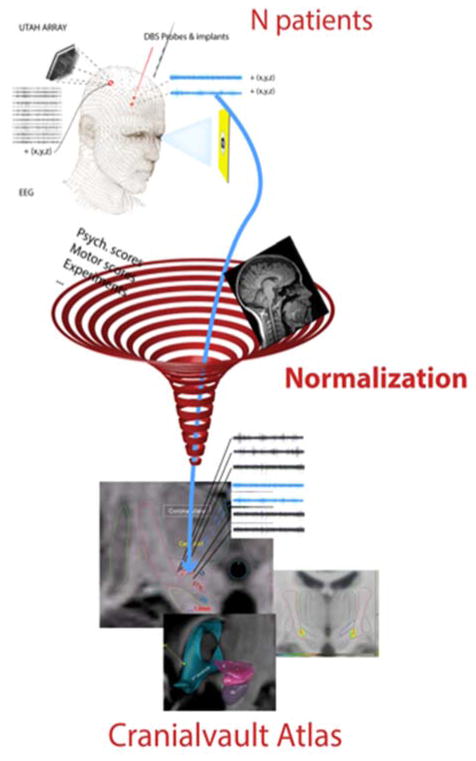
Figure 1.
CranialVault system: any data that can be related to a location within the patient’s MRI is normalized into a common reference system called the atlas for statistical analysis and 3D visualization.
Figure 5.
Each Cranialvault Box/Node acquires the data from the clinical flow, de-identifies and normalizes to prepare it for sharing with other nodes. Each node shares common processing algorithms and reference system that can be updated simultaneously from the Cranialvault Core node.
Figure 7.
Evolution of the number of subjects in the Cranialvault system
Figure 8.
Evolution of the data stored in the CranialVault system in GB.
Acknowledgments
Features and ideas for the CranialVault database were conceived by many individuals over the course of its development. We thank the entire DBS Vanderbilt group including engineers, neurosurgeons, electrophysiologists, and neurologists who contributed to its development. Additionally, this effort would have not been successful without the heavy involvement of our partners from OSU (Dr. A. Rezai), the VA in Richmond, VA (Dr. K. Holloway), Wake Forest University (Dr. S. Tatter) and Jefferson University (Dr. A. Sharan). This research has been supported, in parts, by the grant NIH R01 EB006136.
Footnotes
Conflict of interest: Authors P-F DHaese, P. Konrad and B. Dawant are co-founder and own stocks in Neurotargeting LLC. Authors S. Pallavaram and R.Li own stock in Neurotargeting LLC.
Ethical approval: “All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.”
References
- 1.Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5 (1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 2.Book GA, Anderson BM, Stevens MC, Glahn DC, Assaf M, Pearlson GD. Neuroinformatics Database (NiDB)--a modular, portable database for the storage, analysis, and sharing of neuroimaging data. Neuroinformatics. 2013;11 (4):495–505. doi: 10.1007/s12021-013-9194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das S, Zijdenbos AP, Harlap J, Vins D, Evans AC. LORIS: a web-based data management system for multi-center studies. Frontiers in neuroinformatics. 2011;5:37. doi: 10.3389/fninf.2011.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott A, Courtney W, Wood D, de la Garza R, Lane S, King M, Wang R, Roberts J, Turner JA, Calhoun VD. COINS: An Innovative Informatics and Neuroimaging Tool Suite Built for Large Heterogeneous Datasets. Frontiers in neuroinformatics. 2011;5:33. doi: 10.3389/fninf.2011.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2005;1 (1):55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ascoli GA. The ups and downs of neuroscience shares. Neuroinformatics. 2006;4 (3):213–216. doi: 10.1385/NI:4:3:213. [DOI] [PubMed] [Google Scholar]
- 7.Ascoli GA. A community spring for neuroscience data sharing. Neuroinformatics. 2014;12 (4):509–511. doi: 10.1007/s12021-014-9246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotter R. Online retrieval, processing, and visualization of primate connectivity data from the CoCoMac database. Neuroinformatics. 2004;2 (2):127–144. doi: 10.1385/NI:2:2:127. [DOI] [PubMed] [Google Scholar]
- 9.Van Horn JD, Ishai A. Mapping the human brain: new insights from FMRI data sharing. Neuroinformatics. 2007;5 (3):146–153. doi: 10.1007/s12021-007-0011-6. [DOI] [PubMed] [Google Scholar]
- 10.Teeters JL, Harris KD, Millman KJ, Olshausen BA, Sommer FT. Data sharing for computational neuroscience. Neuroinformatics. 2008;6 (1):47–55. doi: 10.1007/s12021-008-9009-y. [DOI] [PubMed] [Google Scholar]
- 11.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of biomedical informatics. 2009;42 (2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Haese PF, Pallavaram S, Li R, Remple MS, Kao C, Neimat JS, Konrad PE, Dawant BM. CranialVault and its CRAVE tools: a clinical computer assistance system for deep brain stimulation (DBS) therapy. Medical image analysis. 2012;16 (3):744–753. doi: 10.1016/j.media.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohde GK, Aldroubi A, Dawant BM. The adaptive bases algorithm for intensity-based nonrigid image registration. IEEE transactions on medical imaging. 2003;22 (11):1470–1479. doi: 10.1109/tmi.2003.819299. [DOI] [PubMed] [Google Scholar]
- 14.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis. 2008;12 (1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S. Fast free-form deformation using graphics processing units. Computer methods and programs in biomedicine. 2010;98 (3):278–284. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Ardekani BA, Guckemus S, Bachman A, Hoptman MJ, Wojtaszek M, Nierenberg J. Quantitative comparison of algorithms for inter-subject registration of 3D volumetric brain MRI scans. Journal of neuroscience methods. 2005;142 (1):67–76. doi: 10.1016/j.jneumeth.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage. 2009;46 (3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. S1053-8119(08)01297-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tourdias T, Saranathan M, Levesque IR, Su J, Rutt BK. Visualization of intra-thalamic nuclei with optimized white-matter-nulled MPRAGE at 7T. NeuroImage. 2014;84:534–545. doi: 10.1016/j.neuroimage.2013.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, van Zijl P, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. NeuroImage. 2007;36 (3):630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suarez RO, Commowick O, Prabhu SP, Warfield SK. Automated delineation of white matter fiber tracts with a multiple region-of-interest approach. NeuroImage. 2012;59 (4):3690–3700. doi: 10.1016/j.neuroimage.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]