Abstract
Urinary exosomes or microvesicles are being studied intensively to identify potential new biomarkers for renal disease. We sought to identify whether these microvesicles contain nucleic acids. We isolated microvesicles from human urine in the same density range as that previously described for urinary exosomes and found them to have an RNA integrity profile similar to that of kidney tissue, including 18S and 28S rRNA. This profile was better preserved in urinary microvesicles compared with whole cells isolated from urine, suggesting that microvesicles may protect RNA during urine passage. We were able to detect mRNA in the human urinary microvesicles encoding proteins from all regions of the nephron and the collecting duct. Further, to provide a proof of principle, we found that microvesicles isolated from the urine of the V-ATPase B1 subunit knockout mice lacked mRNA of this subunit while containing a normal amount of the B2 subunit and aquaporin 2. The microvesicles were found to be contaminated with extraneous DNA potentially on their surface; therefore, we developed a rapid and reliable means to isolate nucleic acids from within urine microvesicles devoid of this extraneous contamination. Our study provides an experimental strategy for the routine isolation and use of urinary microvesicles as a novel and non-invasive source of nucleic acids to further renal disease biomarker discovery.
Keywords: acute kidney injury, chronic kidney disease, exosome, transcriptional profiling
Exosomes are classically formed from the inward invagination and pinching-off of the late endosomal membrane. This results in the formation of a multivesicular body (MVB) laden with small lipid-bilayered vesicles (~ 40–100 nm in diameter), each of which contains a sample of the parent cell’s cytoplasm.1 Fusion of the MVB with the cell membrane results in the release of these ‘exosomes’ from the cell, and their delivery into the blood, urine, or other body fluids. Exosome-like vesicles including ‘shedding microvesicles’2 may also be formed by the budding-off of the cell’s plasma membrane, and although more heterogeneous in size,2,3 may also contain a snapshot of the parent cell’s RNA. Although the majority of microvesicles isolated from urine are thought to be exosomes,4 both exosomes and other microvesicles do co-isolate during ultracentrifugation and ultrafiltration isolation techniques and will, therefore, be collectively referred to as microvesicles here. Recent pioneering oncology research has shown that exosomes carry mRNA and/or miRNA5–7 that may encode tumor markers, potentially circumventing the need for biopsies and highlighting the enormous diagnostic potential of exosome biology.5
Comprehensive studies have been conducted on the proteomic analysis of urinary microvesicles, revealing that they contain a variety of cell-specific proteins/transporters from the kidney and the urogenital tract.4,8 It was further shown that urinary microvesicles are very stable, highlighting their potential use as a reliable urinary marker.9 However, there have been no in-depth studies analyzing the nature of nucleic acids within the microvesicles or their reliability as a source of nucleic acid biomarkers for renal function. Whole urine is known to contain nucleic acids derived from whole cells and free DNA.10,11 However, such extraneous nucleic acids may not be a reliable source of biomarkers as they may be derived from apoptotic cells, the transcriptional profile of which may not be representative of a functioning cell.
The investigation of new biomarkers for renal disease is currently an important and pressing issue, with renal disease affecting up to 1 in 10 of the US population.12 Urinary microvesicles may provide a unique means to analyze the transcriptional profile of the kidney as they are derived from functioning cells. The isolation of microvesicles usually calls for ultracentrifugation, but recent studies have shown that they may also be rapidly isolated using filtration concentrators,13 increasing the potential for their use in routine diagnostic analysis. Currently, it is not known whether the use of filtration concentrators would yield intact microvesicles for RNA extraction.
Here we conduct an in-depth analysis of the nucleic acids associated with urinary microvesicles, including (i) the potential for extraneous nucleic acid contamination during urinary microvesicle isolation, (ii) the non-invasive identification of renal related transcripts from various regions of the nephron and collecting duct by reverse transcriptase-PCR (RT-PCR), (iii) the application of microvesicle derived RNA analysis in renal pathophysiology, (iv) analysis of RNA integrity in microvesicles versus whole cells in urine, and (v) the nucleic acid analysis of isolated microvesicles using filtration concentrators versus ultracentrifugation. These studies increase our understanding of urinary microvesicles and support their potential as a novel source of new and much needed biomarkers for renal disease analysis.
RESULTS
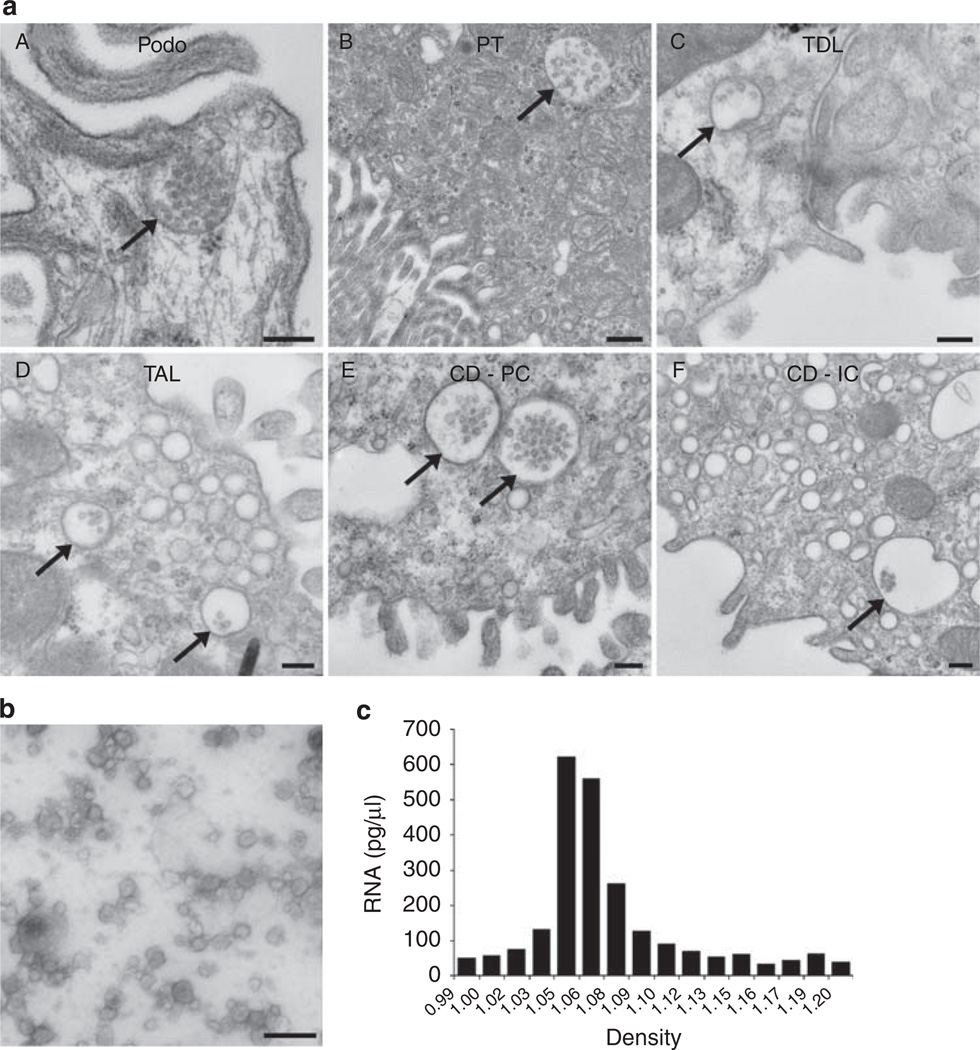
Figure 1a shows transmission electron microscope images of MVBs present in rat renal tissue. This shows that exosomes can indeed be released from various regions of the nephron, as well as from both intercalated and principal cells of the collecting duct. Using transmission electron microscopy we also examined the pellet isolated by differential centrifugation to show that the pellet was indeed rich in microvesicles (Figure 1b). To ensure that the RNA was coming from microvesicles and not large membrane blebs, a Percoll gradient was used to separate the pelleted microvesicles based on density (Figure 1c) and RNA was extracted from each fraction. Results revealed that the RNA obtained was indeed coming from microvesicles within the same density range as that previously described for urinary exosomes.14
Figure 1. Electron microscopy of urinary microvesicles.
(a) Multivesicular bodies (MVBs) can be identified in various regions of the nephron and collecting duct (see arrows). Bar = 200 nm for A, C, D, E, F; 500 nm for B. (b) Human urinary microvesicles isolated using differential ultracentrifugation and imaged via transmission electron microscopy using phosphotungstic acid as a stain. Bar = 200 nm. (c) Percoll gradient analysis of urinary microvesicles shows that RNA-containing microvesicles are within the density range for urinary microvesicles previously characterized as exosomes. CD-IC, collecting duct intercalated cell; CD-PC, collecting duct principal cell; Podo, podocyte; PT, proximal tubule; TAL, thick ascending limb; TDL, thin descending limb.
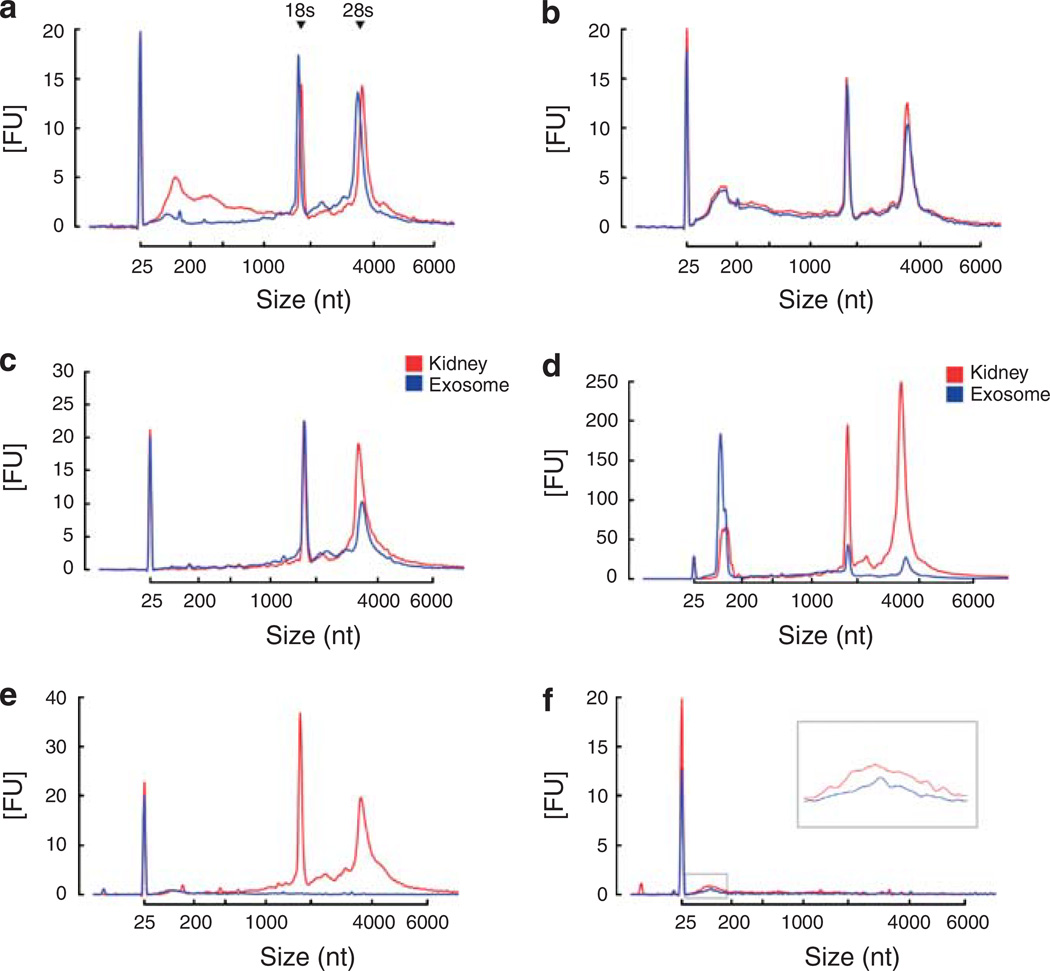
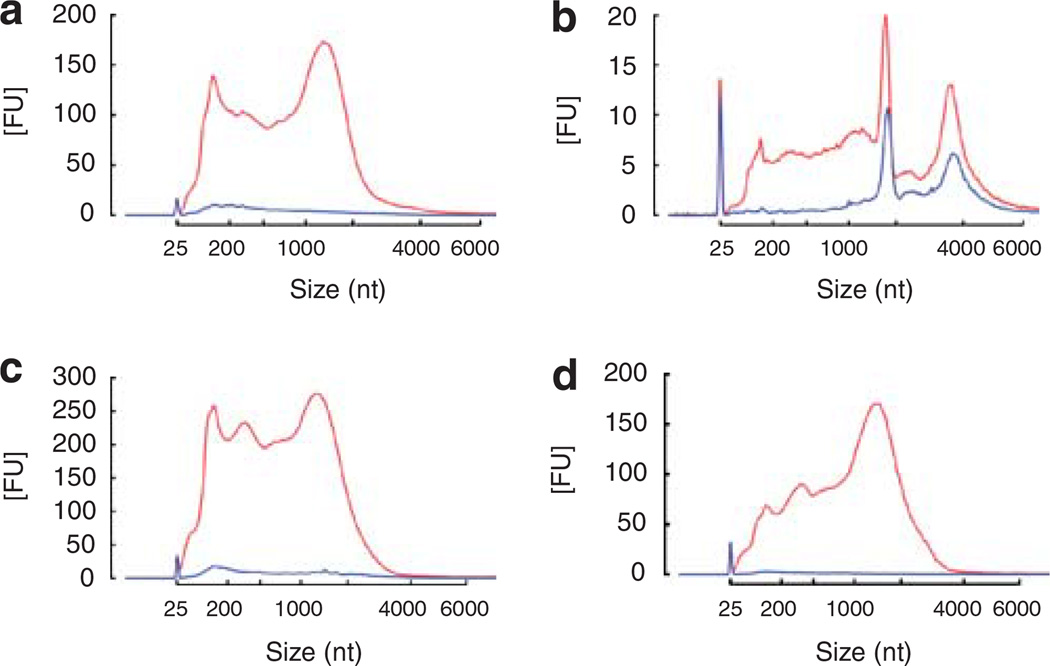
Using differential centrifugation in combination with RNase and DNase digestion of the microvesicle pellet, we determined whether extraneous nucleic acids may co-isolate with urinary microvesicles. Results revealed that extraneous DNA (that is, free DNA) contaminates isolated urinary microvesicles (Figure 2a) and serum-derived microvesicles (see Supplementary Figure S1d). The level of extraneous DNA varied between subjects but was consistently shown to be predominantly in the range of ~25–1500 nucleotides (nt). In contrast, no obvious extraneous RNA could be detected (Figure 2b), consistent with the presence of ribonucleases in urine.15 Analysis of the nucleic acids within microvesicles revealed some striking features, including the presence of prominent 18S and 28S rRNA peaks similar to those seen for RNA isolated from rat kidney tissue (Figure 2c). Such rRNA was not prevalent in previously reported serum-derived or cell culture-derived microvesicles, although they may be detectable depending on isolation technique as shown in the serum RNA isolation profile in Supplementary Figure S1d. Both urinary microvesicles and kidney tissue also contained small RNAs when miRNA isolation methods were used (Figure 2d) as previously reported.16 Various extraction kits are available for the isolation of RNA and removal of DNA. A comparative analysis of RNeasy Qiagen kits and the acid phenol/chloroform-based mirVana kit is shown in Supplementary Figure S1.
Figure 2. Analysis of nucleic acids associated with urinary microvesicles using the Agilent Bioanalyzer.
(a) Plot showing that microvesicles may co-isolate with extraneous DNA that can be removed by DNase digestion of the microvesicle pellet prior to lysis and nucleic acid extraction. Red — profile without DNase digestion, blue — profile with DNase digestion. (b) Plot showing that microvesicles do not co-isolate with detectable levels of extraneous RNA. Red — without RNase digestion, blue — with RNase digestion. (c) RNA isolated from rat kidney (red) and microvesicles (blue) exhibited a very similar profile, including the presence of 18S and 28S rRNA peaks. Both samples underwent processing using the RNeasy Plus Micro kit to remove genomic DNA (gDNA) contamination. (d) Urinary microvesicles contain a prominent ‘small RNA’ peak between 25–200 nt when miRNA isolation techniques are used. Red — kidney RNA isolated using RNeasy Plus Micro kit using the miRNA extraction method, blue — microvesicle RNA isolated with RNeasy Plus Micro kit using the miRNA extraction method. (e) Nucleic acids were isolated from microvesicles that had undergone RNase and DNase digestion on the outside before microvesicle lysis. During RNA extraction using the RNeasy Micro kit, half of the samples underwent on-column RNase digestion (see Materials and methods) while the other half underwent the same on-column incubation without the presence of RNase. Results revealed that RNase digestion was able to remove the majority of the profile, suggesting that RNA is the major nucleic acid within urinary microvesicles. Red — nucleic acid profile without intra-microvesicular RNase digestion, blue — nucleic acid profile with intra-microvesicular RNase digestion. (f) Further digestion with DNase following RNase digestion revealed that the remaining peak could be further reduced, suggesting that some material prone to DNase digestion remained in the sample potentially representing intra-exosomal DNA. Red — nucleic acid profile following intra-microvesicular on-column RNase digestion alone, blue — nucleic acid profile following both intra-microvesicular on-column RNase and DNase digestion. 18S and 28S rRNA peaks are indicated in (a). The peak at 25 nt represents an internal standard.
To determine the nucleic acid content within urinary microvesicles, the pellets were subjected to RNase and DNase digestion to remove extraneous contamination, followed by RNase and/or DNase digestion of intra-microvesicular nucleic acids during column-based nucleic acid isolation. On-column RNase digestion almost completely abolished the nucleic acid profile (Figure 2e), suggesting that RNA represents the most abundant nucleic acid within microvesicles. Further, on-column digestion with DNase revealed that the remaining peak could be further decreased, suggesting that there may be some DNase digestible material within microvesicles (Figure 2f).
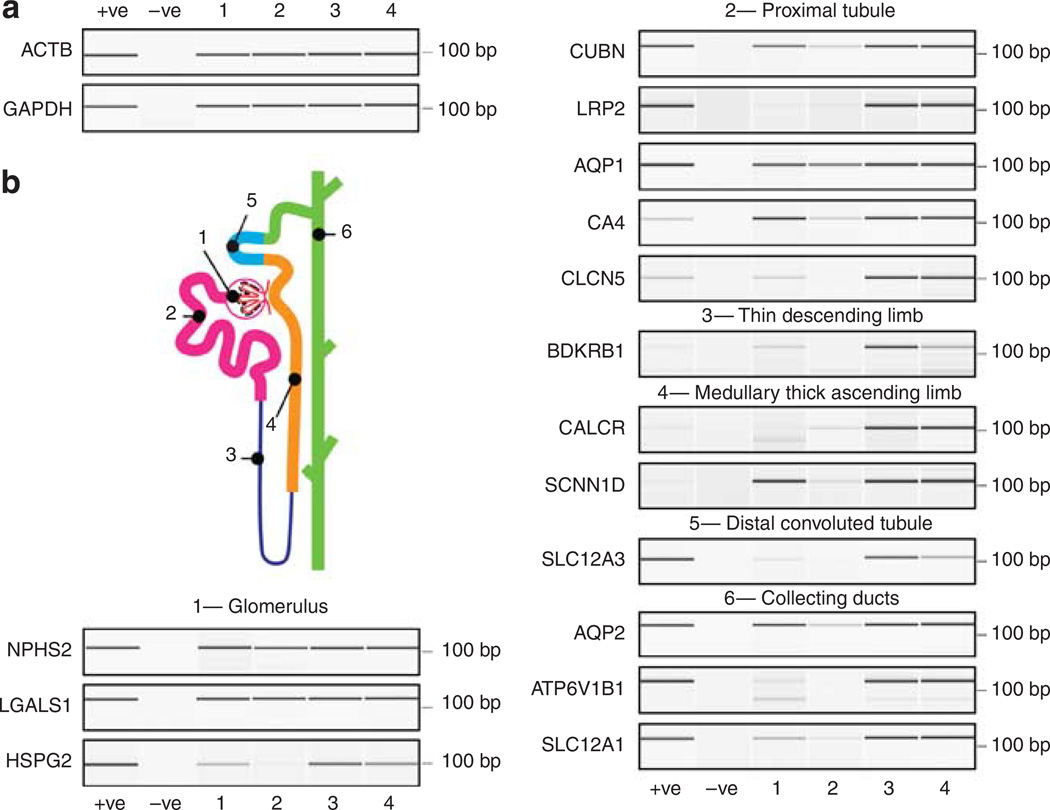
To further determine whether microvesicles contain mRNA transcripts encoding markers from various nephron and collecting duct segments, RNA isolated from urinary microvesicles of four human controls (23–32 years of age) was subjected to RT-PCR. Both glyceraldehyde 3-phosphate dehydrogenase and β-actin genes were identified in all samples (Figure 3a). Next we examined 15 transcripts characteristic of various regions of the nephron and collecting duct (Figure 3b). These included proteins and receptors implicated in various renal diseases: podocin from the glomerulus, cubilin from the proximal tubule, and aquaporin 2 from the collecting duct. Genes from all regions examined could be identified, consistent with the results obtained in Figure 1a. This shows that microvesicles containing mRNA are released from all regions of the nephron and the collecting duct and are, therefore, a novel non-invasive source of potential nucleic acid biomarkers for renal disease.
Figure 3. Urinary microvesicles contain messenger RNA (mRNA) transcripts encoding specific genes from various regions of the nephron and the collecting duct.
(a) Bioanalyzer-generated ‘Pseudo gel’ profiles of the positive identification of RiboAmp-amplified mRNA transcripts for β-actin (ACTB) and glyceraldehyde 3-phosphate (GAPDH) in urinary microvesicles from four subjects by RT-PCR. (b) Cartoon of the nephron and collecting duct highlighting its functionally distinct regions. (c) The positive identification of (1) Glomerulus: NPHS2 — podocin, LGALS1 — Galectin-1, HSPG2 — heparan sulfate proteoglycan; (2) proximal tubule: CUBN — cubilin, LRP2 — megalin, AQP1 — aquaporin 1, CA4 — carbonic anhydrase 4, CLCN5 — chloride channel protein 5, (3) thin descending limb: BDKRB1 — bradykinin B1 receptor; (4) medullary thick ascending limb: CALCR — calcitonin receptor, SCNN1D — amiloride-sensitive sodium channel subunit delta; (5) distal convoluted tubule: SLC12A3 — thiazide-sensitive sodium-chloride cotransporter; (6) collecting ducts: AQP2 — aquaporin 2, ATP6V1B1 — V-ATPase B1 subunit, SLC12A1 — kidney-specific Na–K–Cl symporter via RT-PCR of RiboAmped mRNA from urinary exosomes.
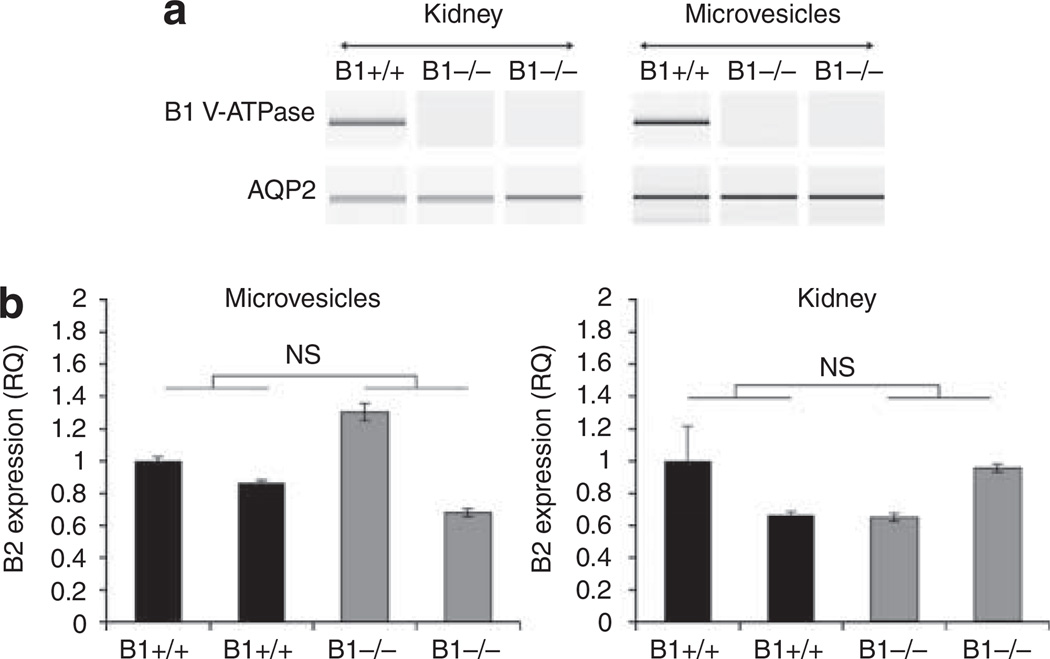
To test the hypothesis that microvesicles may be used to non-invasively examine renal genes in disease, the V-ATPase B1 subunit knockout mouse model of renal acidosis was used.17 Figure 4 shows that expression of the V-ATPase B1 subunit and aquaporin 2 mRNA can be examined noninvasively in the microvesicles using RT-PCR (Figure 4a). In addition, real-time PCR was used to quantitatively examine the expression of the V-ATPase B2 subunit. The renal expression of the V-ATPase B2 subunit is the same in V-ATPase B1 −/− animals as in V-ATPase B1 +/+ mice (Figure 4b) despite the loss of the B1 subunit, confirming the results obtained for the corresponding kidneys as well as previously reported data.18
Figure 4. Analysis of gene expression by RT-PCR and real-time PCR in the V-ATPase B1 knockout model of renal acidosis.
(a) Analysis of the V-ATPase B1 subunit and AQP2 mRNA by RT-PCR in V-ATPase B1 KO (B1 −/−) and wild-type (B1 +/+) mice reveals that similar results can be obtained via both kidney and microvesicle RNA analysis. (b) Using urinary microvesicles, real-time PCR analysis demonstrated that the expression of the V-ATPase B2 subunit could be analyzed non-invasively to demonstrate that the B2 subunit expression is not affected by the absence of the V-ATPase B1. These results were consistent with those obtained via the corresponding kidney-derived RNA analysis. NS, not statistically significant.
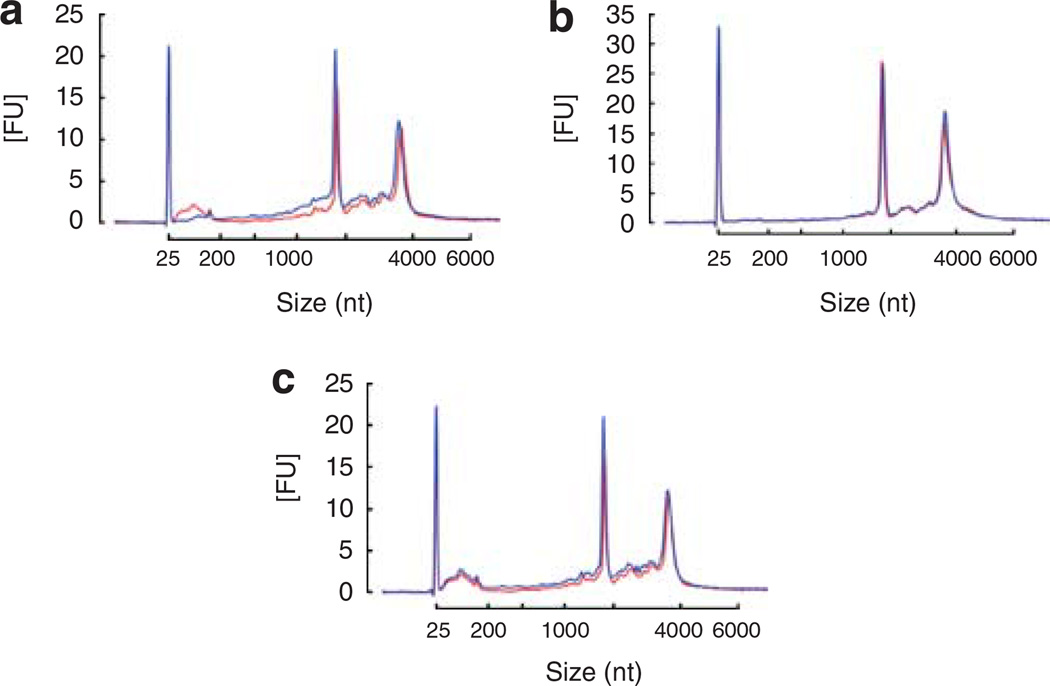
Although mRNA transcripts can be isolated from urinary microvesicles, which are devoid of extraneous DNA and RNA, what advantage does this mRNA source have over RNA derived from cells and cell debris in the urine? To examine this, we utilized a ‘whole’ urine RNA extraction kit (see Materials and methods) and compared the RNA profile with that obtained from microvesicles in the same sample. A large amount of nucleic acid could be isolated using the ZR urine RNA isolation kit (Figure 5a), the majority of which was DNA (Figure 5a). The profile appeared broad and lacked 18S and 28S rRNA peaks in comparison with that normally obtained from tissue or from within microvesicles (see Figure 2c), indicating considerable degradation. In contrast, RNA from microvesicles isolated from the same urine sample had clearly visible 18S and 28S rRNA peaks, indicating good quality RNA (Figure 5b). Analysis of the nucleic acid profile of the various pellets obtained during microvesicle differential centrifugation revealed that pellets from the 300 g (Figure 5c) and 17,000 g (Figure 5d) spins exhibited a nucleic acid profile similar to those obtained from the ZR urine RNA isolation kit and were, indeed, made up of a large proportion of DNA (Figure 5c and d, respectively) along with degraded RNA. This showed that the RNA isolated from urinary cells is less stable than the RNA isolated from urinary microvesicles.
Figure 5. RNA extracted from whole urine cells and debris has a different RNA profile from that of tissue and urinary microvesicles.
(a) Analysis of RNA isolated from whole urine (exclusive of microvesicles that are not captured by the isolation technique) showed that a large yield of nucleic acids can be isolated (see the red profile). Processing of the isolated nucleic acids using the RNeasy Plus Micro kit (which removes gDNA) reveals that the majority of nucleic acids isolated using the ZR urine RNA isolation kit is DNA and the remaining RNA lacks rRNA peaks found in tissue and urinary exosomes. Red — nucleic acids isolated from whole urine without gDNA removal, blue — nucleic acids isolated from whole urine post gDNA removal using the RNeasy Plus Micro kit. (b) Isolation of microvesicles from the same urine sample revealed that the microvesicles retained a normal total RNA profile suggesting that RNA within whole cells may be less stable than that contained in urinary microvesicles. Red — without removal of gDNA, blue — sample processed using the RNeasy Plus Micro kit to remove contaminating gDNA. (c) Isolation of nucleic acids from the pellet formed during the 300 g spin revealed that the nucleic acid profile was different from that of microvesicles and that it contained a large amount of gDNA following processing using the RNeasy Plus Micro kit. Red — nucleic acids isolated from the 300 g pellet without gDNA removal, blue — nucleic acid isolated from the 300 g pellet post gDNA removal using the RNeasy Plus Micro kit. (d) Isolation of nucleic acids from pellets formed during the 17,000 g spin revealed that the nucleic acid profile was different to microvesicles and that it contained a large amount of gDNA following processing using the RNeasy Plus Micro kit. Red — nucleic acids isolated from the 17,000 g pellet without gDNA removal, blue — nucleic acids isolated from the 17,000 g pellet post gDNA removal using the RNeasy Plus Micro kit.
Although urinary microvesicles seem like a promising source of renal biomarkers, their isolation by ultracentrifugation is both time consuming and requires elaborate equipment. We, therefore, investigated whether filtration concentrators, previously used to isolate urinary microvesicles for protein analysis,13 could yield viable microvesicles for RNA extraction. To test this, we processed urine samples up to the 0.8-µm filter step (see Materials and methods) and compared them with samples processed using a 100 kDa MWCO filtration concentrator (Millipore, Bedford, MA, USA) versus ultracentrifugation both with RNase digestion, and with and without DNase digestion to remove extra-microvesicular nucleic acid contamination. Results revealed that the ultracentrifugation method and the filtration concentration method yielded similar RNA concentrations from 75 ml urine samples (ultracentrifugation 410 ± 28 pg/µl, filtration concentrator 381 ± 47 pg/µl mean ± s.d., not statistically significant) with minimal degradation (Figure 6a), suggesting that the use of filtration concentrators may be a reliable method for the isolation of urinary microvesicles for RNA analysis. Finally, we examined whether the urine pre-processing steps could be replaced with just a 0.8-µm filtration step (see Materials and methods). Results using ultracentrifugation (Figure 6b) and filtration concentrators (Figure 6c) revealed that the 0.8 µm filtration could indeed replace the urine pre-processing steps, further decreasing the isolation time.
Figure 6. Isolation of microvesicles using filtration concentrators reveals that this may be a rapid technique to isolate intact microvesicles for nucleic acid extraction.
(a) A sample of 75 ml human urine was subjected to the initial processing steps of 300 g, 17,000 g centrifugation followed by 0.8 µm filtration and was then processed using ultracentrifugation (blue) or using 100 kDa MWCO filters (red). The results revealed that a similar profile was obtained using both extraction methods with minimal degradation of the RNA revealing that filtration concentrators may be a fast and reliable way to isolate urinary microvesicles. Comparison of the ‘normal’ 300 g, 17,000 g spin and 0.8 µm filtration steps (red) with just a 0.8 µm filtration step (blue) followed by (b) ultracentrifugation or (c) filtration concentrators revealed that the 300 g and 17,000 g spins were not crucial for the removal of cell debris and whole cells as no change in profile was observed, further simplifying the isolation technique and the time taken to isolate exosomes. All samples underwent extra-exosomal RNase and DNase digestion before microvesicular lysis.
DISCUSSION
Currently, renal biomarkers are limited to urinary protein analysis and changes in the glomerular filtration rate. Biomarkers at the nucleic acid level are understudied, in part because this requires the invasive and expensive procedure of organ biopsy. However, urinary microvesicles now offer a novel means to obtain this information without the need for invasive and expensive biopsy procedures, potentially taking renal biomarker discovery to a new level.
Analysis of extraneous nucleic acid contamination during urinary microvesicle isolation revealed that there was the potential for DNA contamination. This could be easily removed by (i) digestion of the microvesicle pellet with DNase or (ii) the use of genomic DNA elimination kits such as the RNeasy Plus Micro kit. External DNase digestion should now be part of the standard procedure when isolating microvesicles for nucleic acid analysis. This also appears to remove DNase-susceptible ‘apoptotic DNA ladder’ material, which may contaminate serum microvesicle pellets, as shown in the Supplementary Data. The finding that there was no detectable extraneous RNA contamination was not surprising, because urine contains ribonucleases.15 The fact that microvesicles can resist RNase and DNase digestion and still protect the nucleic acids contained within them is quite remarkable, and adds further support to the previously reported stable nature of urinary exosomes.9
A potential advantage of carrying out extra-microvesicular rather than on-column DNase digestion is that it leaves the nucleic acids within exosomes untouched. This is important, particularly in cases where potential DNase digestible material may be captured from the cytoplasm of a cell and may itself be a source of biomarkers for non-coding sequences, which are now believed to have a potential role in cell regulation.19–21 Other studies have also suggested that mitochondrial DNA is present in exosomes isolated from astrocytes and glioblastoma cell cultures,22 suggesting that cancer-related microvesicles may contain mtDNA. Overall, our studies suggest that microvesicles can be a reliable source of living cell cytoplasm-derived nucleic acids for biomarker discovery, and are devoid of extraneous nucleic acids when processed correctly.
Although urine-derived microvesicles contain an RNA profile similar to whole tissue including prominent 18S and 28S rRNAs, this rRNA material appears less prevalent in microvesicles isolated from the serum5–7 and saliva.23 It is not known whether this is due to differences in microvesicular; (i) yield, (ii) stability, or (iii) origin (that is, whether the RNA profile may be different in shedding microvesicles versus those derived from MVBs) in the various body fluids. A comparison of RNA sources in urine revealed that rRNA peaks appeared better preserved in microvesicles versus cells in urine, suggesting that microvesicles are a reliable source of stable nucleic acids and that they protect their inner content which is extremely important for downstream RNA analysis. Unlike whole cells, microvesicles are quite resistant to freeze–thawing, and nucleic acids can be extracted from the urinary exosomes following freeze–thawing (Russo et al., unpublished data). This suggests that in frozen archived samples, microvesicles may also be a more reliable source of RNA for longitudinal studies than whole cells from urine. Further, it is not known how release of cells from their physiological setting (that is, loss of cell–cell and cell–substrate interaction, and local stimuli) affects gene expression in whole cells found in urine. Urinary microvesicles may also be considered as a unique source of RNA not only because of their stable and non-invasive nature, but also because their RNA represents a snapshot of the whole urinary system. This is unlike the RNA obtained from renal biopsy, which represents a small sample from only one of the two kidneys.
The presence of mRNA transcripts encoding renal genes from various regions of the nephron and the collecting duct was also confirmed. These transcripts were contained within microvesicles and were confirmed as mRNA-derived, because a poly-A tail-specific RNA amplification technique was used. Many of the genes analyzed are disrupted in various renal diseases, including podocin in glomerular diseases such as steroid-resistant nephrotic syndrome,24 cubilin25 associated with proteinuria in Imerslund–Gräsbeck syndrome, and aquaporin 2 associated with diabetes insipidus.26 To further highlight the use of microvesicular RNA analysis in renal pathophysiology, we used both RT-PCR for the mRNA detection of genes as well as real-time PCR for the relative quantitation of renal genes expressed in the V-ATPase B1 knockout mouse. The analysis of mRNA in urinary microvesicles paralleled that obtained in renal tissue from these mice18 (and this study) and adds support for the use of urinary microvesicles for pathophysiological analysis.
The use of filtration concentrators to isolate urinary microvesicles indicates that elaborate ultracentrifugation steps may not be required for the isolation of microvesicles for nucleic acid analysis. The use of a 100 kDa MWCO membrane aided in the removal of DNase I (~ 39 kDa) and RNase A (~ 13.7 kDa) from the sample following extraneous nucleic acid digestion steps. This rapid isolation technique, which reduces 70 min centrifugation steps down to 4 min steps (potentially reducing more than 3.5 h of ultracentrifugation to p<30 min in a bench-top centrifuge), is extremely important for future studies into biomarker discovery and has the potential to move exosome biology into clinical laboratories as a routine diagnostic procedure.
In summary, we have shown that urinary microvesicles-may (i) be complexed with extraneous DNA, highlighting the importance of DNA removal from the sample before nucleic acid analysis; (ii) contain mainly RNA, including prominent 18S and 28S rRNA similar to that seen in tissue-derived RNA; (iii) contain mRNA transcripts representing markers from all regions of the nephron and the collecting duct, suggesting that the mRNA contained within them is stable for RT-PCR analysis; (iv) contain RNA that is more stable than RNA extracted from whole urine; and (v) be rapidly isolated using filtration concentrators without significant loss of RNA integrity, providing a rapid means to isolate microvesicles without the need for elaborate ultracentrifugation. These findings pave the way for the use of urinary microvesicles as a novel source of new and much needed nucleic acid biomarkers for renal disease.
MATERIALS AND METHODS
Urine pre-processing
Human urine was obtained under the approved instititional review board guidelines of the Massachusetts General Hospital. Urine preprocessing by the ‘normal method’ included centrifugation of the urine at 300 g for 10 min at 4 °C, centrifugation of the supernatant at 17,000 g for 20 min at 4 °C, and filtration of the supernatant through a 0.8-µm filter (cellulose nitrate membrane filter unit; Nalgene, Rochester, NY, USA). Alternatively, urine pre-processing using the ‘0.8 µm method’ included filtration of the urine directly through the 0.8 µm filter without any pre-centrifugation steps. For analysis of extraneous nucleic acids ~ 25 ml of duplicate urine samples was used, and ~ 75 ml duplicates were used for the analysis of intra-microvesicular nucleic acids. For comparison of the nucleic acid extraction kits ~ 75 ml of duplicate urine samples was used. For comparison of the RNA from urinary cells extracted using the ZR isolation kit, 300 and 17,000 g pellets, and the corresponding RNA from microvesicles, ~ 75 ml of duplicate urine samples was used. For analysis of filtration concentrators versus ultracentrifugation, ~ 75 ml of duplicate urine samples was used. For RT-PCR analysis 200 ml urine samples were used. Urine samples were collected over a 24-h period and stored at 4 °C.
Microvesicular isolation by ultracentrifugation
Following pre-processing, the filtrate underwent ultracentrifugation at 118,000 g for 70 min at 4 °C; the supernatant was removed and the microvesicle-containing pellet was washed in phosphate buffered saline (PBS) and re-pelleted at 118,000 g for 70 min at 4 °C. The microvesicle pellet was then subjected to RNase and/or DNase digestion to remove extraneous nucleic acids.
Removal of extraneous nucleic acids
The microvesicle pellet derived from ultracentrifugation was resuspended in 1 µl per ml RNase A (DNase and protease free, Fermentas, Glen Burnie, MD, USA) in PBS and incubated for 1 h at 37 °C. The samples were re-pelleted at 118,000 g for 70 min in PBS. The corresponding urine sample was used as control also incubated at 37 °C for 1 h in the presence of the RNase inhibitor cocktail SUPERaseIn (1U/µl) according to the manufacturer’s instructions. For DNase I digestion the pellet was resuspended in 500 µl PBS and DNase I (RNase free, Qiagen, Valencia, CA, USA) diluted in RDD buffer (according to manufacturer’s instructions) was incubated at room temperature for 10 min. The samples were re-pelleted at 118,000 g for 70 min in PBS. For RNase A and DNase I digestion of microvesicles isolated using filtration concentrators, the same concentration of RNase and DNase was used and incubations were carried out in filtration concentrators. RNase A or DNase I was removed by three resuspension/wash steps with 15 ml of PBS.
Microvesicle isolation using filtration concentrators
Filtration concentrators (100 kDa MWCO, Millipore) were prepared according to the manufacturer’s instructions. Pre-processed urine (see above) was added to the filtration concentrator and centrifuged at 4000 g for 4 min at RT. A wash step with 15 ml PBS was included. The isolated microvesicles were then subjected to RNase and/or DNase digestion to remove extraneous nucleic acids as described above.
Nucleic acid extraction and analysis
Once the microvesicles were isolated and digested with nucleases, the pellet was processed by the RNeasy Micro kit (Qiagen) according to the manufacturer’s instructions. In all, 350 µl of RLT buffer (with 10 µl β-mercaptoethanol per ml RLT) was used to lyse microvesicles and 16 µl of nuclease-free water was used for elution. The RNeasy Plus Micro kit (Qiagen) is designed to remove genomic DNA and was used according to the manufacturer’s instructions and eluted in 16 µl nuclease-free water. For small RNA isolation using the RNeasy Micro kit or RNeasy Plus Micro kit, the miRNA isolation method was followed according to the manufacturer’s instructions. RNA was isolated from whole urine using the ZR urine RNA isolation kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions. To remove genomic DNA from the Zymo processed sample, the eluted RNA was resuspended in 350 µl RLT buffer and processed using the RNeasy Plus Micro kit and eluted in 16 µl nuclease-free water. Isolated RNA was analyzed on a RNA Pico 6000 chip (Agilent, Palo Alto, CA, USA) using an Agilent Bioanalyzer (Agilent), which generated an electrophoretic profile and the corresponding ‘pseudo’ gel of the sample.
Percoll gradient analysis of microvesicles
Percoll gradient (Sigma, St Louis, MO, USA) was diluted with 9% saline to form a 0.9% saline/Percoll solution. Pelleted microvesicles from a 600 ml urine sample (RNase and DNase treated as described above) resuspended in 0.5 ml 0.9% saline were gently layered on top of a 14ml Percoll saline solution. A corresponding standard tube was prepared in the same manner except that density standard beads (Sigma) were layered on the top. The two tubes were centrifuged at 50,000 g for 45 min. A standard curve was plotted from the gradient density beads, and the density of each 750 µl fraction removed from the microvesicle tube was determined. For each fraction, the Percoll was separated from the microvesicles by centrifugation for 90 min at 118,000 g followed by three subsequent wash steps in ~ 20 ml PBS at 118,000 g for 70 min. RNA was then extracted from each pellet using the RNeasy Micro kit (Qiagen).
Analysis of nucleic acids within microvesicles
Digestion of nucleic acids within microvesicles was carried out using on-column RNase A (Fermentas) or DNase I (Qiagen) digestion in conjunction with the RNeasy Micro kit (Qiagen). In short, DNA digestion was carried out according to the manufacturer’s guidelines using the RNeasy Micro kit (Qiagen). RNA removal was carried out using the same steps as for DNA digestion except that 700 µl of RNase A (1 µl per ml RW1 wash buffer) was incubated on-column for 1 h at 37 °C. Following incubation, nucleases were removed using RW1 wash steps as outlined in the manufacturer’s guidelines.
Animal studies
All animal experiments were carried out in accordance with approved animal ethics guidelines at the Massachusetts General Hospital. V-ATPase B1 subunit knockout animals have been previously described.17,18 For urine collection animals were caged in metabolic cages in groups of two (n = 4 animals per group) over a period of 72 h (sufficient RNA can also be obtained by caging one animal per cage (Russo et al., unpublished data)) and urine was collected for microvesicle isolation and analysis as described above for human urine. For kidney extraction, animals were anesthetized using pentobarbital sodium (Nembutal) (Abbott Laboratories, Abbott Park, IL, USA) (65 mg per kg body weight i.p.) and kidneys immediately removed and frozen in liquid nitrogen. Using a pestle and mortar in a liquid nitrogen bath, the frozen kidney was ground up, resuspended in RNAlater (Qiagen) and stored in 1ml aliquots at −80 °C. For RNA extraction, an aliquot was thawed on ice and 50 µl lysed in 350 µl RLT buffer (with 10 µl β-mercaptoethanol per ml RLT). The rat kidney samples were processed using the RNeasy Mini kit and the RNeasy Plus kit. To determine the amount of small RNAs in the rat kidney sample they were also processed by both kits using the miRNA isolation method according to the manufacturer’s instructions. Mouse kidney samples were processed using the RNeasy Mini kit (Qiagen) with the inclusion of the DNA digestion step.
Transmission electron microscopy analysis
Rat kidney was fixed by intravascular perfusion with 2% glutaraldehyde in 0.1 mol/l sodium cacodylate buffer, pH 7.4 (Electron Microscopy Sciences, Hatfield, PA, USA), and kidney slices were further fixed overnight at 4 °C. The sample was rinsed in 0.1 mol/l sodium cacodylate buffer, post-fixed in 1% osmium tetroxide in cacodylate buffer for 1 h at room temperature, rinsed in buffer again, then rinsed in distilled water and stained, en bloc, in an aqueous solution of 2% uranyl acetate for 1 h at room temperature. The samples were rinsed in distilled water and dehydrated through a graded series of ethanol to 100%. The samples were infiltrated with Epon resin (Ted Pella, Redding, CA, USA) by overnight immersion in a 1:1 solution of Epon:ethanol. The following day samples were placed in fresh Epon for several hours and embedded in Epon overnight at 60 °C. Thin sections were cut on a Reichert Ultracut E ultramicrotome, collected on formvar-coated grids, and stained with uranyl acetate and lead citrate. For microvesicles, samples were fixed, 1:1 with 4% paraformaldehyde in distilled water. Drops of 10 µl were pipetted onto formvar-coated 200 mesh gold grids and drawn off after 1 min. Samples were rinsed twice with drops of distilled water. Aqueous 2% phosphotungstic acid was applied (10 µl) for 10 s, drawn off and rinsed once with distilled water. Samples were examined in a JEOL JEM 1011 transmission electron microscope (JOEL, Akishima, Tokyo, Japan) at 80 kV. Images were collected using an AMT (Advanced Microscopy Techniques, Danvers, MA, USA) digital imaging system.
RT-PCR analysis
Microvesicles isolated from human or mouse urine were subjected to RNase and DNase digestion on the outside of the exosomes before exosome lysis and RNA extraction. The extracted RNA underwent two rounds of mRNA amplification using RiboAmp (Molecular Devices, Sunnyvale, CA, USA). For the riboamplification in the first round of the in vitro transcription step samples were incubated at 42 °C for 4 h, and for the second in vitro transcription step samples were incubated at 42 °C for 6 h. Amplified RNA was denatured for 5 min at 65 °C and subjected to first-strand cDNA synthesis as described in the Qiagen Omniscript protocol (Qiagen).
For human samples the PCR primers used were: ACTB UTR, forward 5′-GAAGTCCCTTGCCATCCTAA-3′, reverse 5′-GCTATCACCTCCCCTGTGTG-3′; GAPDH EX, forward 5′-ACACCCACTCCTCCACCTTT-3′, reverse 5′-TGCTGTAGCCAAATTCGTTG-3′; NPHS2 UTR, forward 5′-AACTTGGTTCAGATGTCCCTTT-3′, reverse 5′-CAATGATAGGTGCTTGTAGGAAG-3′; LGALS1 EX, forward 5′-GGAAGTGTTGCAGAGGTGTG-3′, reverse 5′-TTGATGGCCTCCAGGTTG-3′; HSPG2 UTR, forward 5′-AAGGCAGGACTCACGACTGA-3′, reverse 5′-ATGGCACTTGAGCTGGATCT-3′; CUBN EX, forward 5′-CAGCTCTCCATCCTCTGGAC-3′, reverse 5′-CCGTGCATAATCAGCATGAA-3′; LRP2 EX, forward 5′-CAAAATGGAATCTCTTCAAACG-3′, reverse 5′-GTCGCAGCAACACTTTCCTT-3′; AQP1 UTR, forward 5′-TTACGCAGGTATTTAGAAGCAGAG-3′, reverse 5′-AGGGAATGGAGAAGAGAGTGTG-3′; CA4 UTR, forward 5′-ATGATGGCTCACTTCTGCAC-3′, reverse 5′-TCATGCCTAAAGTCCCACCT-3′; CLCN5 EX, forward 5′-GTGCCTGGTTACACACAACG-3′, reverse 5′-AGGATCTTGGTTCGCCATCT-3′; BDKRB1 UTR, forward 5′-GTGGTTGCCTTCCTGGTCT-3′, reverse 5′-ATGAAGTCCTCCCAAAAGCA-3′; CALCR UTR, forward 5′-ATTTTGCCACTGCCTTTCAG-3′, reverse 5′-ATTTTCTCTGGGTGCGCTAA-3′; SCNN1D UTR, forward 5′-GCGGTGATGTACCCATGCT-3′, reverse 5′-CTGAGGTGGCTAGGCTTGA-3′; SLC12A3 EX, forward 5′-AGAACAGAGTCAAGTCCCTTCG-3′, reverse 5′-TATGGGCAAAGTGATGACGA-3′; AQP2 UTR, forward 5′-GCAGTTCCTGGCATCTCTTG-3′, reverse 5′-GCCTTTGTCCTTCCCTAACC-3′; ATP6V1B1 EX, forward 5′-AGGCAGTAGTTGGGGAGGAG-3′, reverse 5′-CGAGCGGTTCTCGTAGGG-3′; SLC12A1 EX, forward 5′-CAGATGCAGAACTGGAAGCA-3′, reverse 5′-GGAAGGCTCAGGACAATGAG-3′. UTR refers to primers designed in the UTR and EX refers to primers designed across exons. The PCR protocol was 5 min at 94 °C; 40 s at 94 °C, 30 s at 55 °C, 1 min at 65 °C for 30 cycles; and for 4 min at 68 °C. For mouse samples the primers used were:
AQP2, forward 5′-GCCACCTCCTTGGGATCTATT-3′, reverse 5′-TCATCAAACTTGCCAGTGACAAC-3′, and
V-ATPase B1 subunit, forward 5′-CTGGCACTGACCACGGCTGAG-3′, reverse 5′-CCAGCCTGTGACTGAGCCCTG -3′.
The PCR protocol was 5 min at 94 °C; 40 s at 94 °C, 30 s at 55 °C, 1 min at 65 °C for 30 cycles; 4 min at 68 °C.
Real-time PCR analysis of mouse studies
RNA extracted from mouse urinary microvesicles was denatured for 5 min at 65 °C and subjected to first-strand cDNA synthesis as described in the Qiagen Sensiscript protocol (Qiagen, Maryland, MD, USA). For the Sensiscript reverse transcription oligo-dT primers were used at a final concentration of 1 µm (Applied Biosystems, Foster City, CA, USA). The resulting cDNA was used in the TaqMan PreAmp Master Mix Kit according to the manufacturer’s guidelines using 14 pre-amplification cycles (Applied Biosystems). The pre-amplification product was then diluted 1:20 with 1 × TE buffer (Promega, Madison, WI, USA). The resulting cDNA was then used as a template for real-time PCR according to the Taqman Preamplification guide (Applied Biosystems). Mouse kidney RNA concentration was measured on a SmartSpec 3000 (Bio-Rad, Hercules, CA, USA) and all samples were diluted to 90 ng/µl. Mouse kidney RNA was denatured for 5 min at 65 °C and subjected to first-strand cDNA synthesis as described in the Qiagen Omniscript protocol (Qiagen). In the Omniscript reverse transcription oligo-dT primers were used at a final concentration of 1 µmol/l (Applied Biosystems) and 1 µl of the resulting cDNA was then used per well in the subsequent real-time PCR reaction. The real-time PCR reaction was carried out using TaqMan Gene Expression Master Mix and Expression Assays (Mouse GAPD Part Number 4352339E and mouse Atp6v1b2 assay id Mm00431996_mH) on an ABI 7300 Real-Time PCR System (Applied Biosystems).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Juvenile Diabetes Research Foundation International Innovation Award (LMR, KCM). LMR received a Juvenile Diabetes Research Foundation International Transition Award and a Boston Area Diabetes and Endocrinology Research Center P&F Grant (DK57521). KCM received a National Kidney Foundation Postdoctoral Fellowship Award. TGP was supported by NIH grant DK73266. JS was supported by a Stiftelsen Olle Engkvist Byggmästare Grant. DB was supported by NIH grants DK38452 and DK42956. The microscopy core facility of the MGH Program in Membrane Biology receives additional support from the Boston Area Diabetes and Endocrinology Research Center (DK57521) and the Center for the Study of Inflammatory Bowel Disease (DK43341).
Footnotes
DISCLOSURE
JS and DB consulted for Exosome Diagnostics Inc.
Figure S1. Analysis of various methods for the removal of extraneous DNA contamination.
Supplementary material is linked to the online version of the paper at http://www.nature.com/ki
REFERENCES
- 1.Stoorvogel W, Kleijmeer MJ, Geuze HJ, et al. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 2.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artifacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Marzesco AM, Janich P, Wilsch-Bräuninger M, et al. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci. 2005;118:2849–2858. doi: 10.1242/jcs.02439. [DOI] [PubMed] [Google Scholar]
- 4.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 7.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 8.Gonzales PA, Pisitkun T, Hoffert JD, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, Yuen PS, Pisitkun T, et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471–1476. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cristaudo A, Vivaldi A, Guglielmi G, et al. A simple method to reveal possible RAS mutations in DNA of urinary sediment cells. J Environ Pathol Toxicol Oncol. 1997;16:201–204. [PubMed] [Google Scholar]
- 11.Botezatu I, Serdyuk O, Potapova G, et al. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem. 2000;46:1078–1084. [PubMed] [Google Scholar]
- 12.Coresh J, Selvin E, Stevens LA, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 13.Cheruvanky A, Zhou H, Pisitkun T, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol. 2007;292:F1657–F1661. doi: 10.1152/ajprenal.00434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller S, Rupp C, Stoeck A, et al. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 15.Sugiyama RH, Blank A, Dekker CA. Multiple ribonucleases of human urine. Biochemistry. 1981;20:2268–2274. doi: 10.1021/bi00511a031. [DOI] [PubMed] [Google Scholar]
- 16.Zhou H, Cleary RC, Bogaert YE, et al. Combination of microRNA192 and microRNA27b from Urinary Exosomes Differentiate between Renal Tubular Damage and Glomerular Injury. SA-PO2505] Philadelphia PA: American Society of Nephrology; 2008. (Abstract). [Google Scholar]
- 17.Finberg KE, Wagner CA, Bailey MA, et al. The B1-subunit of the H(+) ATPase is required for maximal urinary acidification. Proc Natl Acad Sci USA. 2005;102:13616–13621. doi: 10.1073/pnas.0506769102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Păunescu TG, Russo LM, Da Silva N, et al. Compensatory membrane expression of the V-ATPase B2 subunit isoform in renal medullary intercalated cells of B1-deficient mice. Am J Physiol Renal Physiol. 2007;293:F1915–F1926. doi: 10.1152/ajprenal.00160.2007. [DOI] [PubMed] [Google Scholar]
- 19.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 20.Mattick JS. Makunin. Non-coding RNA. Human Mol Genet. 2006;15(spec No 1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 21.Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- 22.Guescini M, Genedani S, Stocchi V, et al. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 23.Palanisamy V, Sharma S, Deshpande A, et al. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One. 2010;15:e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boute N, Gribouval O, Roselli S, et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nature Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 25.Kozyraki R, Kristiansen M, Silahtaroglu A, et al. The human intrinsic factor-vitamin B12 receptor, cubilin: molecular characterization and chromosomal mapping of the gene to 10p within the autosomal recessive megaloblastic anemia (MGA1) region. Blood. 1998;91:3593–3600. [PubMed] [Google Scholar]
- 26.Nielsen S, Chou C-L, Marples D, et al. Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Nat Acad Sci. 1995;92:1013–1017. doi: 10.1073/pnas.92.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.