Highlights
-
•
Detection of hantavirus cardiopulmonary syndrome imported into Europe.
-
•
Additional evidence that Choclo hantavirus is currently circulating and causing human disease in Panama.
-
•
Novel diagnostic and sequencing assays for identifying cases of Choclo hantavirus infection.
Keywords: Hantavirus, Imported viral diseases, Molecular methods, Viral infections
1. Why this case is important
We describe a rare case of hantavirus cardiopulmonary syndrome (HCPS) diagnosed in a United Kingdom resident who returned from Panama in July 2014; this represents only the third published case of HCPS imported in Europe to date. Serological analysis identified increasing titres of hantavirus IgG antibodies over the course of illness and detection of hantavirus RNA in serum taken seven days after onset of symptoms. Sequence analysis following a pan-hantavirus diagnostic assay confirmed the presence of Choclo hantavirus, a species of hantavirus known to circulate in Panama and suspected of causing several cases of severe human disease in the Los Santos region of Panama during 2014. A set of molecular assays was designed to characterize the S segment of the genome and to provide rapid molecular identification for future cases.
2. Case description
A 35-year-old British woman returned to the United Kingdom (UK) on 16th July, 2014 following a nine month stay in the Los Santos province of Panama with her partner who worked locally as a rice farmer. They lived in a rural area that they had visited frequently over the last ten years. In the weeks prior to her return, she had swept out a shed that had evidence of rodent infestation and handled packaged horse feed which had evidence of rodent damage.
On 13th July, while in Panama, she developed fever, myalgia, lethargy, headache and rigors. After returning to UK, symptoms progressed to include vomiting, cough, rapidly progressive dyspnoea and chest tightness. She presented at her local Accident and Emergency department on 18th July and was admitted to hospital.
On admission she was pyrexial, hypoxic (oxygen saturation 88%) and tachycardic (heart rate 110 beats per minute). Neither rash nor bleeding were evident. Chest examination revealed bilateral crepitations. Haemoglobin levels were 191 g/L (norm: 120–160 g/L), haematocrit was 0.52 (norm: 0.36–0.46), platelet count was 65 × 109/L (norm: 150–400 × 109/L), neutrophils were 12.76 × 109/L (norm: 2–8 × 109/L) and alanine transferase levels were 160 U/L (norm: 10–40 U/L). Blood gas analysis showed arterial oxygen partial pressure of 8.9 kPa (norm: 11–13 kPa) with a pH of 7.25 (norm: 7.34–7.44); all other indices were within normal parameters.
A chest X-ray showed bilateral perihilar ground-glass opacification and normal cardiac contours. She was admitted to the intensive care unit for aggressive fluid resuscitation, inotropes, and ventilatory support for respiratory distress and severe capillary leak syndrome.
Following discussions between on-call consultants and the UK Imported Fever Service, samples were sent to Public Health England (PHE), Porton Down, to test for potential dengue, hantavirus, leptospirosis, Q-fever, and rickettsial infection.
Doxycycline, ceftriaxone, metronidazole, oseltamivir and high dose co-trimoxazole were commenced immediately after admission; cessation of these regimens was implemented as the clinical picture became increasingly compatible with HPCS and laboratory tests ruled out specific differential diagnoses. Due to the severity of disease, ribavirin was commenced 48 h after admission but was stopped after subsequent agreement that there was lack of supporting data for ribavirin therapy in New World hantavirus infection.
The critical phase lasted 36 h after which diuresis followed. Ventilatory support was weaned over 14 days. The patient was extubated on 1st August and discharged on 5th August after making an uneventful recovery.
Initial diagnosis of HCPS was made using anti-hantavirus IgG indirect immunofluorescence test mosaic 1 (Euroimmun). A serum sample taken on 19th July (day 7 of illness) was positive for Sin Nombre and Puumala IgG at dilutions of 1:100; two days later serum titres had increased to 1:1000 for Sin Nombre IgG and 1:320 for Puumala IgG.
Molecular identification of hantavirus RNA was made in the initial serum sample utilising a modified published pan-hantavirus RT-PCR assay [3]. Sequence analysis of the second round PCR product yielded an amplicon with 96% homology to the Choclo hantavirus (CHOV) L segment (Genbank accession number EF397003). A set of PCR primers was designed to amplify the entire S segment using the only published sequence available on Genbank (Table 1).
Table 1.
Primer information for molecular assays used to diagnose clinical samples, produce complete S segment sequence data and monitor isolation attempts.
| Primer pairs | Sequence | Positiona (strand) | Product size | Comment |
|---|---|---|---|---|
| HAN-L-F1 HAN-L-R1 |
ATGTAYGTBAGTGCWGATGC AACCADTCWGTYCCRTCATC |
L 2940 (sense) L 3391 (antisense) |
451 base pairs | Pan-hanta assayb 1st round (nested) |
| HAN-L-F2 HAN-L-R2 |
TGCWGATGCHACIAARTGGTC GCRTCRTCWGARTGRTGDGCAA |
L 2951 (sense) L 3340 (antisense) |
389 base pairs | Pan-hanta assayb 2nd round (nested) |
| CHOV S 1 F CHOV S 719 R |
TAGTAGTAGACTCCTTGAGAAGC CCAAAACCGATGACACCC |
S 1 (sense) S 719 (antisense) |
719 base pairs | CHOV sequencing Primers set 1 |
| CHOV S 472 F CHOV S 1157R |
AGAGGGAGGCAGACTGTGA CCCATAGACTGTGTCCTTCG |
S 472 (sense) S 1157 (antisense) |
685 base pairs | CHOV sequencing Primers set 2 |
| CHOV S 901 F CHOV S 1480 R |
GCTGAGTCTGAAGGTGCC CCCTTAACCCTAAATTAGTGC |
S 901 (sense) S 1480 (antisense) |
579 base pairs | CHOV sequencing Primers set 3 |
| CHOV S 1260F CHOV S 1972R |
GCAGTTAGCACAGTCCTTAGTTG TAGTAGTATGCTCCTTGAAAAGC |
S 1260 (sense) S 1972 (antisense) |
712 base pairs | CHOV sequencing Primers set 4 |
| CHOV S901 F CHOV S1000R CHOV S933P |
GCTGAGTCTGAAGGTGCCAC ATAGTGCTGTTGGTGGACA FAM- CATTGCTGTCCCTCAC TCTGTGTGG -BHQ1 |
S 901 (sense) S1000 (antisense) S933 (sense) |
100 base pairs | CHOV real-time |
RT-PCR amplification was performed using SuperScript III One-Step RT-PCR System with Platinum Taq (Thermo Fisher Scientific). The final mastermix (25 μL) comprised 12.5 μL of 2 × Reaction Mix 4.5 μL of PCR-grade water, 1 μL of both a forward and reverse primer at 10 μM working concentration, 1 μL of SuperScript III RT/Platinum Taq Mix and 5 μL of template. The cycling conditions used were 50 °C for 15 min, 95 °C for 2 min, followed by 45 cycles of 95 °C for 15 s, 52 °C for 30 s and 68 °C for 45 s, with a final extension step of 68 °C for 5 min. Bands of interest were excised from 2% agarose gel and purified using QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer’s instructions.
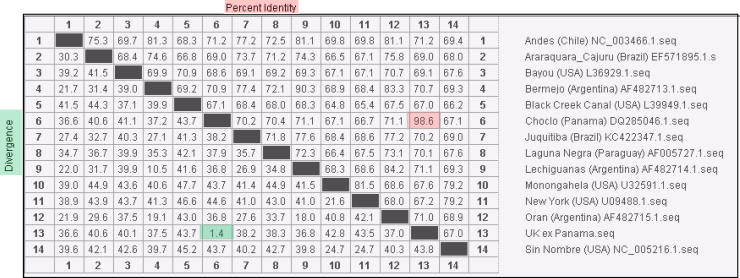
Sequenceanalysis was performed using the same primers used to produce the amplicons. Nucleotide labelling was carried out using Big Dye® Terminator v3.1Cycle Sequencing Kit (Thermo Fisher Scientific), unincorporated dye terminator was removed using DyeEx 2.0 Spin Kit (Qiagen), and sequencing of products was carried out on a 3130xl sequencer (Thermo Fisher Scientific) all according to manufacturer's instructions. Sequence data showed homology to the sole CHOV published on Genbank (98.6% across the entire segment) and significant divergence from other pathogenic New World hantaviruses (Fig. 1).
Fig. 1.
Phylogenetic divergence/identity between full S segment data sequenced from the patient serum sample in relation to other pathogenic New World hantaviruses. Data produced using MegAlign software v11 (Lasergene) following ClustalW alignment.
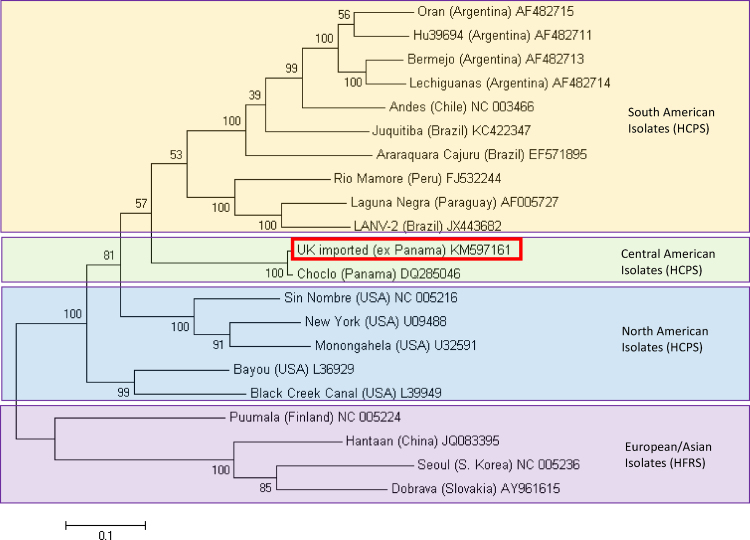
Bootstrapped maximum likelihood analysis of complete hantavirus S segments showed a close relationship between the sequenced virus and the sole CHOV sequence available on GenBank (Fig. 2).
Fig. 2.
Molecular phylogenetic analysis of full S segment hantavirus sequences by maximum likelihood method. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura 3-parameter model conducted in MEGA6. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Analysis includes representative isolates for pathogenic ‘New World’ hantaviruses associated with hantavirus cardiopulmonary syndrome (HCPS) and ‘Old World’ hantaviruses associated with haemorrhagic fever with renal syndrome (HFRS). Branch labels include GenBank accession numbers; the sequence highlighted with a red box designates the sequence in this report.
Attempts to isolate the virus from patient serum samples were unsuccessful, presumably due to the low quantity of sample available for testing from the acute phase of illness and the apparent low levels of viral RNA present in the samples. A real-time RT-PCR assay was designed to detect the CHOV S segment (Table 1) but amplification was only seen in the initial samples. All real-time RT-PCRs were performed on the ABi 7500 (Thermo Fisher Scientific) using the SuperScript III One-Step qRT-PCR System with Platinum Taq (Thermo Fisher Scientific). The final mastermix (20 μL) comprised 10 μL of 2 × Reaction Mix 1.7 μL of PCR-grade water, 1 μL of both a forward and reverse primer at 18 μM working concentration, 0.5 μL of probe at 25 μM working concentration, 0.8 μL of SuperScript III RT/Platinum Taq Mix and 5 μL of template. The cycling conditions used were 50 °C for 10 min, 95 °C for 2 min, followed by 45 cycles of 95 °C for 10 s then 60 °C for 40 s (with quantification analysis of fluorescence performed at the end of each 60 °C step).
3. Other similar and contrasting cases in the literature
HCPS is a severe disease caused by several viruses classified within the Hantavirus genus of the family Bunyaviridae with a case fatality rate of approximately 30–40% [4]. CHOV is the only hantavirus associated with human disease known to circulate in Panama; case fatality rates of approximately 25% have been recorded [11] although seroprevalence studies indicate that a milder form of disease may also be present resulting in undiagnosed cases [1].
Human exposure to hantaviruses occurs following inhalation of aerosolised rodent excreta [5]. The reservoir for CHOV is the pygmy rice rat (Oligoryzomys fulvescens costaricensis) [2]; this rodent is known to thrive in agricultural environments, especially those prone to anthropogenic change [10]. The patient in this report lived near a rice farm in a known CHOV-endemic area of Panama and reported activity in an environment with evidence of rodent infestation.
The clinical course of disease was typical for HCPS and matched well with the limited data available for human infection with CHOV. The patient met all criteria for moderate to severe HCPS resulting from infection with CHOV as previously documented, both in terms of laboratory findings and symptoms of disease [1]. Creatinine levels, which are often elevated following infection with both New World and Old World hantaviruses [7], were normal upon hospitalization but peaked at 127 μmol/L (norm: 50–95 μmol/L) two days after admission. This increase in, combination with transient proteinuria and microalbuminuria observed during the critical phase of infection, indicate a degree of renal impairment.
4. Discussion
Clinical diagnosis of HCPS outside of the Americas is extremely rare. To the best of our knowledge, this case represents only the third report of HCPS imported into Europe following non-fatal importations into France in 2001 [6] and Italy in 2010 [9] from Chile and Cuba respectively; neither previous report had sequence analysis to confirm the causal hantavirus species.
Several cases of HCPS have been reported this year from the Los Santos region of Panama where this patient resided before returning to UK [8]. Reports emanating from Panama have not confirmed the species of hantavirus causing disease although, to date, CHOV is the only pathogenic hantavirus known to circulate in the region.
The diagnostics used to confirm this case show the robustness of the initial molecular and serological assays to detect a rare strain of hantavirus. In addition, the specific molecular assays developed to determine the causative agent for this case report confirmed that CHOV RNA was present in the patient sera. These diagnostics represent the first published real-time RT-PCR and sequencing strategies for the diagnosis of this severe human pathogen and may prove useful for rapid species specific diagnostics in endemic areas and for diagnosing cases in returning travellers worldwide.
Declarations
Funding: Public Health England Grant-In-Aid for research into new and emerging high consequence human pathogens. This funding source had no involvement in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Competing interests
None declared.
Ethical approval
Patient consent was obtained to produce this report.
Authors’ contributions
B.A., B.B., E.A., M.J. and R.H. wrote the paper; B.A., L.J., J.C., S.L., and J.L. performed laboratory diagnostics and viral characterization; E.A., A.S. and T.B. provided clinical advice; B.B., M.J., A.M. and J.A. provided clinical care. All authors reviewed the manuscript before submission.
Disclosure
This report contains work supported by the National Health Service (NHS) and Public Health England Grant-in-Aid. The views expressed are those of the authors and not necessarily those of NHS or the Department of Health.
Acknowledgments
The authors would like to acknowledge the work of staff in the Rare and Imported Pathogens Laboratory for their involvement in processing samples in addition to the work by staff of PHE and the NHS in providing clinical care, infection control and surveillance.
References
- 1.Armien B., Pascale J.M., Muñoz C., Mariñas J., Núñez H., Herrera M., Trujillo J., Sánchez D., Mendoza Y., Hjelle B., Koster F. Hantavirus fever without pulmonary syndrome in Panama. Am. J. Trop. Med. Hyg. 2013;89:489–494. doi: 10.4269/ajtmh.12-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanson J.D., Utrera A., Fulhorst C.F. The delicate pygmy rice rat (Oligoryzomys delicatus) is the principal host of Maporal virus (family Bunyaviridae, genus Hantavirus) Vector Borne Zoonotic Dis. Larchmt. N.Y. 2011;11:691–696. doi: 10.1089/vbz.2010.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klempa B., Fichet-Calvet E., Lecompte E., Auste B., Aniskin V., Meisel H., Denys C., Koivogui L., ter Meulen J., Krüger D.H. Hantavirus in African wood mouse. Guinea Emerg. Infect. Dis. 2006;12:838–840. doi: 10.3201/eid1205.051487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macneil A., Nichol S.T., Spiropoulou C.F. Hantavirus pulmonary syndrome. Virus Res. 2011;162:138–147. doi: 10.1016/j.virusres.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Mills J.N., Corneli A., Young J.C., Garrison L.E., Khan A.S., Ksiazek T.G. Hantavirus pulmonary syndrome–United States: updated recommendations for risk reduction. Centers for disease control and prevention. MMWR Recomm. Rep. Morb. Mortal. Wkly. Rep. Recomm. Rep. Cent. Dis. Control. 2002;51:1–12. [PubMed] [Google Scholar]
- 6.Murgue B., Domart Y., Coudrier D., Rollin P.E., Darchis J.P., Merrien D., Zeller H.G. First reported case of imported hantavirus pulmonary syndrome in europe. Emerg. Infect. Dis. 2002;8:106–107. doi: 10.3201/eid0801.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pergam S.A., Schmidt D.W., Nofchissey R.A., Hunt W.C., Harford A.H., Goade D.E. Potential renal sequelae in survivors of hantavirus cardiopulmonary syndrome. Am. J. Trop. Med. Hyg. 2009;80:279–285. [PMC free article] [PubMed] [Google Scholar]
- 8.ProMED-mail, 2014. PRO/AH/EDR> Hantavirus update – Americas (39): Panama (LS), Argentina (CC) [WWW Document]. Arch. Number 201410112853467. <http://www.promedmail.org/direct.php?id=2853467/> (accessed 11.18.14).
- 9.Rovida F., Percivalle E., Sarasini A., Chichino G., Baldanti F. Imported hantavirus cardiopulmonary syndrome in an Italian traveller returning from Cuba. New Microbiol. 2013;36:103–105. [PubMed] [Google Scholar]
- 10.Ruedas L.A., Salazar-Bravo J., Tinnin D.S., Armién B., Cáceres L., García A., Díaz M.A., Gracia F., Suzán G., Peters C.J., Yates T.L., Mills J.N. Community ecology of small mammal populations in Panamá following an outbreak of hantavirus pulmonary syndrome. J. Vector Ecol. 2004;29:177–191. [PubMed] [Google Scholar]
- 11.Vincent M.J., Quiroz E., Gracia F., Sanchez A.J., Ksiazek T.G., Kitsutani P.T., Ruedas L.A., Tinnin D.S., Caceres L., Garcia A., Rollin P.E., Mills J.N., Peters C.J., Nichol S.T. Hantavirus pulmonary syndrome in Panama: identification of novel hantaviruses and their likely reservoirs. Virology. 2000;277:14–19. doi: 10.1006/viro.2000.0563. [DOI] [PubMed] [Google Scholar]