Abstract
Background. Two adjacent regions upstream CDKN2B on chromosome 9p21 have been associated with type 2 diabetes (T2D) and progression of cardiovascular disease (CVD). The precise location and number of risk variants have not been completely delineated and a possible synergistic relationship between the adjacent regions is not fully addressed. By a population based cross-sectional case-control design, we genotyped 18 SNPs upstream of CDKN2B tagging 138 kb in and around two LD-blocks associated with CVD and T2D and investigated associations with T2D, angina pectoris (AP), myocardial infarction (MI), coronary heart disease (CHD; AP or AMI), and stroke using 5,564 subjects from HUNT2. Results. Single point and haplotype analysis showed evidence for only one common T2D risk haplotype (rs10757282∣rs10811661: OR = 1.19, P = 2.0 × 10−3) in the region. We confirmed the strong association between SNPs in the 60 kb CVD region with AP, MI, and CHD (P < 0.01). Conditioning on the lead SNPs in the region, we observed two suggestive independent single SNP association signals for MI, rs2065501 (P = 0.03) and rs3217986 (P = 0.04). Conclusions. We confirmed the association of known variants within the 9p21 interval with T2D and CHD. Our results further suggest that additional CHD susceptibility variants exist in this region.
1. Introduction
One interesting region associated with type 2 diabetes (T2D) and cardiovascular disease (CVD) is on chromosome 9p21 in a gene desert ~130 kb upstream of CDKN2B. Several SNPs in the 9p21 interval are strongly associated with MI [1–4], vascular disease [5–7], and cancer [8], all highly correlated (r 2 > 0.8) and to be found in a ~60 kb region in high linkage disequilibrium (LD). The 9p21 region also contains two adjacent, but separate, T2D signals; a strong signal mapped to a 2 kb LD-block (represented by rs10811661 and rs10757282) and a putatively independent second signal (rs564398) located ~100 kb from the T2D interval [9–11].
After the initial genome-wide association studies (GWASs), several investigations confirmed the association with the 9p21 candidate SNPs in T2D [12–17] and CVD [18–24] and extended the number of CVD phenotypes associated with the region [25–30]. A shared mechanistic link might therefore exist within this region increasing risk of both CVD and T2D through a common pathway. In patients with T2D, a variant within 9p21 showed significant interaction between poor glycemic control and risk of angiographically verified coronary artery disease (CAD) [31]. However, the effects of the disease susceptibility variants for the two major disease loci have shown to be independent, since T2D risk variants do not seem to confer increased risk of cardiovascular disease or the other way around [5, 32].
A multilocus analysis of the 9p21 region suggested a haplotype-effect on T2D risk rather than an effect from one single SNP [33], indicating that the bona fide locus could be situated somewhere in the vicinity of the test SNPs. However, a comprehensive sequencing study of the 9p21 locus that assessed rare variants and their association with T2D and MI did not discover any variants with stronger association than what was found in the initial GWASs [8]. Thus, we chose to evaluate the distribution of common tagSNPs within the region in individuals with overlapping T2D, angina pectoris (AP), previous MI, or stroke from the Norwegian population-based HUNT2 survey to assess the distribution of T2D and CVD risk alleles in HUNT 2.
2. Materials and Methods
2.1. Study Subjects and Ethics Statement
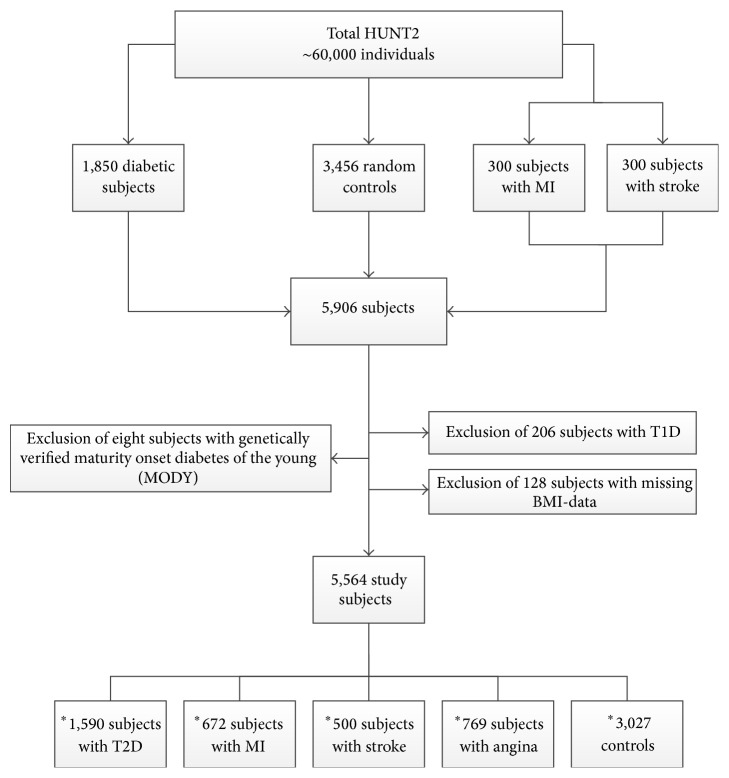
The second Nord-Trøndelag Health Study (HUNT2) is an extensive population-based health survey conducted in a Norwegian county with 127,000 inhabitants of which 60,000 participated [34]. HUNT2 is a subset of HUNT and was carried out in 1995–97. We had access to all subjects with diabetes (n = 1,850), in addition to 600 individuals selected for incident MI and/or stroke, but without diabetes, and 3,456 population-based random controls drawn from the same study population. After excluding 206 subjects with T1D, eight with genetically verified MODY [35], and 128 subjects with missing BMI data, 5,564 subjects were eligible for analysis (Figure 1). Diagnosis of diabetes, angina pectoris, previous MI, and stroke (ischemic or hemorrhagic strokes grouped as one phenotype) was self-reported. Written informed consent was obtained from all participants. This population based cross-sectional case-control study was approved by the Regional Committee for Research Ethics and the Norwegian Data Inspectorate, and was performed according to the latest version of the Helsinki Declaration.
Figure 1.
Flow chart presenting the selection of study subjects. Flow chart presenting the inclusion and exclusion criteria of the study subjects enrolled in the present study. A total of 5,564 subjects were eligible for analysis. ∗Some individuals have more than one outcome (e.g., myocardial infarction (MI) and type 2 diabetes (T2D)); hence, the sum of these counts does not match the total counts of study subjects. T1D denotes type 1 diabetes. The final set of controls was reduced as subjects with MI and stroke were incorporated after the initial controls.
2.2. SNP Selection, Genotyping, and Quality Control
We selected tagSNPs across 9p21 from the interval between Chr9:21,995,330 and 22,133,570 (NCBI Build 36). We selected 18 SNPs tagging a 138 kb region using the Haploview implementation of the Tagger algorithm [36] using the following criteria: minor allele frequency (MAF) of >5% and pairwise r 2 > 0.80. In addition, we added two previously GWAS-identified T2D susceptibility variants (rs564398 and rs10811661) and three confirmed CVD susceptibility variants (rs1333040, rs10757278, and rs1333049). The genotyping was carried out by the multiplex MassARRAY iPLEX System (SEQUENOM Inc., San Diego, CA, USA) at CIGENE, Ås, Norway. Five variants (rs1759417, rs1333049, rs7045889, rs4977761, and rs6475610) did not pass quality control criteria (minimum call rate > 95% and Hardy-Weinberg equilibrium with P > 0.01) and were excluded from analyses. Thus, we assessed a total of 18 SNPs for association with T2D, angina pectoris, previous MI, and stroke.
2.3. Statistical Analysis
We used logistic regression to model single-point and haplotype association for the 18 SNPs with T2D, MI, angina pectoris, coronary heart disease, and stroke positive cases assuming additive effect of allele dosage. Gender, age, and BMI were used as covariates in the regression model in the analysis of T2D. Diabetes status and smoking were added to the list of covariates while analyzing AP, previous MI, CHD, and stroke. Individuals with a history of either AP, previous MI, or stroke were excluded as control subjects in the regression models when analyzing CVD traits. For T2D, AP, MI, and CHD, we carried out tests conditioning on the lead SNPs (MI, angina pectoris, CHD: rs1333040 and rs10757278, T2D: rs10811661) to look for secondary signals of association. Multimarker haplotype analyses, haplotype frequency estimates, and haplotype comparisons for all phenotypes were performed using PLINK [37]. The sliding window approach used for multimarker haplotype analysis associates direct neighboring SNPs, generating 17 pairs of SNPs in the two-point analysis. All SNPs frequencies were consistent with Hardy-Weinberg equilibrium (HWE, P > 0.01). All analyses were carried out using PLINK version 1.07 software [37] and/or Stata SE v10.0 for Windows (Stata Corp LP, Brownsville, TX, USA). Figures displaying regional information such as the strength and extent of the association signals relative to genomic position, local linkage disequilibrium (LD), and recombination patterns and the positions of genes in the region were created using a combination of LocusZoom web interface [38], R package SNP Plotter [39], and Haploview [36]. We had >80% power to detect high-frequency alleles with ORs of 1.20 to 1.30 for both T2D and CVD phenotypes, but only around 50% and 30% power for T2D and CVD phenotypes, respectively, if the true ORs were 1.10. These estimates were performed using the Genetic Power Calculator [40]. All P values are presented without correction for multiple testing.
3. Results
Table 1 shows the clinical characteristics for the 5564 individuals enrolled in the present study.
Table 1.
Clinical characteristic of the 5564 subjects included in the study and eligible for analysis.
| All | T2D | AP | MI | Stroke | No T2D and/or CVD | |
|---|---|---|---|---|---|---|
| Individuals (n) | 5,564 | 1,590a | 769a | 672a | 500a | 3,027a |
| Gender (male/female) | 2,754/2,810 | 754/836 | 435/334 | 475/197 | 256/244 | 1,424/1,603 |
| Age (years at examination) | 60.4 ± 17.1 | 68.1 ± 12.0 | 72.4 ± 9.2 | 70.7 ± 10.3 | 70.8 ± 11.0 | 53.2 ± 17.6 |
| BMI (kg/m2) | 27.3 ± 4.4 | 29.2 ± 4.8 | 28.0 ± 4.3 | 27.5 ± 3.9 | 27.4 ± 3.9 | 26.4 ± 4.1 |
| Ever smoked (yes/no) | 2,600/2,964 | 647/943 | 345/424 | 367/305 | 241/259 | 1,468/1,559 |
| Nonfasting serum glucoseb (mmol/L) | 6.6 ± 3.1 | 9.6 ± 4.2 | 7.6 ± 3.6 | 7.2 ± 3.5 | 6.6 ± 2.7 | 5.4 ± 1.2 |
| Serum triglyceride (mmol/L) | 2.0 ± 1.3 | 2.5 ± 1.6 | 2.4 ± 1.6 | 2.3 ± 1.3 | 2.2 ± 1.5 | 1.8 ± 1.1 |
| Serum cholesterol (mmol/L) | 6.1 ± 1.3 | 6.2 ± 1.3 | 6.3 ± 1.3 | 6.2 ± 1.3 | 6.4 ± 1.3 | 6.0 ± 1.3 |
| Serum HDL cholesterol (mmol/L) | 1.3 ± 0.4 | 1.2 ± 0.4 | 1.2 ± 0.4 | 1.2 ± 0.4 | 1.3 ± 0.4 | 1.4 ± 0.4 |
| Heart rate (bpm) | 73.6 ± 13.6 | 75.5 ± 14.5 | 6.8 ± 13.6 | 67.6 ± 13.3 | 72.1 ± 13.4 | 74.1 ± 12.8 |
| Type 2 diabetes (n, %) | 1,590 (28.6%) | 1,590 (100%) | 326 (42.4%) | 212 (31.5%) | 110 (22%) | n/a |
| Myocardial infarction (n, %) | 672 (12.1%) | 212 (13.3%) | 357 (46.9%) | 672 (100%) | 83 (16.6%) | n/a |
| Stroke (n, %) | 500 (9.0%) | 110 (6.9%) | 115 (15.1%) | 357 (53.1%) | 500 (100%) | n/a |
| Angina pectoris (n, %) | 769 (13.7%) | 326 (20.5%) | 769 (100%) | 83 (12.4%) | 115 (23%) | n/a |
Values are presented as means ± SD or number (%). aSome individuals have more than one outcome (for example MI + diabetes); hence, the sum of these column counts does not match the total counts of individuals. bOnly nonfasting glucose measures were available for participants in the HUNT2 cohort. MI denotes previous myocardial infarction. Abbreviations: T2D, Type 2 diabetes; AP, angina pectoris; MI, myocardial infarction; CVD, cardiovascular disease; bpm, beats per minute.
3.1. Type 2 Diabetes
Regression analysis for association with T2D revealed only modest evidence for a single-point association for rs10811661 (P = 0.058) after correction for age, gender, and BMI (Figure 2, Table 2). No SNP outside the previously implicated T2D block (LD-block 4 in Figure 2) showed evidence for an association with T2D.
Figure 2.

Plot summaries for single point association results. Plot summary of association results for 18 SNPs tagging the 138 kb CVD and T2D region on chromosome 9p21 for association with T2D, myocardial infarction, stroke, angina pectoris, or CHD (both MI and angina) using 5564 subjects from the HUNT2 study. The plot show local association results for all phenotypes together with the location and orientation of the genes it includes, local estimates of recombination rates and LD heat map with defined blocks (Gabriel et al.). The plots were created using the R-package SNP Plotter [39].
Table 2.
Single point and two-point haplotype association results for T2D.
| SNP | Minor allele | Single point | Two-point | Haplotype | |||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | SNPs | Omnibus P |
Haplotype | Frequency | OR | P | ||
| rs3217986 | C | 1.03 (0.86–1.22) | 7.74 × 10−1 | rs3217986/rs523096 | 4.69 × 10−1 | AC/CT/AT | 0.48/0.08/0.44 | 1.05/1.05/0.95 | 0.31/0.61/0.22 |
| rs523096 | C | 1.04 (0.95–1.14) | 3.97 × 10−1 | rs523096/rs10965215 | 4.70 × 10−1 | CA/TA/CG/TG | 0.03/0.40/0.46/0.12 | 0.83/0.97/1.06/0.98 | 0.23/0.53/0.24/0.76 |
| rs10965215 | A | 0.96 (0.88–1.05) | 3.63 × 10−1 | rs10965215/rs564398 | 5.64 × 10−1 | GG/AA/GA | 0.46/0.42/0.12 | 1.05/0.96/0.98 | 0.29/0.36/0.82 |
| rs564398 | G | 1.05 (0.96–1.15) | 2.75 × 10−1 | rs564398/rs1333034 | 5.69 × 10−1 | AG/GA/AA | 0.12/0.46/0.43 | 0.97/1.05/0.96 | 0.71/0.29/0.41 |
| rs1333034 | G | 0.97 (0.84–1.12) | 6.96 × 10−1 | rs1333034/rs8181047 | 9.32 × 10−1 | AA/GG/AG | 0.34/0.11/0.55 | 1.02/0.98/1.00 | 0.76/0.74/0.97 |
| rs8181047 | A | 1.01 (0.92–1.12) | 7.78 × 10−1 | rs8181047/rs10811647 | 6.02 × 10−1 | GG/AC/GC | 0.40/0.34/0.26 | 0.96/1.01/1.04 | 0.35/0.78/0.44 |
| rs10811647 | G | 0.96 (0.87–1.05) | 3.51 × 10−1 | rs10811647/rs1333039 | 6.49 × 10−1 | CG/GC/CC | 0.44/0.40/0.17 | 1.04/0.96/1.01 | 0.44/0.36/0.86 |
| rs1333039 | G | 1.04 (0.95–1.14) | 4.35 × 10−1 | rs1333039/rs16905599 | 5.47 × 10−1 | CA/GG/CG | 0.06/0.44/0.50 | 1.05/1.04/0.95 | 0.62/0.40/0.28 |
| rs16905599 | A | 1.05 (0.87–1.26) | 6.20 × 10−1 | rs16905599/rs1333040 | 9.28 × 10−1 | AC/GC/GT | 0.06/0.39/0.56 | 1.03/1.00/0.99 | 0.78/0.99/0.86 |
| rs1333040 | C | 1.00 (0.91–1.09) | 9.72 × 10−1 | rs1333040/rs10757278 | 5.95 × 10−1 | CG/TG/CA/TA | 0.04/0.44/0.40/0.12 | 1.06/0.96/1.00/1.09 | 0.66/0.42/0.92/0.24 |
| rs10757278 | G | 0.97 (0.89–1.07) | 5.67 × 10−1 | rs10757278/rs10811658 | 3.46 × 10−1 | GA/AA/GG/AG | 0.16/0.14/0.32/0.38 | 0.89/0.94/1.04/1.06 | 0.10/0.42/0.49/0.28 |
| rs10811658 | A | 0.92 (0.83–1.02) | 9.85 × 10−1 | rs10811658/rs10811659 | 4.60 × 10−1 | AC/GC/AT/GT | 0.19/0.03/0.11/0.67 | 0.93/0.98/0.95/1.08 | 0.20/0.87/0.49/0.11 |
| rs10811659 | C | 0.93 (0.83–1.04) | 1.89 × 10−1 | rs10811659/rs10757282 | 1.98 × 10−1 | TC/CT/TT | 0.44/0.21/0.35 | 1.07/0.92/0.98 | 0.14/0.14/0.72 |
| rs10757282 | C | 1.08 (0.99–1.18) | 9.88 × 10−2 | rs10757282/rs10811661 | 2.05 × 10−3 | CT/TT/CC | 0.16/0.28/0.56 | 1.19/0.93/0.89 | 7.63 × 10−4/0.11/5.71 × 10−2 |
| rs10811661 | C | 0.89 (0.78–1.00) | 5.76 × 10−2 | rs10811661/rs1333051 | 8.58 × 10−2 | CT/CA/TA | 0.11/0.05/0.84 | 0.95/0.81/1.12 | 0.45/0.04/6.48 × 10−2 |
| rs1333051 | T | 0.95 (0.82–1.10) | 5.04 × 10−1 | rs1333051/rs2065501 | 5.05 × 10−1 | TA/AA/TC/AC | 0.03/0.29/0.08/0.60 | 0.92/1.08/0.95/0.96 | 0.61/0.15/0.57/0.38 |
| rs2065501 | A | 1.06 (0.96–1.17) | 2.36 × 10−1 | rs2065501/rs10757287 | 6.15 × 10−1 | AT/CT/AA/CA | 0.09/0.04/0.23/0.64 | 1.07/1.02/1.05/0.94 | 0.41/0.91/0.36/0.19 |
| rs10757287 | T | 1.05 (0.92–1.21) | 4.43 × 10−1 | n/a | n/a | n/a | n/a | n/a | n/a |
Association results for T2D from single and two-point haplotype analysis after correction for gender, age, and BMI. Top associated haplotype rs10757282 and rs10811661 is outlined.
Next, we performed a two-point sliding-window haplotype analysis and observed an increase in the association for this locus (rs10757282∣rs10811661) with T2D (P = 2.0 × 10−3) (Table 2). The association seemed to be driven by the C-T risk haplotype (OR = 1.19, P = 7.6 × 10−4), compared to the two other common two-marker haplotypes (Table 3). Further haplotype analysis in this LD-block revealed that rs10757282 and rs10811661 completely tagged one distinct risk haplotype spanning four consecutive markers in a 2-kb region (LD block 4 in Figure 2). We observed a breakup of the haplotype at markers rs10811658 and rs2065501, which confines a candidate region, located 117–128 kb upstream of CDKN2B. The risk haplotype had a frequency of 29 versus 26% in cases and controls (Table 3). HapMap data indicated similar boundaries and frequencies for the haplotype (not shown). An exploratory analysis of increasing haplotype window sizes were performed but did only produce less significant results; the strongest association was found for haplotypes incorporating both rs10757282 and rs10811661.
Table 3.
T2D association results for haplotype rs10757282/rs10811661.
| Haplotype | Frequency | OR | P | |
|---|---|---|---|---|
| Cases | Controls | |||
| Overall evidence | — | — | — | 2.05 × 10−3 |
| CT | 0.29 | 0.26 | 1.19 | 7.63 × 10−4 |
| TT | 0.56 | 0.57 | 0.93 | 1.06 × 10−1 |
| CC | 0.15 | 0.17 | 0.87 | 5.71 × 10−2 |
Association results for haplotypes defined by rs10757282 and rs10811661 in individuals with type 2 diabetes.
3.2. Cardiovascular Diseases: Angina Pectoris, Myocardial Infarction, and Stroke
Figure 2 and Additional file 1 (in Supplementary Material available online at http://dx.doi.org/10.1155/2014/164652) show the association results for each of the 18 SNPs with AP, previous MI, CHD (AP or previous MI), and stroke positive cases after adjustment for age, gender, BMI, diabetes status, and smoking. We report replication of the strong association between SNPs in the 60 kb CVD region (defined by rs8181047 to rs10757278, Figure 2) with AP (rs10757278: OR = 1.22; P = 1.1 × 10−3, Figure 2), MI (rs1333040: OR = 1.23, P = 1.8 × 10−3, Figure 2), and CHD (rs10757278: OR = 1.37; P = 2.0 × 10−4, Figure 2). Subanalyses showed that the effect of the CHD-associated SNPs was strongest in those having the most severe phenotype including both AP and previous MI. None of the SNPs in the CVD region demonstrated association with stroke, but one marker (rs10757282) in the previously implicated T2D region did show nominal evidence for association with stroke (OR = 1.2 (1.04–1.38), P = 0.01, Figure 2).
In exploratory analysis, we observed several nominally significant potentially novel single SNP associations for angina pectoris, previous MI, individuals with both AP and previous MI, and stroke in the 138 kb interval (Additional file 1). After conditioning upon the highly confirmed CVD susceptibil0ity SNPs rs1333040 and rs10757278, only two remaining SNPs (rs206550, OR = 1.32, P = 0.04; and rs3217986, OR = 1.15, P = 0.04) showed nominal P values <0.05 and only for MI (Table 4).
Table 4.
Top five association results for CVD after conditioning upon lead SNPs.
| SNP | Minor allele | AP | MI | Both MI and AP | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | ||
| rs3217986 | C | 1.21 (0.95–1.53) | 0.13 | 1.32 (1.01–1.71) | 0.04 | 1.25 (0.89–1.75) | 0.19 |
| rs2065501 | A | 1.07 (0.94–1.21) | 0.33 | 1.15 (1.01–1.32) | 0.04 | 1.11 (0.94–1.33) | 0.23 |
| rs10757282 | C | 1.09 (0.96–1.24) | 0.18 | 1.14 (1.00–1.31) | 0.05 | n/a | n/a |
| rs10811647 | G | n/a | 0.84 (0.69–1.03) | 0.09 | n/a | n/a | |
| rs16905599 | A | 1.25 (0.95–1.63) | 0.11 | 1.27 (0.95–1.71) | 0.10 | 1.31 (0.89–1.94) | 0.17 |
| rs1333051 | T | 0.87 (0.71–1.07) | 0.20 | n/a | n/a | 0.85 (0.64–1.13) | 0.27 |
| rs8181047 | A | n/a | n/a | n/a | n/a | 1.16 (0.89–1.53) | 0.28 |
Association results for the top five associated SNPs after conditioning upon the lead CVD SNPs rs1333040 and rs10757278 for individuals with angina pectoris (AP), myocardial infarction (MI), and both MI and AP.
4. Discussion
Our findings highlight the genetic complexity of the chromosome 9p21 region. We found a weak but consistent single-point association between marker rs10811661 and T2D, as previously found in several studies [9–11, 41]. This was in agreement with our former results obtained for this marker in a replication study performed in the same material from the HUNT2 population [13]. However, in the present study, we demonstrate a stronger association with a haplotype tagged by rs10811661 and rs10757282 and T2D. These results are in line with other studies [8]. Thus, these SNPs may tag a risk haplotype harboring an allele important for development of T2D. Alternatively, the 11 kb candidate region could harbor several variants associated with the disease.
Published data are conflicting regarding any additional T2D-associated signals in the 9p21 region [9–11]. Our data do not support the existence of additional signals. The role of rs564398 as a T2D susceptibility variant is disputed [9, 12, 42]. Ethnicity may play a role, although our data are not supporting that this marker has a particularly strong effect in Caucasians [43].
The 9p21 risk variants are located in non-protein coding regions; their effects possibly influencing expression of nearby genes. The region contains two cyclin-dependent kinase inhibitors, CDKN2A (p16 INK4a) and CDKN2B (p15 INK4b), and CDKN2BAS, a large antisense noncoding RNA gene. Expression of these genes is coregulated and most of the confirmed CVD risk variants correlate with decreased expression of CDKN2BAS and furthermore to atherosclerosis [44, 45]. Recent follow-up studies show correlation between the number of risk alleles and atherosclerotic CAD progression, but no predisposition to MI in patients with preexisting atherosclerotic CAD nor increased reoccurrence of MI [46–48]. This suggests 9p21 risk variants promote atherosclerosis rather than triggering MI [49]. Our associations with angina pectoris as well as MI and with the strongest associations in those having both AP and previous MI at the time of screening may thus likely be mediated through increased propensity for atherosclerosis.
The rs10757278 SNP has been highlighted as a potential functional variant for the association with atherosclerotic disease based on effects on expression of the INK4/ARF locus (p15INK4b, p16INK4a, ARF and CDKN2BAS) [50–52]. In the present study, we confirmed the associations for SNPs in the CVD region with AP and MI. The associations were strongest among subjects having both AP and previous MI. This could be a marker for early progression of atherosclerotic CAD, supporting the aforementioned association between 9p21 risk variants and early progression. Moreover, the rs10757278 SNP has been mapped to one of 33 identified enhancers in the 9p21 interval, in which the risk variant disrupts a transcription factor binding site, which could have functional relevance for an atherosclerosis-associated pathway in human endothelial cells [53].
We found no association between SNPs in the CVD region and stroke. Our results are in accordance with some studies [5, 54], but not with others [52, 55]. Several investigations aiming to address this discrepancy have confirmed 9p21 as a risk factor for stroke, but with evidence for heterogeneity of effect across stroke subtypes. The strongest association has been shown for large vessel stroke [56]. Thus, lacking stroke subtyping in our study may be the reason we did not find this association. Participants of the HUNT2 survey were identified having stroke through a self-administered questionnaire, hence details regarding type of stroke, hemorrhagic versus ischemic, or subtypes like atherothrombotic or cardioembolic were not available. One could anticipate that SNPs in the 9p21 region associated with ischemic, but not hemorrhagic stroke. Studies have indicated that sequence variation in 9p21 influences atherosclerosis development and progression; the strongest association being seen for large vessels [29]. On the other hand, rs1333040 has recently been linked to sporadic brain arteriovenous malformations known to increase hemorrhagic stroke risk [7]. Moreover, the adjacent rs10757278 has been linked to hemorrhagic stroke [52]. These results might suggest different pathways for ischemic and hemorrhagic stroke sharing common mechanisms linked to the same SNPs in the 9p21 region. Interestingly, when restricting the analysis to subjects with T2D, several SNPs in the 60 kb CVD region appeared associated with stroke, with the most significant being rs1333040 (OR = 1.44; P = 0.01). This association was not seen in stroke subjects without T2D. Interaction between variants within the 9p21 region and poor glycemic control increasing risk of CVD in patients with T2D has been suggested [31]. If similar associations were to be found for stroke risk in diabetics, it would be interesting to see whether poor glycemic control also affects different types of stroke differently.
Our exploratory results also highlights two potential novel CVD susceptibility variants, rs3217986 and rs2065501, which are located close to, but not in strong LD with the former and well-confirmed CVD region. The rs3217986 is located in the 3′ UTR of CDKN2B as well as in intron 1 of the non-protein coding CDKN2B antisense RNA, CDKN2BAS. Although speculative, it could be hypothesized that the risk variant of rs3217986 might exert an effect on atherosclerotic CAD susceptibility by influencing expression of one or both of these two genes. To our knowledge, there are no reports on whether the risk variant of rs3217986 is correlated with expression of CDKN2B and/or CDKN2BAS; thus, this hypothesis needs to be further resolved.
The study must be viewed in light of its limitations. Although previous studies have confirmed highly significant associations between SNPs in the region and CVD and T2D, the many tests performed in this study could lead to a risk of false positive findings. Thus, while the primary single SNP associations and the T2D-risk haplotype are supported by previous studies, the more explorative findings of putative secondary signals need to be further investigated in much larger cohorts. The sparse risk increase associated with these common variants also renders our findings inadequate for clinical prediction. Fine-mapping studies of disease associated regions may still prove important to guide further investigation towards understanding the disease pathogenesis and possibly providing tools for cost-efficient risk stratification in the future.
Despite the close proximity between the CVD and T2D risk regions, our study is in line with previous studies and indicates that there is no apparent overlap between the two risk regions. Theories with reference to the concrete disease mechanism mediated by the risk variants of the 9p21 interval have increased in numbers the last years. However, since most of them still remain exploratory, the exact nature of the disease associated variants and their targets require further elucidation. They may possibly differ between CVD and T2D. It is possible that large-scale genome sequencing efforts may aid by identifying the underlying risk variants in the 9p21 region.
5. Conclusions
In conclusion, we confirm the association between variants in the 9p21 interval with T2D and CHD. Our results suggest that there exist additional CVD susceptibility variants in this region, highlighting the genetic complexity of the 9p21 region and human disease.
Supplementary Material
Sheet AP: Association results for angina pectoris (AP) from single and two-point haplotype analysis after correction for gender, age, BMI, diabetes and smoking.
Sheet MI: Association results for myocardial infarction (MI) from single and two-point haplotype analysis after correction for gender, age, BMI, diabetes and smoking.
Sheet MI+AP: Association results for both angina pectoris and myocardial infarction from single and two-point haplotype analysis after correction for gender, age, BMI, diabetes and smoking.
Sheet Stroke: Association results for stroke from single and two-point haplotype analysis after correction for gender, age, BMI, diabetes and smoking.
Acknowledgments
The study was supported in part by funds from the University of Bergen, Haukeland University Hospital, the Western Norway Regional Health Authority, Innovest, European Research Council (AdG to PR Njølstad), KG Jebsen Foundation, and the Research Council of Norway. Genotyping was in part provided by the CIGENE technology platform (Ås, Norway), which is supported by the Functional Genomics Programme (FUGE) of the Research Council of Norway. The Nord-Trøndelag Health Study (HUNT) is a collaboration between the HUNT Research Center at the Norwegian University of Science and Technology, Levanger, the Norwegian Institute for Public Health, and the Nord-Trøndelag County Council. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the paper. No additional external funding was received for this study.
Abbreviations
- AP:
Angina pectoris
- CAD:
Coronary artery disease
- CHD:
Coronary heart disease
- CVD:
Cardiovascular disease
- MI:
Myocardial infarction
- GWAS:
Genome-wide association study
- HUNT:
Helseundersøkelsen Nord-Trøndelag
- LD:
Linkage disequilibrium
- MODY:
Maturity onset diabetes of the young
- SNP:
Single-nucleotide polymorphism
- T2D:
Type 2 diabetes
- UTR:
Untranslated region.
Conflict of Interests
The authors declare that they have no competing interests.
Authors' Contribution
Øyvind Helgeland wrote the paper with assistance from Jens K. Hertel, Helge Ræder, Anders Molven, Pål R. Njølstad, and Stefan Johansson. Carl G. P. Platou, Kristian Midthjell, and Ottar Nygård reviewed and edited the paper. Øyvind Helgeland and Jens K. Hertel performed statistical analysis and interpreted the data with assistance from Anders Molven, Helge Ræder, Ottar Nygård, Pål R. Njølstad, and Stefan Johansson. Pål R. Njølstad and Stefan Johansson conceived the study design with contribution from Øyvind Helgeland, Jens K. Hertel, and Anders Molven. Kristian Midthjell collected background data. Stefan Johansson directed genotyping and statistical analysis. Øyvind Helgeland, Jens K. Hertel, Helge Ræder, Carl G. P. Platou, Kristian Midthjell, Ottar Nygård, Pål R. Njølstad, and Stefan Johansson contributed to discussion. All authors read and approved the final paper.
References
- 1.Burton P. R., Clayton D. G., Cardon L. R., et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helgadottir A., Thorleifsson G., Manolescu A., et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 3.McPherson R., Pertsemlidis A., Kavaslar N., et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samani N. J., Erdmann J., Hall A. S., et al. Genomewide association analysis of coronary artery disease. The New England Journal of Medicine. 2007;357(5):443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helgadottir A., Thorleifsson G., Magnusson K. P., et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nature Genetics. 2008;40(2):217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 6.Newton-Cheh C., Cook N. R., Vandenburgh M., Rimm E. B., Ridker P. M., Albert C. M. A common variant at 9p21 is associated with sudden and arrhythmic cardiac death. Circulation. 2009;120(21):2062–2068. doi: 10.1161/CIRCULATIONAHA.109.879049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sturiale C. L., Puca A., Sebastiani P., et al. Single nucleotide polymorphisms associated with sporadic brain arteriovenous malformations: where do we stand? Brain. 2013;136(2):665–681. doi: 10.1093/brain/aws180. [DOI] [PubMed] [Google Scholar]
- 8.Shea J., Agarwala V., Philippakis A. A., et al. Comparing strategies to fine-map the association of common SNPs at chromosome 9p21 with type 2 diabetes and myocardial infarction. Nature Genetics. 2011;43(8):801–805. doi: 10.1038/ng.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saxena R., Voight B. F., Lyssenko V., et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 10.Scott L. J., Mohlke K. L., Bonnycastle L. L., et al. A genome-wide association study of type 2 diabetes in finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeggini E., Weedon M. N., Lindgren C. M., et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duesing K., Fatemifar G., Charpentier G., et al. Strong association of common variants in the CDKN2A/CDKN2B region with type 2 diabetes in French Europids. Diabetologia. 2008;51(5):821–826. doi: 10.1007/s00125-008-0973-4. [DOI] [PubMed] [Google Scholar]
- 13.Hertel J. K., Johansson S., Ræder H., et al. Genetic analysis of recently identified type 2 diabetes loci in 1,638 unselected patients with type 2 diabetes and 1,858 control participants from a Norwegian population-based cohort (the HUNT study) Diabetologia. 2008;51(6):971–977. doi: 10.1007/s00125-008-0982-3. [DOI] [PubMed] [Google Scholar]
- 14.Horikawa Y., Miyake K., Yasuda K., et al. Replication of genome-wide association studies of type 2 diabetes susceptibility in Japan. Journal of Clinical Endocrinology and Metabolism. 2008;93(8):3136–3141. doi: 10.1210/jc.2008-0452. [DOI] [PubMed] [Google Scholar]
- 15.Ng M. C. Y., Park K. S., Oh B., et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008;57(8):2226–2233. doi: 10.2337/db07-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Li H., Loos R. J. F., et al. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57(10):2834–2842. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voight B. F., Scott L. J., Steinthorsdottir V., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schunkert H., Götz A., Braund P., et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117(13):1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assimes T. L., Knowles J. W., Basu A., et al. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Human Molecular Genetics. 2008;17(15):2320–2328. doi: 10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiura Y., Fukushima Y., Yuno M., et al. Validation of the association of genetic variants on chromosome 9p21 and 1q41 with myocardial infarction in a Japanese population. Circulation Journal. 2008;72(8):1213–1217. doi: 10.1253/circj.72.1213. [DOI] [PubMed] [Google Scholar]
- 21.Larson M. G., Atwood L. D., Benjamin E. J., et al. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Medical Genetics. 2007;8(1, article S5) doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen G.-Q., Li L., Rao S., et al. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28(2):360–365. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- 23.Shen G.-Q., Rao S., Martinelli N., et al. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. Journal of Human Genetics. 2008;53(2):144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L., Zhang X., He M., et al. Associations between single nucleotide polymorphisms on chromosome 9p21 and risk of coronary heart disease in Chinese Han population. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28(11):2085–2089. doi: 10.1161/ATVBAHA.108.176065. [DOI] [PubMed] [Google Scholar]
- 25.Abdullah K. G., Li L., Shen G.-Q., et al. Four SNPS on chromosome 9p21 confer risk to premature, familial CAD and MI in an American Caucasian Population (GeneQuest) Annals of Human Genetics. 2008;72(5):654–657. doi: 10.1111/j.1469-1809.2008.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z., Qian Q., Ma G., et al. A common variant on chromosome 9p21 affects the risk of early-onset coronary artery disease. Molecular Biology Reports. 2009;36(5):889–893. doi: 10.1007/s11033-008-9259-7. [DOI] [PubMed] [Google Scholar]
- 27.Matarin M., Brown W. M., Singleton A., Hardy J. A., Meschia J. F. Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke. 2008;39(5):1586–1589. doi: 10.1161/STROKEAHA.107.502963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahlstrand B., Orho-Melander M., Delling L., et al. The myocardial infarction associated CDKN2A/CDKN2B locus on chromosome 9p21 is associated with stroke independently of coronary events in patients with hypertension. Journal of Hypertension. 2009;27(4):769–773. doi: 10.1097/HJH.0b013e328326f7eb. [DOI] [PubMed] [Google Scholar]
- 29.Ye S., Willeit J., Kronenberg F., Xu Q., Kiechl S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. Journal of the American College of Cardiology. 2008;52(5):378–384. doi: 10.1016/j.jacc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- 30.Zee R. Y. L., Ridker P. M. Two common gene variants on chromosome 9 and risk of atherothrombosis. Stroke. 2007;38(10, article e111) doi: 10.1161/STROKEAHA.107.497669. [DOI] [PubMed] [Google Scholar]
- 31.Doria A., Wojcik J., Xu R., et al. Interaction between poor glycemic control and 9p21 locus on risk of coronary artery disease in type 2 diabetes. Journal of the American Medical Association. 2008;300(20):2389–2397. doi: 10.1001/jama.2008.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broadbent H. M., Peden J. F., Lorkowski S., et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Human Molecular Genetics. 2008;17(6):806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 33.Browning B. L., Browning S. R. Haplotypic analysis of wellcome trust case control consortium data. Human Genetics. 2008;123(3):273–280. doi: 10.1007/s00439-008-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romundstad S., Holmen J., Hallan H., Kvenild K., Krüger Ø., Midthjell K. Microalbuminuria, cardiovascular disease and risk factors in a nondiabetic/nonhypertensive population. The Nord-Trøndelag Health Study (HUNT, 1995–97), Norway. Journal of Internal Medicine. 2002;252(2):164–172. doi: 10.1046/j.1365-2796.2002.01025.x. [DOI] [PubMed] [Google Scholar]
- 35.Eide S. Å., Ræder H., Johansson S., et al. Prevalence of HNF1A (MODY3) mutations in a Norwegian population (the HUNT2 Study) Diabetic Medicine. 2008;25(7):775–781. doi: 10.1111/j.1464-5491.2008.02459.x. [DOI] [PubMed] [Google Scholar]
- 36.Barrett J. C., Fry B., Maller J., Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Purcell S., Neale B., Todd-Brown K., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruim R. J., Welch R. P., Sanna S., et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luna A., Nicodemus K. K. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics. 2007;23(6):774–776. doi: 10.1093/bioinformatics/btl657. [DOI] [PubMed] [Google Scholar]
- 40.Purcell S., Cherny S. S., Sham P. C. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 41.Cugino D., Gianfagna F., Santimone I., et al. Type 2 diabetes and polymorphisms on chromosome 9p21: a meta-analysis. Nutrition, Metabolism and Cardiovascular Diseases. 2012;22(8):619–625. doi: 10.1016/j.numecd.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Bao X. Y., Xie C., Yang M. S. Association between type 2 diabetes and CDKN2A/B: a meta-analysis study. Molecular Biology Reports. 2012;39(2):1609–1616. doi: 10.1007/s11033-011-0900-5. [DOI] [PubMed] [Google Scholar]
- 43.Peng F., Hu D., Gu C., et al. The relationship between five widely-evaluated variants in CDKN2A/B and CDKAL1 genes and the risk of type 2 diabetes: a meta-analysis. Gene. 2013;531(2):435–443. doi: 10.1016/j.gene.2013.08.075. [DOI] [PubMed] [Google Scholar]
- 44.Cunnington M. S., Koref M. S., Mayosi B. M., Burn J., Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genetics. 2010;6(4) doi: 10.1371/journal.pgen.1000899.e1000899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Congrains A., Kamide K., Oguro R., et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220(2):449–455. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 46.Dandona S., Stewart A. F. R., Chen L., et al. Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. Journal of the American College of Cardiology. 2010;56(6):479–486. doi: 10.1016/j.jacc.2009.10.092. [DOI] [PubMed] [Google Scholar]
- 47.Patel R. S., Su S., Neeland I. J., et al. The chromosome 9p21 risk locus is associated with angiographic severity and progression of coronary artery disease. European Heart Journal. 2010;31(24):3017–3023. doi: 10.1093/eurheartj/ehq272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ardissino D., Berzuini C., Merlini P. A., et al. Influence of 9p21.3 genetic variants on clinical and angiographic outcomes in early-onset myocardial infarction. Journal of the American College of Cardiology. 2011;58(4):426–434. doi: 10.1016/j.jacc.2010.11.075. [DOI] [PubMed] [Google Scholar]
- 49.Anderson J. L., Horne B. D. The 9p21 locus and coronary heart disease: initiator, promoter, or precipitator? Journal of the American College of Cardiology. 2010;56(6):487–489. doi: 10.1016/j.jacc.2010.02.056. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y., Sanoff H. K., Cho H., et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS ONE. 2009;4(4) doi: 10.1371/journal.pone.0005027.e5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holdt L. M., Beutner F., Scholz M., et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arteriosclerosis, Thrombosis, and Vascular Biology. 2010;30(3):620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 52.Zhang W., Chen Y., Liu P., et al. Variants on chromosome 9p21.3 correlated with ANRIL expression contribute to stroke risk and recurrence in a large prospective stroke population. Stroke. 2012;43(1):14–21. doi: 10.1161/STROKEAHA.111.625442. [DOI] [PubMed] [Google Scholar]
- 53.Harismendy O., Notani D., Song X., et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ 3 signalling response. Nature. 2011;470(7333):264–270. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikram M. A., Seshadri S., Bis J. C., et al. Genomewide association studies of stroke. The New England Journal of Medicine. 2009;360(17):1718–1728. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson C. D., Biffi A., Rost N. S., Cortellini L., Furie K. L., Rosand J. Chromosome 9p21 in ischemic stroke: population structure and meta-analysis. Stroke. 2010;41(6):1123–1131. doi: 10.1161/STROKEAHA.110.580589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traylor M., Farrall M., Holliday E. G., et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE Collaboration): a meta-analysis of genome-wide association studies. The Lancet Neurology. 2012;11(11):951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet AP: Association results for angina pectoris (AP) from single and two-point haplotype analysis after correction for gender, age, BMI, diabetes and smoking.
Sheet MI: Association results for myocardial infarction (MI) from single and two-point haplotype analysis after correction for gender, age, BMI, diabetes and smoking.
Sheet MI+AP: Association results for both angina pectoris and myocardial infarction from single and two-point haplotype analysis after correction for gender, age, BMI, diabetes and smoking.
Sheet Stroke: Association results for stroke from single and two-point haplotype analysis after correction for gender, age, BMI, diabetes and smoking.