Abstract
Mammalian cells have many membranous organelles that require proper composition of proteins and lipids. Cargo sorting is a process required for transporting specific proteins and lipids to appropriate organelles, and if this process is disrupted, organelle function as well as cell function is disrupted. ArfGAP family proteins have been found to be critical for receptor sorting. In this review, we summarize our recent knowledge about the mechanism of cargo sorting that require function of ArfGAPs in promoting the formation of transport vesicles, and discuss the involvement of specific ArfGAPs for the sorting of a variety of receptors, such as MPR, EGFR, TfR, Glut4, TRAIL-R1/DR4, M5-muscarinic receptor, c-KIT, rhodopsin and β1-integrin. Given the importance of many of these receptors to human disease, the studies of ArfGAPs may provide novel therapeutic strategies in addition to providing mechanistic insight of receptor sorting.
Introduction
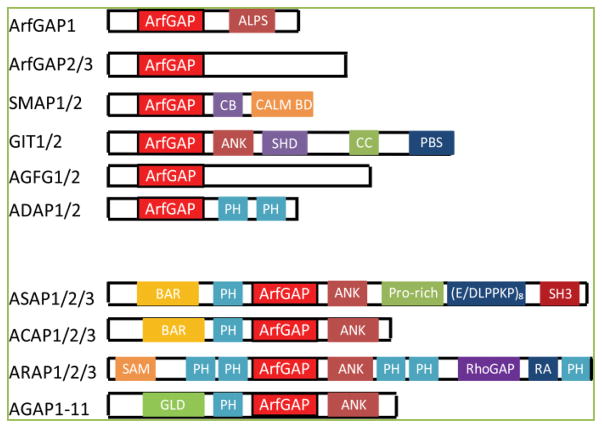
ArfGAPs are a protein family containing the ArfGAP domain. There are 31 genes encoding ArfGAPs in human[1]. ArfGAPs are structurally diverse, having a variety of domains other than the ArfGAP domain (Figure 1), consistent with the idea that each ArfGAP has a different function. They have the common function of catalyzing the hydrolysis of GTP that is bound to Arf, thereby converting Arf•GTP to Arf•GDP. The activity is essential for the function of Arf proteins, a family of proteins within the Ras superfamily of gunanine nucleotide binding proteins[2–3]. There are six Arfs (Arf1-Arf6) in mammals, though human lacks Arf2. Arf in GDP bound form is in cytosol with weak affinity for membrane. When GDP on Arf is converted to GTP by ArfGEF (Arf Guanine Nucleotide Exchange Factor) on the Golgi membrane, Arf•GTP binds to the Golgi membrane as well as coat protein, COPI. COPI is a coat protein that is recruited to the Golgi membrane by Arf•GTP and binds to cargos to produce COPI-coated vesicles from the Golgi apparatus[4]. Arf by itself cannot hydrolyze GTP, but the function of Arf requires GTP hydrolysis.
Figure 1. Schematic of domain structure of human ArfGAPs (modified from [15]).
Not drawn to scale. Abbreviations are; ArfGAP, ArfGAP domain; ALPS, ArfGAP1 Lipid-Packing Sensor; CB, clathrin-box; CALM BD, CALM Binding Domain; ANK, ankyrin repeat; SHD, Spa-Homology Domain; CC, Coiled Coil; PBS, Paxillin Binding Site; PH, Preckstrin Homolody; (E/DLPPKP)8, eight times repeat of E/DLPPKP sequence; SH3, Src-Homology 3; SAM, Sterile Alpha-Motif; RhoGAP, Rho Gap domain; RA, Ras-association domain; GLD, GTP-binding protein-Like Domain
ArfGAP1 functions in COPI-mediated transport in pre-Golgi
The first insight into the role of ADP-ribosylation factor (Arf) and GTP hydrolysis in cargo sorting came up from the study of COPI-coated vesicle transport in pre-Golgi transport. Several groups reported that GTPγS, a non-hydrolyzable analogue of GTP and GTP-locked mutant of Arf (Arf1Q71L) blocked cargo incorporation into COPI vesicles, supporting the idea that GTP hydrolysis by Arf is critical for cargo sorting [5–8]. GTP bound to Arf is hydrolyzed by ArfGAPs (Arf GTPase activating protein), thereby converting Arf•GTP to Arf•GDP. ArfGAP1 is known to be involved in transport from the Golgi[9,10], therefore, ArfGAP1 has been thought to be a critical molecule to regulate cargo sorting in COPI transport[11,12]. ArfGAPs were also thought to be important for vesicle uncoating, which occurs after cargo sorting. Several models were proposed to explain the dual functions of ArfGAPs, which are discussed in recent reviews[13–15].
Although most studies have focused on the role of Arf GAPs in uncoating transport vesicles, function in promoting the formation of coated transport vesicles has also been proposed. Initial evidence was generated using assays for in vitro generation of transport vesicles from purified Golgi membranes. Production of vesicles required functional ArfGAP1 able to hydrolyze GTP[12]. In cells, overexpression of ArfGAP1 increased the number of intracellular vesicular structures with the dimensions of COPI coated vesicles[16]. Overexpression of the GAP-dead mutant of ArfGAP1 (R50K) that still binds to Arf•GTP also results in producing vesicles. However, more STxB-KDEL, a COPI cargo, remained in the Golgi, implying that ArfGAP1(R50K) partially inhibits transport of a COPI cargo to the endoplasmic reticulum (ER)[16]. The concept that GAP activity of ArfGAP1 links vesicle production with cargo sorting is supported by the finding that peptide from the cytoplasmic tail of COPI cargos stimulates the GAP activity of ArfGAP1 in the presence of COPI in vitro[17]. Taking these results together, a model was proposed in which ArfGAP1 is required for the coupling of COPI and COPI cargo and promote COPI polymerization to produce COPI vesicles [15]. This model provides perspective for our discussion of the emerging roles of the function of ArfGAPs in receptor sorting in post-Golgi trafficking.
ArfGAPs in Post-Golgi traffic
ArfGAP3 in MPR and EGFR traffic
Mannose 6-phosphate receptor (MPR) is the critical receptor required for the transport of lysosomal enzyme from the Golgi to the lysosome. A screen using siRNA targeting all ArfGAPs, in which effects on the localization of MPR were examined, identified ArfGAP3 as a regulator of transport of MPR from the early endosomes to the late endosomes[18]. ArfGAP3 had previously been reported to have a redundant role with ArfGAP1 and ArfGAP2 in COPI-mediated transport[19]. In contrast, ArfGAP3 effect on MPR traffic was found to be specific for ArfGAP3: ArfGAP1 or ArfGAP2 depletion did not affect the localization of MPR. Epidermal growth factor receptor (EGFR) transport from the early endosomes to the late endosomes was also perturbed in ArfGAP3 knock down cells whereas other transport pathways such as transferrin receptor recycling and Vesicular Stomatitis Virus Glycoprotein ts045 transport from the ER to the plasma membrane (PM) were not detectably affected. The results supported the idea that ArfGAP3 is specifically required for transport of MPR and EGFR from the early endosomes to the late endosomes. In the same paper, ArfGAP3 was reported to bind to Golgi-localizing, Gamma-adaptin ear domain, Arf-binding proteins (GGAs), which are coat proteins known to medicating transport of MPR. Therefore GGAs are likely the targets of ArfGAP3. These data support the idea that ArfGAP3 is required for promoting MPR transport by regulating GGAs. It was also discovered that ArfGAP3 regulates full-length MPR, but not luminal and transmembrane domain depleted MPR. It is possible that ArfGAP3 recognizes clustered MPR dependent on its luminal and transmembrane domain. Further study is required for revealing the mechanism by which ArfGAP3 regulates MPR sorting.
ArfGAP3 was reported to be an androgen target gene, and its overexpression promotes cell proliferation and migration of prostate cancer cells[20]. ArfGAP3 binds to Paxillin, an adaptor protein in focal adhesion and localizes to focal adhesions[20]. It will be interesting to examine whether ArfGAP3 regulates trafficking of integrin receoptors as well as EGFR and androgen receptor, which could account for its effects on migration and proliferation of the prostate cancer cells.
ACAP1 in tranferrin receptor, β1-integrin, and Glut4 recycling
ACAP family is the protein family that has BAR, PH, ArfGAP and ankyrin repeat domains. ACAP1 and ACAP2 have GAP activity towards Arf6 over Arf1 or Arf5 [21]. ACAP1 functions in cargo sorting by recognizing recycling cargos, such as transferrin receptor (TfR), β1-integrin and glucose transporter type 4 (Glut4), controlling translocation to the PM. ACAP1 binds to the cytoplasmic tail of TfR, and TfR recycling is slowed in ACAP1 knock down cells[22]. TfR is a marker for constitutive recycling pathway, however, ACAP1 also functions in regulated recycling of β1-integrin[23] and Glut4[24]. The recycling of β1-integrin to the PM upon serum stimulation is inhibited in ACAP1 knock down cells. ACAP1 binds to β1-integrin upon serum stimulation, and the binding is dependent on phosphorylation of ACAP1 by Akt. Depletion of ACAP1 or Akt inhibits cell migration[23]. The binding site between ACAP1 and β1-integrin was biochemically defined. The cytoplasmic tail of β1-integrin binds to C-terminal portion of ACAP1 that includes ArfGAP and ankyrin repeat domains [25]. Phosphorylation of ACAP1 enhances this binding. ACAP1 also seems to affect Glut4 transport to the PM under insulin stimulation in 3T3-L1 adipocytes, as ACAP1 depletion inhibits glucose uptake under insulin stimulation[24]. ACAP1 binds to clathrin, a major coat protein in post-Golgi, and showed colocalization with clathrin and Arf6 in Glut4 positive compartment. Depletion of clathrin and Arf6 also inhibits glucose uptake, therefore it is proposed that ACAP1 mediates recycling of Glut4 as a coat component of clathrin and Arf6.
ARAP1 in EGFR and DR4 traffic
ARAP1-3 are the family of proteins that have SAM, five PH, ArfGAP, ankyrin repeats, RhoGAP, and RA (Ras-associating) domains. ARAP1 (also known as Centaurin delta 2) has GAP activity towards Arf1 and Arf5 over Arf6 [26]. ARAP1 was reported to regulate EGFR transport [27–28]. There is a discrepancy between two reports. Yoon et al. reported that EGFR degradation is accelerated and phosphorylation of Extracellular- signal-Related Kinase (ERK) and c-Jun-N-terminal Kinase (JNK) is rapidly diminished in ARAP1 knock down cells. Daniele et al. reported that EGFR accumulated in sorting/late endosomes and EGFR degradation was slower in ARAP1 knock down cells compared to controls. The reason for this difference is unknown. In both reports, ARAP1 localized to the Golgi, an endosomal compartment and the PM, and internalization and recycling of transferrin were not perturbed by decreased expression of ARAP1. Kang et al. reported that ARAP1 binds to PTK6, a non-receptor tyrosine kinase and found that PTK6 binds and phosphorylates ARAP1 upon EGF stimulation [29]. The phosphorylation-defective mutant of ARAP1 accelerates EGFR degradation compared with wild type ARAP1, and silencing of PTK6 in breast cancer cells down-regulates EGFR. These results support the idea that ARAP1 inhibits EGF/EGFR degradation dependent on PTK6. One possibility is that ARAP1 promotes EGFR recycling to the PM upon EGF stimulation; therefore depletion of ARAP1 or expression of a phosphorylation-deficient mutant of ARAP1 accelerates EGF/EGFR degradation. Further studies are required for determining the mechanism by which ARAP1 specifically regulates EGFR upon EGF stimulation.
ARAP1 was also reported to regulate death receptor, TRAIL-R1/DR4[30]. ARAP1 binds to DR4 and TRAIL-R2/DR5 upon stimulation of apoptosis-inducing ligand, TRAIL, in human immortalized keratinocytes (NCTC). ARAP1 c-terminal region that lacks fifth PH domain (Δexon30) binds to DR4, and the presence of exon30 abolished the binding. This splicing variant of ARAP1 (Δexon30) is the predominantly expressed form in a wide variety of cell lines. The binding site on DR4 resides in the first two α-helices of the death domain of DR4. This region showed the highest binding to ARAP1 by co-immunoprecipitation. The knock down of ARAP1 results in the down-regulation of DR4 and slower initial phase of TRAIL-induced apoptosis. Total amount of DR4 is not changed upon ARAP1 knock down. It is possible that recycling of DR4 from an intracellular compartment to the PM is inhibited in ARAP1 knock down cells, similar to EGFR transport.
AGAP1 in M5 muscarinic receptor traffic
AGAPs are the family of proteins that have GLD (GTP-binding protein-like domain), PH, ArfGAP and ankyrin repeats domains. There are 11 genes identified for AGAPs, however, AGAP4 to AGAP10 genes encode almost the same mRNA sequence[18], while the gene loci of these genes are different[1]. Probably these genes arose by gene duplication. AGAP1 (also known as centaurin gamma 2) has GAP activity towards Arf1 and Arf5 over Arf6[31], and was reported to regulate M5 muscarinic receptor transport[32]. Muscarinic receptors (MRs) are a family of G-protein coupled receptors (GPCRs) for acetylcholine. Activation of MR5 (M5) receptors potentiates dopamine release in neurons. Dysfunction of M5 may contribute to the pathophysiology of schizophrenia and drug addiction. The precise function and regulation of M5, particularly compared to other subtypes, has not been defined. Overall, there is significant sequence similarity among MR family members and the subtypes have not been found to have significant selectivity for MR ligands. Functional differences should be related to the large third intracellular loop (i3 loop) whose sequence is highly divergent between subtypes. Bendor et al. found that AGAP1 is an interacting partner of the i3 loop of M5 muscarinic receptor, and showed that the binding between AGAP1 and M5 is specific among other MRs and AGAP family. M5 mutant lacking AGAP1 binding site showed the inhibition of recycling of M5 to the PM in cultured neurons, and the decrease of presynaptic M5-mediated dopamine release potentiation in mice brain slices. AGAP1 has been reported to bind to AP-3[33], a coat-like adaptor complex that is known to be involved in the biogenesis of lysosomal related organelles such as synaptic vesicles[34]. M5 was not detected in synaptic vesicles. Further studies are required for identifying the compartment from where M5 is recycled to the PM. Both AP-3 and M5 bind the PH domain of AGAP1, but it is not known whether the bindng sites are overlapping. It is possible that AGAP1 mediates complex formation of AP-3 and M5. Whether the M5 binding to PH domain affects the GAP activity of AGAP1 has yet to be studied. Further studies are required for understanding the mechanism by which AGAP1 regulates M5 transport through AP-3.
AGAP2 in STxB transport
AGAP2 (also known as PIKE-A, Centaurin gamma 1 and GGAP2) has a very similar sequence as AGAP1 (71% identities in nucleotide level, 55% in amino acid) and has GAP activity towards Arf1 and Arf5 over Arf6 similar to AGAP1. However, its function seems to be different than AGAP1. AGAP2 depletion inhibits the retrograde transport of Shiga Toxin B subunit (STxB) from early endosomes to the trans-Golgi network (TGN) [35]. STxB binds to a glycosphingolipid, globotriaosylceramide (Gb3) [36] therefore AGAP2 regulates lipid-based transport. Gb3 does not have a cytoplasmic domain that can be recognized by cytosolic proteins. The mechanism how AGAP2 recognizes Gb3 is unknown.
AGAP2 was reported to bind to β-arrestins, which in turn bind to β2 adrenergic receptor (β2AR) and affects the intracellular localization of β2AR[37]. β-arrestins bind to PH domain of AGAP2. These results raise the possibility that PH domain of AGAP family is the binding site for cargos and the binding is coupled with regulation of GAP activity of AGAPs.
AGAP3 in AMPA receptor transport
AGAP3 (also known as centaurin gamma3, MRIP-1, and GGAP3) is another AGAP family protein whose amino acid sequence has 72% identity with AGAP1 and 52% identity with AGAP2. AGAP3 was reported to be a binding partner of SynGAP, a component of the N-methyl-D- aspartate receptor (NMDA) receptor complex [38]. NMDA receptor mediates long-term potentiation (LTP) in synapses. Following the activation of NMDA receptor, synaptic connection between two cells is strengthened by up-regulation of trafficking of AMPA-type glutamate receptors to the synapse. Downstream of the NMDA receptor, Ras/ERK signaling pathway is key to the induction of LTP, however, the link between Ras/ERK signaling and AMPA receptor trafficking is unknown. AGAP3 is co-precipitated with SynGAP and NR2A NMDA receptor complex. AGAP3 knock down increases the basal level of phospho-ERK signal, and decreases the phospho-ERK signal upon stimulation. AGAP3 binds to the GTP-locked mutant of Arf6, and AGAP3 overexpression decreases cellular Arf6-GTP levels. Whether AGAP3 has direct GAP activity towards Arf6 is unknown. AGAP3 overexpression also decreases Ras-GTP levels. Upon knock down of AGAP3, AMPA receptor on the cell surface in rat hippocampal cultures slightly increases (~20%), and this phenotype is rescued by GDP-locked mutant of Arf6. Upon stimulation, cell surface AMPA receptor decreases in AGAP3 knock down cells; in contrast, AMPA receptor increases in control cells. NMDA receptor transport is not affected by AGAP3 knock down. It is proposed that AGAP3 plays a role as a component of the NMDA receptor signaling complex that links activation of NMDA receptor to AMPA receptor trafficking.
ASAP1 in β1-integrin and rhodopsin transport
ASAP1 (also known as AMAP1, DDEF1, DEF1 or centaurin β4) is the most studied ArfGAP of the PH domain-containing ArfGAPs. ASAPs has BAR, PH, ArfGAP, ankyrin repeat, Proline-rich, (E/DLPPKP) repeat and SH3 domains. ASAP1 uses Arf1 and Arf5 as substrates in a 200-fold preference over Arf6[39]. ASAP1 localizes at the focal adhesions[40], and ASAP1 is implicated in tumor invasion and malignancy[41, 42]. Chromosomal amplification of ASAP1 has been found in uveal melanoma [41]. Overexpression of ASAP1 was found in prostate cancer[43], ovarian cancer[44], breast cancer[45], hepatocellular carcinoma[46], and colorectal cancer[47]. ASAP1 binds to a number of signaling proteins including PRKD2[39, 48–55]. PRKD2 binds to β1-integrin, and ASAP1 or PRKD2 knock down inhibits the recycling of β1-integrin upon EGF stimulation in breast cancer cells[54]. Overexpression of a fragment of PRKD2 that inhibits endogenous binding between ASAP1 and PRKD2 also inhibits β1-integrin recycling upon EGF stimulation, therefore the binding between ASAP1 and PRKD2 is required for β1-integrin recycling. The depletion of ASAP1 or PRKD2, or overexpression of a fragment of PRKD2 that inhibits the binding between ASAP1 and PRKD2, inhibits matrigel invasion but not adhesion to collagen of breast cancer cells.
ASAP1 is proposed to be an effecter of Arf6[42, 56], as ASAP1 binds to GTP-locked mutant of Arf6 over Arf1 and Arf5[57]; however, the interaction does not appear to be direct. Inoue et al. reported that ASAP1 binds to the Arf6/Rab11 binding proteins Fip3 and Fip4, and that there is a likely indirect interaction with Arf6 mediated by Fip3/4[52]. As ASAP1 GAP activity is towards Arf1 and Arf5 in vitro[39], and ASAP1 knock down leads to up-regulation of Arf1 GTP levels in fibroblast cells[58], the direct substrate of ASAP1 is likely Arf1. It is possible that there is a signaling cascade such as GTP hydrolysis on Arf1 by ASAP1 occurs in the downstream of Arf6 binding to ASAP1.
ASAP1 has also been reported to function in ciliary transport of rhodopsin in photoreceptor cells and polycystin-1 in retinal cortical tubular epithelia (RCTE) and Madin-Darby canine kidney (MDCK) cells[59–61]. The VxPx motif in the cytoplasmice tail of rhodoposin binds to Arf4[62], and makes a complex with ASAP1. Depletion of ASAP1 or inhibition of GAP activity of ASAP1 inhibits incorporation of rhodopsin into rhodopsin-transport carriers[60], and ciliary targeting of rhodopsin[61]. ASAP1 also binds to another ciliary targeting motif, the FR motif in cytoplasmic tail of rhodoposin separately from Arf4. Wang et al. proposed that ASAP1 serves as a platform for Rab11- FIP3 and Rabin8-Rab8 complexes sequentially. Another ciliary targeting cargo, polycystin-1 also has the VxPx motif, and binds to Arf4 and ASAP1[59].
SMAP1 in c-KIT transport
SMAP1/2 are ArfGAPs with a clathrin-binding motif, and their overexpression inhibits clathrin dependent transport [63–65]. SMAP1 has GAP activity towards Arf6[63], and SMAP2 towards Arf1 and Arf6 [64]. C-KIT(CD117) is a surface marker for hematopoietic progenitor cells. C-KIT is a receptor tyrosine kinase and its ligand is stem cell factor (SCF). In SMAP1 knock out mouse, c-KIT degradation upon SCF stimulation is inhibited in bone marrow-derived mast cells (BMMCs) and transport of c-KIT to the lysosome is inhibited in mouse embryo fibroblast (MEF) cells[66]. In SMAP1−/− MEF cells, c-KIT colocalizes with multi-vesicular bodies (MVBs) markers such as Hrs and Rab7, therefore it is thought that SMAP1 plays a role in transport of c-KIT from MVBs to the lysosome. Interestingly, EGFR transport is not affected in SMAP1−/− MEF cells, suggesting the function of SMAP1 in c-KIT transport to the lysosome is specific. This result is consistent the general model that we have proposed that ArfGAPs are cargo specific regulators of membrane traffic.
Concluding remarks
The role of ArfGAPs in post-Golgi traffic is just beginning to emerge. Recent work indicates that ArfGAPs are necessary for specific intracellular trafficking of receptors. The specificity between cargos and ArfGAPs, and the relationship between cargo recognition and regulation of GAP activity, are still poorly understood. The spatial, temporal, and molecular level studies are required for understanding these processes. These studies will reveal the importance of ArfGAPs in physiology and disease states that lacks proper regulation of intracellular trafficking.
Acknowledgments
The work was funded by the intramural program of the National Cancer Institute (project BC007365).
References
- 1.Kahn RA, Bruford E, Inoue H, Logsdon JM, Jr, Nie Z, Premont RT, et al. Consensus nomenclature for the human ArfGAP domain-containing proteins. J Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. http://dx.doi.org/10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. http://dx.doi.org/10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 3.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. http://dx.doi.org/10.1038/nrm3159 http://dx.doi.org/10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glick BS, Nakano A. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol. 2009;25:113–132. doi: 10.1146/annurev.cellbio.24.110707.175421. http://dx.doi.org/10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nickel W, Malsam J, Gorgas K, Ravazzola M, Jenne N, Helms JB, et al. Uptake by COPI-coated vesicles of both anterograde and retrograde cargo is inhibited by GTPgammaS in vitro. J Cell Sci. 1998;111:3081–3090. doi: 10.1242/jcs.111.20.3081. [DOI] [PubMed] [Google Scholar]
- 6.Lanoix J, Ouwendijk J, Lin CC, Stark A, Love HD, Ostermann J, et al. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COP I vesicles. EMBO J. 1999;18:4935–4948. doi: 10.1093/emboj/18.18.4935. http://dx.doi.org/10.1093/emboj/18.18.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pepperkok R, Whitney JA, Gomez M, Kreis TE. COPI vesicles accumulating in the presence of a GTP restricted arf1 mutant are depleted of anterograde and retrograde cargo. J Cell Sci. 2000;113:135–144. doi: 10.1242/jcs.113.1.135. [DOI] [PubMed] [Google Scholar]
- 8.Yang JS, Lee SY, Gao M, Bourgoin S, Randazzo PA, Premont RT, et al. ARFGAP1 promotes the formation of COPI vesicles, suggesting function as a component of the coat. J Cell Biol. 2002;159:69–78. doi: 10.1083/jcb.200206015. http://dx.doi.org/10.1083/jcb.200206015 http://dx.doi.org/10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. http://dx.doi.org/10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 10.Poon PP, Cassel D, Spang A, Rotman M, Pick E, Singer RA, et al. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 1999;18:555–564. doi: 10.1093/emboj/18.3.555. http://dx.doi.org/10.1093/emboj/18.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanoix J, Ouwendijk J, Stark A, Szafer E, Cassel D, Dejgaard K, et al. Sorting of Golgi resident proteins into different subpopulations of COPI vesicles: a role for ArfGAP1. J Cell Biol. 2001;155:1199–1212. doi: 10.1083/jcb.200108017. http://dx.doi.org/10.1083/jcb.200108017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SY, Yang JS, Hong W, Premont RT, Hsu VW. ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J Cell Biol. 2005;168:281–290. doi: 10.1083/jcb.200404008. http://dx.doi.org/10.1083/jcb.200404008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009;583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. http://dx.doi.org/10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 14.East MP, Kahn RA. Models for the functions of Arf GAPs. Semin Cell Dev Biol. 2011;22:3–9. doi: 10.1016/j.semcdb.2010.07.002. http://dx.doi.org/10.1016/j.semcdb.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiba Y, Randazzo PA. ArfGAP1 function in COPI mediated membrane traffic: currently debated models and comparison to other coat-binding ArfGAPs. Histol Histopathol. 2012;27:1143–1153. doi: 10.14670/HH-27.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiba Y, Luo R, Hinshaw JE, Szul T, Hayashi R, Sztul E, et al. ArfGAP1 promotes COPI vesicle formation by facilitating coatomer polymerization. Cell Logist. 2011;1:139–154. doi: 10.4161/cl.1.4.18896. http://dx.doi.org/10.4161/cl.1.4.18896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo R, Ha VL, Hayashi R, Randazzo PA. Arf GAP2 is positively regulated by coatomer and cargo. Cell Signal. 2009;21:1169–1179. doi: 10.1016/j.cellsig.2009.03.006. http://dx.doi.org/10.1016/j.cellsig.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiba Y, Kametaka S, Waguri S, Presley JF, Randazzo PA. ArfGAP3 regulates the Transport of Cation-Independent Mannose 6-phosphate Receptor in the post-Golgi compartment. Current Biology. 2013;23:1945–1951. doi: 10.1016/j.cub.2013.07.087. http://dx.doi.org/10.1016/j.cub.2013.07.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saitoh A, Shin HW, Yamada A, Waguri S, Nakayama K. Three homologous ArfGAPs participate in coat protein I-mediated transport. J Biol Chem. 2009;284:13948–13957. doi: 10.1074/jbc.M900749200. http://dx.doi.org/10.1074/jbc.M900749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obinata D, Takayama K, Urano T, Murata T, Ikeda K, Horie-Inoue K, et al. ARFGAP3, an androgen target gene, promotes prostate cancer cell proliferation and migration. Int J Cancer. 2011;130:2240–2248. doi: 10.1002/ijc.26224. http://dx.doi.org/10.1002/ijc.26224. [DOI] [PubMed] [Google Scholar]
- 21.Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, et al. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J Cell Biol. 2000;151:627–638. doi: 10.1083/jcb.151.3.627. http://dx.doi.org/10.1083/jcb.151.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai J, Li J, Bos E, Porcionatto M, Premont RT, Bourgoin S, et al. ACAP1 promotes endocytic recycling by recognizing recycling sorting signals. Dev Cell. 2004;7:771–776. doi: 10.1016/j.devcel.2004.10.002. http://dx.doi.org/10.1016/j.devcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Ballif BA, Powelka AM, Dai J, Gygi SP, et al. Phosphorylation of ACAP1 by Akt regulates the stimulation-dependent recycling of integrin beta1 to control cell migration. Dev Cell. 2005;9:663–673. doi: 10.1016/j.devcel.2005.09.012. http://dx.doi.org/10.1016/j.devcel.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Peters PJ, Bai M, Dai J, Bos E, Kirchhausen T, et al. An ACAP1-containing clathrin coat complex for endocytic recycling. J Cell Biol. 2007;178:453–464. doi: 10.1083/jcb.200608033. http://dx.doi.org/10.1083/jcb.200608033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai M, Pang X, Lou J, Zhou Q, Zhang K, Ma J, et al. Mechanistic insights into regulated cargo binding by ACAP1 protein. J Biol Chem. 2012;287:28675–28685. doi: 10.1074/jbc.M112.378810. http://dx.doi.org/10.1074/jbc.M112.378810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, et al. ARAP1: a point of convergence for Arf and Rho signaling. Mol Cell. 2002;9:109–119. doi: 10.1016/s1097-2765(02)00428-8. http://dx.doi.org/10.1016/S1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- 27.Yoon HY, Lee JS, Randazzo PA. ARAP1 regulates endocytosis of EGFR. Traffic. 2008;9:2236–2252. doi: 10.1111/j.1600-0854.2008.00839.x. http://dx.doi.org/10.1111/j.1600-0854.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniele T, Di Tullio G, Santoro M, Turacchio G, Yoon HY, De Matteis MA. ARAP1 regulates EGF receptor trafficking and signalling. Traffic. 2008;9:2221–35. doi: 10.1111/j.1600-0854.2008.00823.x. http://dx.doi.org/10.1111/j.1600-0854.2008.00823.x. [DOI] [PubMed] [Google Scholar]
- 29.Kang SA, Lee ES, Yoon HY, Randazzo PA, Lee ST. PTK6 inhibits down-regulation of EGF receptor through phosphorylation of ARAP1. J Biol Chem. 2010;285:26013–26021. doi: 10.1074/jbc.M109.088971. http://dx.doi.org/10.1074/jbc.M109.088971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simova S, Klima M, Cermak L, Sourkova V, Andera L. Arf and Rho GAP adapter protein ARAP1 participates in the mobilization of TRAIL-R1/DR4 to the plasma membrane. Apoptosis. 2008;13:423–436. doi: 10.1007/s10495-007-0171-8. http://dx.doi.org/10.1007/s10495-007-0171-8. [DOI] [PubMed] [Google Scholar]
- 31.Nie Z, Stanley KT, Stauffer S, Jacques KM, Hirsch DS, Takei J, et al. AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J Biol Chem. 2002;277:48965–48975. doi: 10.1074/jbc.M202969200. http://dx.doi.org/10.1074/jbc.M202969200. [DOI] [PubMed] [Google Scholar]
- 32.Bendor J, Lizardi-Ortiz JE, Westphalen RI, Brandstetter M, Hemmings HC, Jr, Sulzer D, et al. AGAP1/AP-3-dependent endocytic recycling of M5 muscarinic receptors promotes dopamine release. EMBO J. 2010;29:2813–2826. doi: 10.1038/emboj.2010.154. http://dx.doi.org/10.1038/emboj.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie Z, Boehm M, Boja ES, Vass WC, Bonifacino JS, Fales HM, et al. Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev Cell. 2003;5:513–521. doi: 10.1016/s1534-5807(03)00234-x. http://dx.doi.org/10.1016/S1534-5807(03)00234-X. [DOI] [PubMed] [Google Scholar]
- 34.Dell’Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Cell Biol. 2009;21:552–559. doi: 10.1016/j.ceb.2009.04.014. http://dx.doi.org/10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Shiba Y, Romer W, Mardones GA, Burgos PV, Lamaze C, Johannes L. AGAP2 regulates retrograde transport between early endosomes and the TGN. J Cell Sci. 2010;123:2381–2390. doi: 10.1242/jcs.057778. http://dx.doi.org/10.1242/jcs.057778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandvig K, Bergan J, Kavaliauskiene S, Skotland T. Lipid requirements for entry of protein toxins into cells. Prog Lipid Res. 2014;54C:1–13. doi: 10.1016/j.plipres.2014.01.001. http://dx.doi.org/10.1016/j.plipres.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y, Zhao Y, Ma X, Zhu Y, Patel J, Nie Z. The Arf GAP AGAP2 interacts with beta-arrestin2 and regulates beta2-adrenergic receptor recycling and ERK activation. Biochem J. 2013;452:411–421. doi: 10.1042/BJ20121004. http://dx.doi.org/10.1042/BJ20121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oku Y, Huganir RL. AGAP3 and Arf6 regulate trafficking of AMPA receptors and synaptic plasticity. J Neurosci. 2013;33:12586–12598. doi: 10.1523/JNEUROSCI.0341-13.2013. http://dx.doi.org/10.1523/JNEUROSCI.0341-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown MT, Andrade J, Radhakrishna H, Donaldson JG, Cooper JA, Randazzo PA. ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol Cell Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Randazzo PA, Andrade J, Miura K, Brown MT, Long YQ, Stauffer S, et al. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc Natl Acad Sci U S A. 2000;97:4011–4006. doi: 10.1073/pnas.070552297. http://dx.doi.org/10.1073/pnas.070552297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehlers JP, Worley L, Onken MD, Harbour JW. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin Cancer Res. 2005;11:3609–3613. doi: 10.1158/1078-0432.CCR-04-1941. http://dx.doi.org/10.1158/1078-0432.CCR-04-1941. [DOI] [PubMed] [Google Scholar]
- 42.Sabe H, Onodera Y, Mazaki Y, Hashimoto S. ArfGAP family proteins in cell adhesion, migration and tumor invasion. Curr Opin Cell Biol. 2006;18:558–564. doi: 10.1016/j.ceb.2006.08.002. http://dx.doi.org/10.1016/j.ceb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Lin D, Watahiki A, Bayani J, Zhang F, Liu L, Ling V, et al. ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res. 2008;68:4352–4359. doi: 10.1158/0008-5472.CAN-07-5237. http://dx.doi.org/10.1158/0008-5472.CAN-07-5237. [DOI] [PubMed] [Google Scholar]
- 44.Hou T, Yang C, Tong C, Zhang H, Xiao J, Li J. Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. Int J Clin Exp Pathol. 2013;7:280–287. [PMC free article] [PubMed] [Google Scholar]
- 45.Onodera Y, Hashimoto S, Hashimoto A, Morishige M, Mazaki Y, Yamada, et al. Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J. 2005;24:963–973. doi: 10.1038/sj.emboj.7600588. http://dx.doi.org/10.1038/sj.emboj.7600588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okabe H, Furukawa Y, Kato T, Hasegawa S, Yamaoka Y, Nakamura Y. Isolation of development and differentiation enhancing factor-like 1 (DDEFL1) as a drug target for hepatocellular carcinomas. Int J Oncol. 2004;24:43–48. [PubMed] [Google Scholar]
- 47.Muller T, Stein U, Poletti A, Garzia L, Rothley M, Plaumann D, et al. ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene. 2010;29:2393–2403. doi: 10.1038/onc.2010.6. http://dx.doi.org/10.1038/onc.2010.6. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Loijens JC, Martin KH, Karginov AV, Parsons JT. The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol Biol Cell. 2002;13:2147–2156. doi: 10.1091/mbc.E02-01-0018. http://dx.doi.org/10.1091/mbc.E02-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oda A, Wada I, Miura K, Okawa K, Kadoya T, Kato T, et al. CrkL directs ASAP1 to peripheral focal adhesions. J Biol Chem. 2003;278:6456–6460. doi: 10.1074/jbc.M210817200. http://dx.doi.org/10.1074/jbc.M210817200. [DOI] [PubMed] [Google Scholar]
- 50.Hashimoto S, Hirose M, Hashimoto A, Morishige M, Yamada A, Hosaka H, et al. Targeting AMAP1 and cortactin binding bearing an atypical src homology 3/proline interface for prevention of breast cancer invasion and metastasis. Proc Natl Acad Sci U S A. 2006;103:7036–7041. doi: 10.1073/pnas.0509166103. http://dx.doi.org/10.1073/pnas.0509166103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nam JM, Onodera Y, Mazaki Y, Miyoshi H, Hashimoto S, Sabe H. CIN85, a Cbl-interacting protein, is a component of AMAP1-mediated breast cancer invasion machinery. EMBO J. 2007;26:647–656. doi: 10.1038/sj.emboj.7601534. http://dx.doi.org/10.1038/sj.emboj.7601534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue H, Ha VL, Prekeris R, Randazzo PA. Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol Biol Cell. 2008;19:4224–4237. doi: 10.1091/mbc.E08-03-0290. http://dx.doi.org/10.1091/mbc.E08-03-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiba Y, Randazzo PA. GEFH1 binds ASAP1 and regulates podosome formation. Biochem Biophys Res Commun. 2011;406:574–579. doi: 10.1016/j.bbrc.2011.02.093. http://dx.doi.org/10.1016/j.bbrc.2011.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Onodera Y, Nam JM, Hashimoto A, Norman JC, Shirato H, Hashimoto S, et al. Rab5c promotes AMAP1-PRKD2 complex formation to enhance beta1 integrin recycling in EGF-induced cancer invasion. J Cell Biol. 2012;197:983–996. doi: 10.1083/jcb.201201065. http://dx.doi.org/10.1083/jcb.201201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. http://dx.doi.org/10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- 56.Sabe H, Hashimoto S, Morishige M, Ogawa E, Hashimoto A, Nam JM, et al. The EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer invasion and metastasis. Traffic. 2009;10:982–993. doi: 10.1111/j.1600-0854.2009.00917.x. http://dx.doi.org/10.1111/j.1600-0854.2009.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hashimoto S, Hashimoto A, Yamada A, Onodera Y, Sabe H. Assays and properties of the ArfGAPs, AMAP1 and AMAP2, in Arf6 function. Methods Enzymol. 2005;404:216–231. doi: 10.1016/S0076-6879(05)04021-8. http://dx.doi.org/10.1016/S0076-6879(05)04021-8. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, Yerushalmi GM, Grigera PR, Parsons JT. Mislocalization or reduced expression of Arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling. J Biol Chem. 2005;280:8884–8892. doi: 10.1074/jbc.M412200200. http://dx.doi.org/10.1074/jbc.M412200200. [DOI] [PubMed] [Google Scholar]
- 59.Ward HH, Brown-Glaberman U, Wang J, Morita Y, Alper SL, Bedrick EJ, et al. A conserved signal and GTPase complex are required for the ciliary transport of polycystin-1. Mol Biol Cell. 2011;22:3289–3305. doi: 10.1091/mbc.E11-01-0082. http://dx.doi.org/10.1091/mbc.E11-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mazelova J, Astuto-Gribble L, Inoue H, Tam BM, Schonteich E, Prekeris R, et al. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28:183–192. doi: 10.1038/emboj.2008.267. http://dx.doi.org/10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Morita Y, Mazelova J, Deretic D. The Arf GAP ASAP1 provides a platform to regulate Arf4- and Rab11-Rab8-mediated ciliary receptor targeting. EMBO J. 2012;31:4057–4071. doi: 10.1038/emboj.2012.253. http://dx.doi.org/10.1038/emboj.2012.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc Natl Acad Sci U S A. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. http://dx.doi.org/10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanabe K, Torii T, Natsume W, Braesch-Andersen S, Watanabe T, Satake M. A novel GTPase-activating protein for ARF6 directly interacts with clathrin and regulates clathrin-dependent endocytosis. Mol Biol Cell. 2005;16:1617–1628. doi: 10.1091/mbc.E04-08-0683. http://dx.doi.org/10.1091/mbc.E04-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Natsume W, Tanabe K, Kon S, Yoshida N, Watanabe T, Torii T, et al. SMAP2, a novel ARF GTPase-activating protein, interacts with clathrin and clathrin assembly protein and functions on the AP-1-positive early endosome/trans-Golgi network. Mol Biol Cell. 2006;17:2592–2603. doi: 10.1091/mbc.E05-10-0909. http://dx.doi.org/10.1091/mbc.E05-10-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kon S, Tanabe K, Watanabe T, Sabe H, Satake M. Clathrin dependent endocytosis of E-cadherin is regulated by the Arf6GAP isoform SMAP1. Exp Cell Res. 2008;314:1415–1428. doi: 10.1016/j.yexcr.2007.11.006. http://dx.doi.org/10.1016/j.yexcr.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 66.Kon S, Minegishi N, Tanabe K, Watanabe T, Funaki T, Wong WF, et al. Smap1 deficiency perturbs receptor trafficking and predisposes mice to myelodysplasia. J Clin Invest. 2013;123:1123–1137. doi: 10.1172/JCI63711. http://dx.doi.org/10.1172/JCI63711. [DOI] [PMC free article] [PubMed] [Google Scholar]