Abstract
The down regulation of glutamic acid decarboxylase67 (GAD1), reelin (RELN), and BDNF expression in brain of schizophrenia (SZ) and bipolar (BP) disorder patients is associated with overexpression of DNA methyltransferase1 (DNMT1) and ten-eleven translocase methylcytosine dioxygenase1 (TET1). DNMT1 and TET1 belong to families of enzymes that methylate and hydroxymethylate cytosines located proximal to and within cytosine phosphodiester guanine (CpG) islands of many gene promoters, respectively. Altered promoter methylation may be one mechanism underlying the down-regulation of GABAergic and glutamatergic gene expression. However, recent reports suggest that both DNMT1 and TET1 directly bind to unmethylated CpG rich promoters through their respective Zinc Finger (ZF-CXXC) domains. We report here, that the binding of DNMT1 to GABAergic (GAD1, RELN) and glutamatergic (BDNF-IX) promoters is increased in SZ and BP disorder patients and this increase does not necessarily correlate with enrichment in promoter methylation. The increased DNMT1 binding to these promoter regions is detected in the cortex but not in the cerebellum of SZ and BP disorder patients, suggesting a brain region and neuron specific dependent mechanism. Increased binding of DNMT1 positively correlates with increased expression of DNMT1 and with increased binding of MBD2. In contrast, the binding of TET1 to RELN, GAD1 and BDNF-IX promoters failed to change. These data are consistent with the hypothesis that the down-regulation of specific GABAergic and glutamatergic genes in SZ and BP disorder patients may be mediated, at least in part, by a brain region specific and neuronal-activity dependent DNMT1 action that is likely independent of its DNA methylation activity.
Keywords: epigenetics, methylation, schizophrenia, bipolar disorder, glutamate decarboxylase, DNMT1, TET1
1. Introduction
When post-mortem brains of schizophrenia (SZ) and bipolar disorder (BP) patients are compared with those of non-psychiatric subjects (NPS), GABAergic and glutamatergic neuropathologies are detected in the hippocampus and cortex (Akbarian et al.,1995, Benes et al.,1992, 2001, 2007, Fatemi et al., 2000, Ikegame et al.2013, Impagnatiello et al.,1998, Guidotti et al., 2000; Lewis et al., 2005, Mill et al., 2008, Weickert et al., 2003). These neuropathologies are characterized by a decrease in the expression of glutamic acid decarboxylase 67 (GAD1), reelin (RELN), nicotinic acetylcholine receptors, glutamate receptors, and tyrosine kinase receptors in GABAergic neurons (for a review see Guidotti et al., 2011; Grayson and Guidotti 2013), and brain derived neurotrophic factor (BDNF) and vesicular glutamate transporter 1 (VGLUT1) in glutamatergic neurons (Weickert et al., 2003; Mill et al., 2008; Ray et al., 2014).
Population, family, and twin studies indicate that SZ and BP are highly heritable, neuropsychiatric diagnoses. Single alleles conferring increased risk have been identified but only account for small proportion of observed phenotypic variants (Li et al., 2010, Sullivan et al., 2008, Richards et al., 2012). Hence, it appears that these disorders are the consequence of synergistic interactions of multiple susceptibility genes with environmental neuroepigenetic factors (Costa et al., 2002; Ptak and Petronis 2008). In support of a role for aberrant epigenetic mechanisms in the pathogenesis of altered GABAergic and glutamatergic gene regulation in SZ and BP disorders, we have recently reported that the down-regulation of GAD1, RELN, and BDNF-IX expression in brains of SZ and BP patients is associated with increased expression of DNA methyltransferases 1 (DNMT1) and Tet-methylcytosine dioxygenase 1 (TET1), and additional downstream alterations in the DNA demethylation pathway associated with various target genes (for a review see Grayson and Guidotti, 2013).
DNMTs and TETs each represent a distinct family of enzymes that methylate and hydroxymethylate the five position of cytosines, respectively, when present in shores of so-called CpG islands (CpGIs). Studies suggest that changes in methylation and/or hydroxymethylation, when associated with promoter domains, modulate transcription by altering local chromatin organization and nucleosome positioning (for a review see Guidotti et al., 2011, Grayson and Guidotti 2013). In SZ post-mortem brain, altered DNA methylation and hydroxymethylation marks at the promoters of RELN (Abdolmaleky et al., 2005; Grayson et al., 2005), GAD1 (Dong et al 2012), COMT (Abdolmaleky et al 2011), BDNF (Mill et al., 2008; Gavin et al., 2012; Ikegame et al., 2013), and glucocorticoid receptors (NR3C1, Zhang et al., 2013) have been reported. Genome-wide promoter methylation analyses of post-mortem cortical DNA showed altered methylation associated with some 817 promoters including nitric oxide synthase (NOS1), v-akt thymoma viral oncogene homologue-1 (AKT1), DNMT1, dystrobrevin binding protein 1 (DTNBP10, protein phosphatase 3, catalytic subunit gamma isozyme (PPP3CC), and SRY (sex determining region Y) box 10 (SOX10) (Wockner et al., 2014).
These alterations are the product of a dynamic balance between DNA methylation and demethylation. In fact, the regulation of both hyper- and hypo-methylated genomic DNA is under the control of complex networks of methylating, hydroxymethylating and demethylating enzymes and proteins. For example, 5-methylcytosine (5mC) at specific promoters can be oxidized forming 5-hydroxymethylcytosine (5hmC) by members of the TET family of proteins in mammalian brains (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). In addition, 5hmC is further oxidized by TET family members forming 5-formylcytosine (5fC) and 5- carboxycytosine (5caC) (Ito et al., 2011; Yu et al., 2012; Cadet and Wagner, 2013). Both 5-fC and 5-caC are specifically recognized by thymine deglycosylase (TDG) producing abasic sites which are replaced by base excision repair (BER) enzymes forming unmodified cytosine (He et al., 2011; Maiti and Drohat, 2011; Hashimoto et al., 2012; Shen et al., 2014). The sequential deamination and repair of 5hmC by activation-induced cytidine deaminase (AID)/apolipoprotein B editing complex (APOBEC) and BER enzymes has been proposed (Guo et al., 2011), although AID/APOBEC enzymes do not appear to use double-stranded 5hmC-containing DNA as a substrate (Wu and Zhang, 2011; Shen et al., 2014). The growth and arrest and DNA damage inducible (GADD45) proteins have been implicated in the targeting of gene-specific DNA demethylation to specific genes in response to neuronal activity (Ma et al., 2007). While DNA demethylation is critical during neurodevelopment, the extent and frequency of active demethylation and the pathways utilized in adult brain are incompletely understood.
Although increases in promoter methylation/hypermethylation catalyzed by the overexpression of DNMT1 or TET1, respectively, may be one mechanism underlying the downregulation of GABAergic, glutamatergic and other gene targets in SZ and BP patient brain, the inhibitory action of DNMT1 and TET1 on gene expression could be the consequence of an interaction between the ZF-CXXC (zinc finger-CXXC) domains of DNMT1 and TET1 binding CpG dinucleotides as recognition sites (Long et al., 2013). The ZF-CXXC domain is a short (35–42 amino acid) polypeptide stretch found in numerous Zn-finger proteins that bind non-methylated CpGs at CpG islands (Long et al., 2013). In addition to DNMT1 and TET1, the domain is present in several additional chromatin modifiers, such as histone lysine demethylases (KDM2A and 2B), histone H3K4 methyltransferase (MLL1), methyl-binding domain protein 1 (MBD1) and the CXXC finger protein 1 (CFP1), that couple various DNA and histone modifications to CpG islands. For example, TET1 acts as a maintenance DNA demethylase that does not decrease methylation levels per se, but specifically prevents aberrant gene-specific methylation spreading into CpG islands in differentiated cells (Williams et al., 2012; Jin et al 2014). Moreover, DNMT1 and TET1 target additional chromatin-modifying activities, including methyl CpG binding protein 2 (MeCP2) and methyl binding domain protein 2 (MBD2) to CpG rich promoter regions at selected genes through protein interacting domains. The ability of DNMT1 and TET1 to bind to candidate risk genes in post-mortem brain of SZ patients or to form complexes with other chromatin remodeling proteins such as MBD2 has not, until now, been systemically studied.
2. Methods and Materials
2.1 Demographic Characteristics
We obtained fresh-frozen PFC (BA9) and cerebellar tissue from the Harvard Brain Tissue Resource Center, McLean Hospital (Belmont, MA, USA). All samples were obtained from family-referred, community-based cases, and none were referred by a medical examiner’s office. The demographics associated with each patient population are presented in Table 1. The methods, of tissue harvest, preparation and storage have been described in detail elsewhere (Veldic et al., 2004; 2005; 2007). As shown in Table 1 we find no significant diagnostic differences in post-mortem interval, pH, or age. The psychiatric diagnoses were established by two senior psychiatrists based on clinical and family histories and according to criteria in the Diagnostic and Statistical Manual of Mental Disorders IV. From the available neuropathology reports there were no signs of infarction, hemorrhage or inflammatory lesions. In addition, all control cases were free of neurological disorders, seizures, mental retardation, dementia, and metabolic disorders based on medical records.
Table 1.
Demographic Characteristics of the Study Groups A
| NPS (n = 32) | Patient Cohort
|
Statistical Analysis | ||
|---|---|---|---|---|
| BP (n = 20) | SZ (n = 22) | |||
| M/F ratio | 20/12 | 5/15 | 15/7 | χ22 = 9.49, P = .009 |
| Age, y | 59.2 ± 21.4 | 59.6 ± 14.8 | 61.0 ± 11.7 | F2, 73 = .06, P = .94B |
| Postmorterm interval, h | 21.8 ± 3.6 | 22.2 ± 5.1 | 23.9 ± 7.3 | F2, 73 = .45, P = .64B |
| Brain pH | 6.4 ± 0.3 | 6.6 ± 0.3 | 6.5 ± 0.3 | F2, 73 = .47, P = .63B |
| Cause of death | ||||
| Cardiopulmonary | 29 | 6 | 9 | χ26 = 66.6, P = .000 |
| Other | 3 | 14 | 13 | χ26 = 61.2, P = .000 |
| Antipsychotic drug useC, D | 0 | 17 | 18 | χ26 = 85.4, P = .000 |
| AntidepressantC, E | 0 | 1 | 0 | |
| Mood stabilizersC,F | 0 | 12 | 5 | χ26 = 48.4, P = .000 |
| Abuse or dependence | ||||
| Alcohol | 3 | 8 | 5 | χ26 = 93.2, P = .000 |
| Cocaine | 2 | 3 | 2 | χ26 = 87.2, P = .000 |
| Tobacco | 11 | 7 | 6 | χ26 = 87.1, P = .000 |
Values are expressed as mean ± SD
One-way analysis of variance among patient cohorts
Present to the time of death
Includes the following (Typical neuroleptics): haloperidol, fluphenazine, thioridazine, perphenazine, trifluoperazine, chlorpromazine and molindone; (Atypical neuroletptics): clozapine, olanzapine, aripiprazole, risperidone and quetiapine.
Includes: the following: trazodone, bupropion and venlafaxine.
Includes: lithium and valproic acid.
2.2 Quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR)
QPCR was carried out using the Applied Biosystems Real-Time PCR System with a SYBR green master mix (Fermentas, Glen Burnie, MD). Total RNA was isolated from brain samples using TRIZOL reagent (Life Technologies, Grand Island, NY), which was further purified using the Qiagen RNeasy kit (Qiagen, Valencia, CA). Primer sequences for the genes analyzed are shown (Table S1 in Supplemental information). Each sample was run in duplicate and repeated twice. For normalizing mRNA expression, several housekeeping genes: Enolase 2 (ENO2), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and Beta-actin (ACTB) were chosen as internal controls. For each control, we measured the gene stability ranking using the NormFinder algorithm (Dong et al., 2012). This protocol allows for the identification of the housekeeping gene best suited for normalization. Because each of the genes studied yielded similar results when normalized to either ENO2, GAPDH, or ACTB, and because ACTB had the highest housekeeping gene stability (NormFinder), we normalized our data to ACTB.
2.3 Western blot analysis
For protein quantification we conducted measurements as described in detail elsewhere (Dong et al., 2012). Anti-DNMT1 monoclonal antibodies (0.5 μg/ml, Imagenex, San Diego, CA), or anti-TET1 monoclonal antibodies (Zymo Rearch Irvine, CA), or MBD2 polyclonal antibody (Millipore, Billerica, MA) were used to detect the corresponding proteins. The levels of these proteins in NPS, BP or SZ were normalized to β-actin protein levels.
2.4 Chromatin immunoprecipitation assays
We performed chromatin immunoprecipitation (ChIP) based on protocols previously described (Dong et al., 2012). The percentages of immunoprecipitated DNA were calculated using the following: % (IP/total input)=2(Ct(10% input)−3.32)−Ct(IP) × 100%. ChIP grade anti-DNMT1 (Imgenex, San Diego, CA), anti-TET1 monoclonal antibody (Zymo, Irving, CA, USA) and anti-MBD2 (Millipore, Billerica, MA) were used to precipitate cross-linked chromatin. The primer sequences for RELN, GAD1, GAD2, BDNF, GAPDH are summarized in Table S1, supplemental materials.
2.5 Statistical analysis
Data were analyzed using the Boxplot of PASW v.18 software (SPSS, Chicago, IL, USA) to identify outliers, which were excluded for further statistical analysis. One-way ANOVA and analysis of covariance with Bonferroni post-hoc comparisons were performed for determining significance of mRNA content; binding of DNMT1, TET1 and MBD2 at selected gene promoters; DNMT1, TET1 and MBD2 protein content; medication use, age, gender, post-mortem interval, substance use and alcohol abuse, between the three groups (NPS, BP and SZ), using PASW v.18 software. In all samples, P-values were two-tailed, and comparisons were considered to be statistically significant at P<0.05 level using Bonferroni’s multiple comparison correction. Relationships between DNMT1 mRNA and DNMT1 ChIP at the GAD1, RELN, or BDNF-IX promoters were analyzed with Pearson correlations and scatter plots.
3. RESULTS
3.1 The expression (mRNA and protein levels) of DNMT1 and TET1 is increased in PFC of SZ and BP disorder patients
In previous studies, we showed that DNMT1 and TET 1 mRNAs and proteins are increased in the neocortex of SZ and BP disorder patients (Dong et al., 2012, Ruzicka, et al., 2007, Veldic et al., 2004). Using a completely new cohort of post-mortem brain tissue, patients diagnosed with SZ (n= 22), BP disorder (n=20), and a group of non-psychiatric subjects (NPS), (n=32), we confirmed previous reports showing DNMT1 mRNA (overall ANOVA, F2,71; p = 0.002, followed by Bonferroni comparison: p= 0.003 SZ vs NPS; p=0.031 BP vs NPS), and TET1 mRNA (overall ANOVA, p= 0.01, followed by Bonferroni comparison: p= 0.023 SZ vs NPS; p= 0.042 BP vs NPS) are increased by 30–40% in the PFC of these patients (Table 2). DNMT1 and TET1 protein levels are also increased in SZ and BP patients (Table 2).
Table 2.
Expression of DNMT1 and TET1 in the PFC (Brodmann area 9) of schizophrenia (SZ) and bipolar disorder (BP) patients
| Gene | NPS (32) |
Patient cohort
|
Overall ANOVA
|
||||
|---|---|---|---|---|---|---|---|
| SZ (22) | BP (20) | F | d.f. | P-value | |||
| DNMT1 | mRNA | 0.062 ± 0.000 | 0.086 ± 0.007* | 0.082 ± 0.001* | 6.965 | 2 | 0.002 |
| protein | 0.019 ± 0.004 | 0.039 ± 0.005* | 0.030 ± 0.006 | 5.098 | 2 | 0.010 | |
| TET1 | mRNA | 0.065 ± 0.001 | 0.094 ± 0.013* | 0.093 ± 0.001* | 4.969 | 2 | 0.010 |
| protein | 0.28 ± 0.02 | 0.50 ± 0.07* | 0.48 ± 0.05* | 5.035 | 2 | 0.011 | |
mRNA = The values were normalized to β-actin; Protein = The values were normalized by β-actin.
Denotes: significant difference from NPS. One-way ANOVA followed by Boferroni comparison.
Note: 2 sample from NPS and 1 sample from SZ were excluded from statistical analysis because they were identified as outliers when analyzed using the Boxplot of PASW v18 software (see method).
3.2 Higher DNMT1 but not TET1 binding at GAD1, RELN, and BDNF IX promoters in PFC of SZ and BP disorder patients
We and others (Abdolmaleky et al., 2005; Dong et al., 2012 Gavin et al., 2012; Guidotti et al 2000, Grayson et al., 2005; Ikegame et al., 2013) have shown that GAD1, RELN, and BDNF-IX CpG rich promoter regions exhibit hypermethylation or hydroxymethylation in the neocortex of SZ and BP disorder patients. Moreover, the expression of these genes is also reduced.
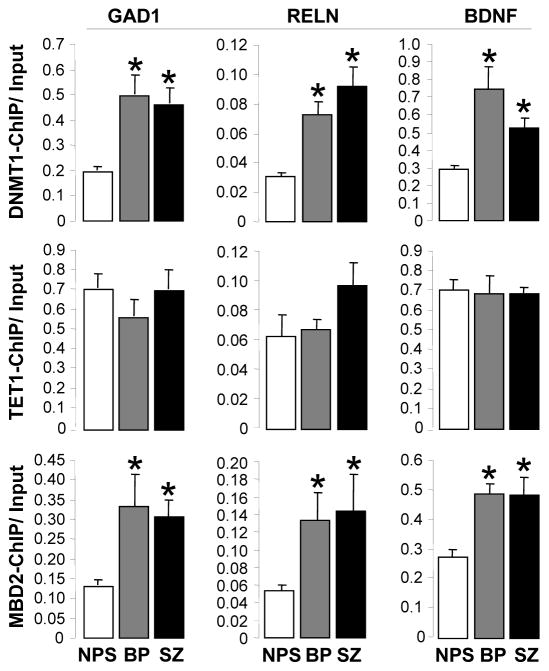
Using ChIP assays with specific antibodies (see methods), we demonstrated that the down-regulation of GAD1, RELN, and BDNF-IX expression is accompanied by an increased binding of DNMT1 and TET1 to the promoter domains of these genes. In Fig. 1, we show that there is a significant increase in DNMT1 binding to GAD1 (overall ANOVA, F2,67, p<0.0001, followed by Bonferroni comparison: p< 0.0001, SZ vs NPS; p< 0.0001, BP vs NPS), RELN (overall ANOVA, p<0.0001, followed by Bonferroni comparison: p< 0.0001; SZ vs NPS; p=0.001, BP vs NPS), and BDNF-IX (overall ANOVA, p= 0.0001, followed by Bonferroni comparison: p= 0.04, SZ vs NPS; p< 0.001 BP vs NPS) promoters in the PFC of SZ and BP disorder patients. In contrast, DNMT1 binding to GAD2 and GAPDH promoters was one to two orders of magnitude lower than the binding of DNMT1 and failed to increase in SZ and BP disorder patients (DNMT1 bound to GAD2: NPS= 0.0047± 0.006; BP= 0.0021± 0.0009; SZ= 0.0024± 0.001), (DNMT1 bound to GAPDH: NPS= 0.012 ± 0.07; BP= 0.018 ± 0.015; SZ= 0.010± 0.007).
Fig. 1.
DNMT1, TET1, and MBD2 binding to GAD1, RELN, and DBNF-IX promoters in PFC (BA9) of non-psychiatric subjects (NPS), schizophrenia (SZ), and bipolar (BP) disorder patients. Chromatin immune precipitation was performed as described in Materials and methods using the indicated antibodies. Top panels show DNMT1 ChIP results at the GAD1, RELN and BDNF IX promoters. Middle panels correspond to TET1 ChIP data and bottom panels show data obtained with MBD2 antibody. Denotes significant difference from NPS (*P< 0.05). One-way ANOVA followed by Bonferroni comparison. Note: 1 sample from NPS and 1 sample from SZ were excluded from statistical analysis because they were identified as outliers when analyzed using the Boxplot of PASW v18 software (see method).
As shown in Fig 2, mRNA levels corresponding to DNMT1 significantly and positively correlated with DNMT1 binding to the RELN and GAD1 promoters. The binding of DNMT1 to BDNF promoter IX also positively correlated with DNMT1 mRNA levels, but due to considerable variability in the measurements of BDNF-IX in the BP disorder group, this correlation was statistically significant only when the SZ and NPS groups were considered separately. Importantly, increased binding of DNMT1 to GAD1 and RELN was not observed in the cerebellum of SZ and BP disorder patients (GAD1: NPS= 0.20 ± 0.014, BP=0.215 ± 0.060, SZ= 0.231± 0.030; n=7 per group), (RELN: NPS= 0.068 ± 0.012, BP= 0.054 ± 0.038, SZ= 0.040 ± 0.016; n=7 per group).
Fig. 2.
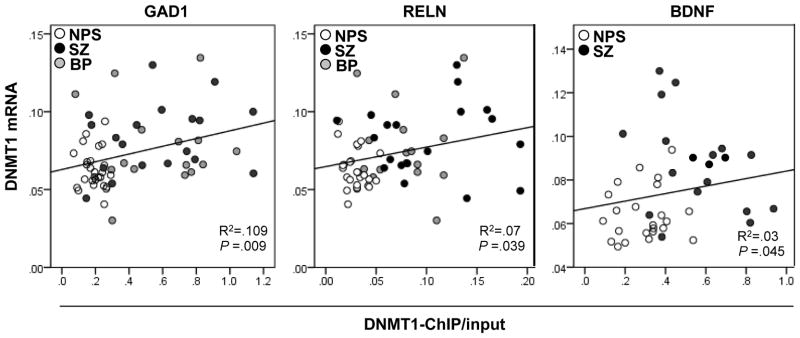
Pearson correlation between DNMT1 mRNA expression and DNMT1 binding to GAD1, RELN, and BDNF-IX promoters. The panels (left, RELN; middle GAD1; right, BDNF IX) show the relationship between DNMT1 mRNA and DNMT1 ChIP binding at the respective promoters. Note: Two samples from NPS and two samples from SZ were excluded from statistical analysis because they were identified as outliers when analyzed using the Boxplot of PASW v18 software (see method).
While there is significant binding of TET1 to GAD1, RELN, and BDNF-IX promoter regions, the binding did not increase (Fig. 1) in psychiatric patients in spite of the significant increase in TET1 mRNA and protein in the neocortex of SZ and BP patients (Table 2). The lack of increased TET1 binding to GAD1, RELN, and BDNF-IX promoters in SZ and BP disorder patients is not the consequence of methodological considerations because in a recent study using the same technique, we demonstrated an increase in TET1 binding to GAD1 and RELN in cerebellum of autistic spectrum disorder patients (Zhubi et al., 2014). It is interesting to note that binding of MBD2 (a reader of promoter methylation and/or a member of a co-repressor complex interacting with DNMT) to GAD1, RELN, and BDNF-IX in the same patients was increased (Fig. 1).
3.3 Potential Confounding variables
To show that the observed results on the levels of DNMT1 and MBD2 binding to the GAD1, RELN, and BDNF-IX promoters cannot be attributed to potential confounding variables (age, brain pH, post-mortem interval), we analyzed the data with one-way ANOVA and analyses of covariance (ANCOVA). We found no significant differences in demographic variables among the three groups (SZ, BP and NPS) by ANOVA (Table 1), with the exception of gender (BP disorder group) in which females were twice as prevalent as males. Hence, we investigated whether gender might be a factor responsible for the changes observed in BP disorder patients. ANCOVA did not reveal a statistically significant correlation between gender and changes in DNMT1 (P= 0.117), and TET1 (P= 0.544) mRNA, between gender and DNMT1 binding to GAD1 (P=0.151), RELN (P= 0.953), and BDNF-IX (P= 0.737), or between gender and TET1 binding to GAD1 (P= 0.736), RELN (P= 0.568), and BDNF-IX (P= 0.935) in the BP disorder group. When the effects of demographic variables such as cause of death, tobacco, alcohol, and cocaine abuse, entered as covariates, were examined on group differences of DNMT1 or MBD2 binding to target genes, we found the observed differences among the three groups were still significant. In addition, similar analyses of history of antipsychotic medication, antidepressants, or treatment with mood stabilizers, entered as covariates, failed to influence group differences observed for DNMT1 and MBD2 binding to GAD1, RELN, and BDNF-IX.
Hence, our analyses of covariance suggest that the increased binding of DNMT1 at RELN, GAD1, BDNF-IX promoters in PFC of SZ and BP disorder patients could not be attributed to confounding demographic variables including the use of antidepressants, mood stabilizers, antipsychotics, or drugs of abuse. However, before excluding the existence of an interaction between the increase in DNMT1 binding and medications, we need to study a larger number of SZ and BP disorder patients who have not been treated with antipsychotic medications.
4. Discussion
Studies in post-mortem brains of SZ and BP disorder patients (Gavin et al., 2012, Dong et al., 2012), and in experimental animal models of SZ (Matrisciano et al., 2012, 2013) show that altered DNA methylation and hydroxymethylation of promoter proximal regions of actively expressed genes is associated with reduced gene expression. Typically, enriched methylation or hydroxymethylation of CpG rich promoter regions of GAD1, RELN, and BDNF is accompanied by an increased expression of DNMT1 and TET1, enzymes that play an important role in the dynamic regulation of gene promoter methylation (for a review see Grayson and Guidotti 2013). In this study, we confirm previous observations that the expression of DNMT1 and TET 1 is increased in the PFC of SZ and BP disorder patients. Consistent with the increase in TET1, we previously reported that the levels of 5hmC are elevated in both total DNA and in the promoters of GAD1 and BDNF-IX of the inferior parietal lobule of SZ and BP disorder patients (Dong et al., 2012, Gavin et al., 2012). Moreover, the increase in 5hmc levels at GAD1 and BDNF-IX promoters correlates with reduced mRNA expression (Dong et al., 2012). The increase of DNMT1, particularly evident in PFC GABAergic neurons of SZ patients (Ruzicka et al., 2007), also correlates with reduced expression of GAD1 and RELN. However, studies showing the levels of 5mC at the RELN and GAD1 promoter regions, point to either an increase (Grayson et al., 2005, Abdolmaleky et al., 2005, Lintas and Persico, 2010), no change (Mill et al., 2008, Huang et al., 2007) or no methylation (Tochigi et al., 2008).
We (Dong et al., 2012) and others (Huang et al., 2007) have reported that 5mC levels at the GAD1 promoter are not increased in PFC of SZ patients even though DNMT1 expression is increased in GABAergic neurons and the expression of GAD1 mRNA is decreased (Akbarian et al., 1995; Guidotti et al., 2000; Lewis et al., 2005). Hence, these results suggest that cytosine methylation might not be the only mechanism by which DNMT1 regulates gene expression. Here, we used chromatin immune precipitation to test the relationship between DNMT1 and TET1 levels and binding of these proteins to target gene promoters in SZ and BP disorder patients. Our studies revealed, for the first time, that the binding of DNMT1 at promoter regions of GAD1, RELN, and BDNF IX is markedly increased in the PFC of SZ and BP disorder patients, compared to NPS.
We also observed a positive correlation between DNMT1 mRNA expression and binding of DNMT1 to the GAD1 and RELN promoters in the PFC but not in the cerebellum of the same patients. The differential responsiveness of genes in GABAergic neurons of PFC and cerebellum did not come as a surprise. In fact, GABAergic neurons in PFC are inhibitory interneurons, whereas GABAergic Purkinje neurons in cerebellum are principal neurons projecting from cerebellar cortex to the deep cerebellar nuclei. Furthermore, RELN, which is expressed in GABAergic interneurons in the PFC, is expressed in excitatory granular neurons in the cerebellar cortex. Hence, the data suggest that the binding of DNMT1 to GABAergic promoters may be a neuron-specific and perhaps neuronal activity dependent mechanism. Because the levels of DNMT1 mRNA positively correlate with binding of DNMT1 to the GAD1 and RELN promoters in the absence of any clear alterations in promoter methylation, it suggests that DNMT1 might regulate the expression of GABAergic genes using a mechanism which is, at least in part, independent form cytosine methylation.
At present we do not have data to suggest the precise mechanism by which DNMT1 binding to GAD1, RELN, and BDNF-IX promoters is increased in SZ and BP disorder patients. While we presume that DNMT1 binds DNA directly, it may also bind indirectly to promoters. That is, DNMT1 may interact with MBD2, or other ancillary chromatin remodeling proteins, and form a chromatin repressor complex which facilitates transitions between active and inactive chromatin states.
One possibility that should be considered is that DNMT1 and TET1 each encode ZF-CXXC zinc finger domains as part of their primary structure which contains eight conserved cysteine residues that bind two zinc ions. The CXXC protein domain, like the methyl binding domain (MBD), binds specifically to CpGs, but in the unmethylated state (Lee et al., 2001). In addition to DNMT1 and TET1, CXXC domains are present in a variety of chromatin-associated proteins, including CFP1, MLL1 and 2, KDM2A and 2B, and TET3 (Long et al., 2013). These independently folded structures facilitate binding of the associated proteins with high affinity to clusters of unmethylated CpGs and are thought to target these proteins to non-methylated CpG islands (Long et al., 2013; Rose and Klose, 2014). The CXXC domains of DNMT1 and TET1 are very similar with the exception that the TET1 domain lacks the conserved motif KFGG (Lys-Phe-Gly-Gly). Recent in vitro binding data show that the CXXC domain of TET1 prefers binding unmethylated over methylated CpGs by ~3:1 (Zhang et al., 2010). In contrast, DNMT1 binds strongly to unmethylated and less so to methylated CpG dinucleotides with a preference for unmethylated dinucleotides of 48 to 1 (Zhang et al., 2010). Another study, using transient expression assays, shows that the CXXC domain of TET1 fails to bind DNA and is dispensable for catalytic activity (Frauer et al., 2011). The above studies support the concept that while the CXXC domain of DNMT1 is a functionally independent binding motif, the corresponding domain of TET1 likely requires additional regions of the protein for full biological function. Moreover, the presence of a CXXC domain may not be sufficient per se to confer DNA binding in isolation. Available data indicate that both DNMTs and TET proteins have the ability to modify the methylation status of CpG dinucleotides and to act as transcriptional repressors but while DNMT1 acts as a context-dependent transcriptional repressor in addition to its normal cytosine methylating activity, TET1 may function primarily as an active 5-methylcytosine hydroxylase.
The increased binding of DNMT1 to SZ and BP disorder risk genes appears to be specific because the promoters of genes whose expression is not altered, such as GAD2 and GAPDH, fail to show increased DNMT1 binding. Most importantly, we found that in contrast to DNMT1, TET1 chromatin immune precipitation assays fail to show a positive correlation between TET1 mRNA expression and promoter binding. In conclusion, the data are consistent with the hypothesis that the reduced expression of genes observed in SZ and BP disorder patients may be mediated, at least in part, by region specific variations in DNMT1 action that is likely independent of promoter methylation mechanisms.
An important point implicit in this study is the observation that SZ and BP disorder, although distinct clinical entities, often share common epigenetic features. In both disorders, patients exhibit reduced levels of GAD1, RELN, BDNF, and NMDA receptor expression. Similarly, in both SZ and BP disorder, the expression of DNMTs and TETS are increased and 5mC and 5hmC are enriched at several GABAergic and glutamatergic target genes (reviewed in Grayson and Guidotti, 2013). Here we show that the binding of DNMT1 to GAD1, RELN and BDNF is equally increased in PFC of SZ and BP disorder patients suggesting that the two disorders share common pathophysiological epigenetic features.
Supplementary Material
Footnotes
Contributors
E. Dong performed the experiments presented and was primarily responsible for data analysis. Dr. Ruzicka was also involved with data analysis. Drs. Guidotti and Grayson were involved with experimental design, data analysis and manuscript preparation. All authors approved the final manuscript.
Author Disclosure
This work was supported by 5R01MH093348-03 to A. Guidotti.
References
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, Smith CL, Shafa R, Aeali B, Carnevale J, Pan H, Papageorgis P, Ponte JF, Sivaraman V, Tsuang MT, Thiagalingam S. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, Thiagalingam S, Tsuang MT. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am J Med Genet B Genet. 2005;134B:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, Jr, Jones EG. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- Benes FM, Beretta S. GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci USA. 2007;104:10164–10169. doi: 10.1073/pnas.0703806104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Vincent SL, Alsterberg G, Bird ED, SanGiovanni JP. Increased GABAA receptor binding in superficial layers of cingulate cortex in schizophrenics. J Neurosci. 1992;12:924–929. doi: 10.1523/JNEUROSCI.12-03-00924.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadet J, Wagner JR. TET enzymatic oxidation of 5-methylcytosine, 5-hydroxymethylcytosine and 5-formylcytosine. Mutat Res. 2013;764–765:18–35. doi: 10.1016/j.mrgentox.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Costa E, Chen Y, Davis J, Dong E, Noh JS, Tremolizzo L, Veldic M, Grayson DR, Guidotti A. Reelin and schizophrenia: a disease at the interface of the genome and the epigenome. Mol Interv. 2002;2:47–57. doi: 10.1124/mi.2.1.47. [DOI] [PubMed] [Google Scholar]
- Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and down-regulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry. 2012;2:e159. doi: 10.1038/tp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Earle JA, McMenomy T. Reduction in reelin immunoreactivity in hippocampus of subjects with schizophrenia, bipolar disorder and major depression. Mol Psychiatry. 2000;5:654–665. doi: 10.1038/sj.mp.4000783. [DOI] [PubMed] [Google Scholar]
- Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenöder S, Wang M, Qin W, Söding J, Spada F, Leonhardt H. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One. 2011;6:e16627. doi: 10.1371/journal.pone.0016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth Arrest and DNA-Damage-Inducible, Beta (GADD45b)-Mediated DNA Demethylation in Major Psychosis. Neuropsychopharmacology. 2012;2:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38:138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CO, Guidotti A, Costa E. Reelin promoter hypermethylation in schizophrenia. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Chen Y, Davis JM, Dong E, Gavin DP, Grayson DR, Matrisciano F, Pinna G, Satta R, Sharma RP, Tremolizzo L, Tueting P. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology. 2011;60:1007–1016. doi: 10.1016/j.neuropharm.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, Impagnatiello F, Pandey G, Pesold C, Sharma R, Uzunov D, Costa E. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Zhang X, Cheng X. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012;40:8276–8284. doi: 10.1093/nar/gks628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21:442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegame T, Bundo M, Sunaga F, Asai T, Nishimura F, Yoshikawa A, Kawamura Y, Hibino H, Tochigi M, Kakiuchi C, Sasaki T, Kato T, Kasai K, Iwamoto K. DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci Res. 2013;77:208–214. doi: 10.1016/j.neures.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Impagnatiello F, Guidotti A, Pesold C, Dwivedi Y, Caruncho H, Pisu M, Uzunov D, Smalheiser N, Davis J, Pandey G, Pappas G, Tueting P, Sharma R, Costa E. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci USA. 1998;95:15718–15723. doi: 10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Lu Y, Jelinek J, Liang S, Estecio MR, Barton MC, Issa JP. TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res. 2014;42:6956–6971. doi: 10.1093/nar/gku372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Voo KS, Skalnik DG. Identification and characterization of the DNA binding domain of CpG-binding protein. J Biol Chem. 2001;276:44669–44676. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Li MD. Grand challenges and opportunities for molecular psychiatry research: a perspective. Front Psychiatry. 2010;1:1–3. doi: 10.3389/fpsyt.2010.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lintas C, Persico AM. Neocortical RELN promoter methylation increases significantly after puberty. Neuroreport. 2010;2:114–118. doi: 10.1097/WNR.0b013e328334b343. [DOI] [PubMed] [Google Scholar]
- Long HK, Blackledge NP, Klose RJ. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans. 2013;41:727–40. doi: 10.1042/BST20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45β promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Tueting P, Dalal I, Kadriu B, Grayson DR, Davis JM, Nicoletti F, Guidotti A. Epigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice. Neuropharmacology. 2013;68:184–194. doi: 10.1016/j.neuropharm.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Tueting P, Maccari S, Nicoletti F, Guidotti A. Pharmacological activation of group-II metabotropic glutamate receptors corrects a schizophrenia-like phenotype induced by prenatal stress in mice. Neuropsychopharmacology. 2012;37:929–938. doi: 10.1038/npp.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J, Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Ann Rev Pharmacol Toxicol. 2008;48:257–276. doi: 10.1146/annurev.pharmtox.48.113006.094731. [DOI] [PubMed] [Google Scholar]
- Ray MT, Shannon Weickert C, Webster MJ. Decreased BDNF and TrkB mRNA expression in multiple cortical areas of patients with schizophrenia and mood disorders. Transl Psychiatry. 2014;4:e389. doi: 10.1038/tp.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, Sanders AR, Purcell S, Visscher PM, Craddockm N, Owen MJ, Holmans P, O’Donovan MC Molecular Genetics of Schizophrenia Collaboration (MGS); International Schizophrenia Consortium (ISC) Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17:193–1201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.02.007. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;12:385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
- Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev, Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochigi M, Iwamoto K, Bundo M, Komori A, Sasaki T, Kato N, Kato T. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63:530–533. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Veldic M, Caruncho JH, Liu WS, Davis J, Satta R, Grayson DR, Guidotti A, Costa E. DNA methyltransferase-1 (DNMT1) is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc Natl Acad Sci USA. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc Natl Acad Sci U S A. 2005;102:2152–2157. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidottim A, Davims JM, Costa E. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizophr Res. 2007;91:51–61. doi: 10.1016/j.schres.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wockner LF, Noble EP, Lawford BR, Young RM, Morris CP, Whitehall VL, Voisey J. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl Psychiatry. 2014;4:e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- Zhang L, Szulwach KE, Hon GC, Song CX, Park B, Yu M, Lu X, Dai Q, Wang X, Street CR, Tan H, Min JH, Ren B, Jin P, He C. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nat Commun. 2013;4:1517. doi: 10.1038/ncomms2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Transl Psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.